Abstract
Background
Adiponectin, secreted mainly by mature adipocytes, is a protein with insulin-sensitising and anti-atherogenic effects. Human adiponectin is encoded by the ADIPOQ gene on the chromosomal locus 3q27. Variations in ADIPOQ are associated with obesity, type 2 diabetes (T2DM) and related phenotypes in several populations. Our aim was to study the association of the ADIPOQ variations with body weight, serum adiponectin concentrations and conversion to T2DM in overweight subjects with impaired glucose tolerance. Moreover, we investigated whether ADIPOQ gene variants modify the effect of lifestyle changes on these traits.
Methods
Participants in the Finnish Diabetes Prevention Study were randomly assigned to a lifestyle intervention group or a control group. Those whose DNA was available (n = 507) were genotyped for ten ADIPOQ single nucleotide polymorphisms (SNPs). Associations between SNPs and baseline body weight and serum adiponectin concentrations were analysed using the univariate analysis of variance. The 4-year longitudinal weight data were analysed using linear mixed models analysis and the change in serum adiponectin from baseline to year four was analysed using Kruskal-Wallis test. In addition, the association of SNPs with the risk of developing T2DM during the follow-up of 0-11 (mean 6.34) years was analysed by Cox regression analysis.
Results
rs266729, rs16861205, rs1501299, rs3821799 and rs6773957 associated significantly (p < 0.05) with body weight at baseline and in the longitudinal analyses. The rs266729 C allele and the rare minor alleles of rs2241766 and rs2082940 were associated with an increased adjusted hazard ratio of developing T2DM. The differences in baseline serum adiponectin concentrations were seen according to rs16861210, rs17366568, rs2241766, rs6773957 and rs2082940 and differences in the change of serum adiponectin levels from baseline to the four year examination were seen according to rs16861205, especially in subjects who were able to lose weight during the first year of intervention.
Conclusions
These results from the Finnish Diabetes Prevention Study support the concept that genetic variation in ADIPOQ locus contributes to variation in body size and serum adiponectin concentrations and may also modify the risk of developing T2DM.
Trial registration number
ClinicalTrials.gov NCT00518167
Similar content being viewed by others
Background
Type 2 diabetes (T2DM) and obesity are increasing worldwide. Although this is mainly due to environmental factors, such as changes in diet and lifestyle, much evidence for genetic predisposition to these complex traits exist [1].
Adiponectin is an adipokine, and its plasma levels are decreased in obesity [2], T2DM [3], insulin resistance [4], dyslipidemia [5], and coronary artery disease (CAD) [6]. In humans, weight reduction increases serum adiponectin levels [3, 7], and in mice, chronic administration of recombinant adiponectin leads to enhanced fatty acid oxidation, and weight loss [8] with beneficial effects on lipid metabolism and insulin sensitivity [9]. Furthermore, adiponectin knock-out mice are highly sensitive to diet induced insulin resistance [10].
Adiponectin is encoded by the ADIPOQ gene on chromosome 3q27, a region identified as susceptibility locus for the metabolic syndrome and T2DM by genome wide scans [11–13]. In various study populations, ADIPOQ SNPs and haplotypes associate with phenotypes related to obesity [14–18], insulin resistance and T2DM [14–16, 19–27] and serum adiponectin levels [14, 16, 18, 21, 23, 28–34]. However, possibly due to differences in metabolic or ethnic backgrounds of the participants in different studies, the results of previous genetic association studies are conflicting. In addition, most of the studies performed so far have been cross-sectional and have only included few SNPs in the region of ADIPOQ. The aim of the present study was to provide supportive evidence for the involvement of ADIPOQ variation in T2DM and obesity related phenotypes and serum adiponectin levels in the prospective Finnish Diabetes Prevention Study (DPS). Moreover, the aim was to examine whether ADIPOQ SNPs modify the effect of lifestyle changes on these traits.
Methods
Subjects and study design
The DPS is a randomised, controlled multicentre study with five participating clinics in Finland. The study design has been described in detail earlier [35, 36]. The main inclusion criteria were BMI > 25 kg/m2, age 40-64 years, and impaired glucose tolerance (IGT) based on the mean value of two consecutive oral glucose tolerance tests (OGTTs). The diagnosis of IGT and T2DM were based on WHO 1985 criteria [37]. IGT was defined as fasting plasma glucose < 7.8 mmol/l and a 2-h plasma glucose 7.8-11.0 mmol/l (OGTT, glucose load 75 g). A total of 522 subjects (BMI 31.1 ± 4.6 kg/m2) were randomly allocated into one of the two groups: an intensive diet and exercise intervention group or a control group. Randomisation was stratified according to the clinic attended, sex and 2-hour plasma glucose concentration. The study protocol was approved by Ethics Committee of the National Public Health Institute in Helsinki, Finland and a written informed consent was received from all subjects [35, 36]. We certify that all applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during this research.
Intervention
The subjects in the intervention group received individualized counselling on diet and exercise [38]. The five goals of the intervention were: ≥ 5% or more reduction in body weight, reduction in the intake of total fat to ≤ 30% and of saturated fat to ≤ 10% of daily energy intake, increase of the intake of dietary fibre to at least 15 g per 1000 kcal and moderate-to-vigorous exercise at least 30 min per day. The subjects in the control group received general information about healthy diet and exercise at baseline.
Examinations
A medical history was recorded and a physical examination with anthropometric measurements was performed at baseline and at each annual follow-up visit. Measurements recorded at baseline and 1, 2, 3 and 4 year examinations were used in the present study.
Assessment of glucose and insulin metabolism
A 2-h OGTT was performed at baseline and annually. Plasma glucose and insulin samples were drawn at 0 min and 120 min during the OGTT with a glucose load of 75 g. Plasma glucose was measured locally by standard methods as previously described [36]. Serum insulin was measured by RIA (Phadaseph Insulin RIA 100, Pharmacia Diagnostica, Uppsala, Sweden). The intra-assay coefficient of variation was 5.3% and the interassay coefficient of variation was 7.6%. The diagnosis of IGT was based on the mean value of two oral glucose tolerance tests. A new diagnosis of diabetes (fasting plasma glucose ≥7.8 mmol/l or 2-h plasma glucose ≥11.0 mmol/l) was confirmed by a second oral glucose tolerance test.
Determination of adiponectin concentrations
Fasting serum adiponectin levels were measured using an enzyme-linked immunosorbent assay (ELISA) (B-Bridge International, Inc., San Jose, CA, USA), on whole plasma stored at -80°C. The intra-assay and inter-assay coefficients of variation were 5.5-7.9% and 6.5%, respectively. Frozen serum samples for adiponectin measurements were only available from subset of participants from three study clinics (n = 243 at baseline, and n = 209 at 4-year examination). In altogether 190 subjects both baseline and year four serum adiponectin concentrations were measured.
Selection of SNPs and genotyping
DNA sample was available from 507 subjects (166 men and 341 women). Tagging SNPs with minimum minor allele frequency of 5% were selected based on genotype data of the Hapmap CEU population [39] by using the Tagger algorithm [40] with rs2241766 and rs1501299, selected based on previous literature, being forced into the selection. Altogether, the ten SNPs selected for genotyping covered 73% of the variation within ADIPOQ region (20 kbp) with r2 ≥ 0.8. Rs2241766 and rs1501299 were genotyped using PCR followed by SNaPshot ddNTP Primer Extension Kit technique (ABI Prism; Applied Biosystems) as described earlier [24]. Rs17366568 was genotyped using PCR-RFLP method. The genomic DNA was amplified by PCR by using the following primers: forward 5'-CCCAATAGTCAAACATGTGC-3' and reverse 5'-TCATCCTTGGAAGACCAACC-3' followed by digestion with restriction enzyme MseI (New England Biolabs). Other SNPs were genotyped with TaqMan Allelic Discrimination assays according to manufacturer's instructions by using the ABI PRISM 7000 sequence detector (Applied Biosystems, Foster City, CA). For a subset of randomly selected samples (6.3%) genotyping was repeated in order to calculate success rate.
Statistical analysis
The data were analysed with SPSS for Windows 14.0 (SPSS Inc, Chicago, IL). Chi square test was used to test the departure of genotypes from Hardy-Weinberg equilibrium (HWE) and the distribution of genotypes between study groups. Skewed variables were transformed for analysis and are reported as back-transformed geometric means and 95% confidence intervals (CIs), or were analysed with nonparametric tests. Normally distributed data are presented as mean ± SE. All analyses were carried out using three inheritance models: additive, dominant (common allele homozygotes compared with heterozygotes and minor allele homozygotes), and recessive (common allele homozygotes and heterozygotes compared with minor allele homozygotes). The baseline differences in continuous variables between genotypes were evaluated with the univariate ANOVA, general linear model (GLM). Longitudinal weight data were examined using the linear mixed models analysis. Normality was assessed by plotting the residuals. Association of the SNPs with conversion to T2DM was analysed by using Cox regression. Adjustments for age, sex, BMI, waist-to-hip ratio (WHR), fasting plasma glucose and study group (intervention or control) were made when appropriate. A multiple SNP analysis was performed for groups of SNPs that associated with baseline weight, baseline serum adiponectin levels and the risk of T2DM by including all significant SNPs (coded as major allele homozygotes vs. minor allele carriers) in the same statistical model and then removing SNPs sequentially starting from the least significant one. Sex-genotype or study group-genotype interaction term was included in analyses when appropriate. When significant interaction (p < 0.05) was observed, men and women or the study groups were analysed separately. Change in serum adiponectin concentration was calculated by subtracting the baseline values from the fourth year values. Correction for multiple hypothesis testing for single SNP analyses was performed with false discovery rate (FDR) method using Q value 1.0 software [41]. The q-values were calculated separately for each trait, but all SNPs and inheritance models were included in the same calculations. The pair wise linkage disequilibrium (LD) between individual SNPs was evaluated with Haploview software (version 3.32; Broad institute, Cambridge, MA) [42]. Haploblocks were defined using the default algorithm [43].
Results
Table 1 shows the clinical and metabolic characteristics of the study population.
Genotype frequencies and linkage disequilibrium (LD) patterns
Ten SNPs were genotyped from the 507 DNA samples of the DPS participants with an error rate of 0% and a call rate of 100% in replicated samples for all markers. The locations of the analysed variants are presented in Figure 1 with pairwise LD measures (D' and r2). As previously reported, we observed two LD blocks [31] with rs266729 and rs16861205 constituting the block 1, and rs1501299, rs3821799 and rs6773957 constituting the block 2, while other SNPs remained between or outside the blocks.
ADIPOQ gene, locations of the genotyped variants and their pairwise linkage disequilibrium (LD) patterns. Schematic presentation of the ADIPOQ, indicating the locations of the analysed variants, the two observed haploblocks and the pairwise LD measures D' (above) and r2 (below). Grey boxes, untranslated region; black boxes, coding region; MAF, minor allele frequency.
Genotype counts and allele frequencies are shown in Table 2. All SNPs, except rs16861210, were in HWE (p < 0.05) in the entire DPS population. The deviation from HWE could be explained as a chance finding due to low frequency of the minor allele and limited sample size of DPS. Thus, rs16861210 was included in further analyses. Genotype frequencies of rs16861205, rs3821799 and rs2082940 differed significantly (p < 0.05) between the intervention and control group of DPS. Again, this is probably due to chance and these SNPs were included in analyses, but all longitudinal data analyses were performed separately in both study groups in addition to analyses in the total study population.
Genotype associations with body weight
Rs266729, rs16861205, rs1501299, rs3821799 and rs6773957 associated with baseline body weight, when adjusted for age and sex (table 3). Furthermore, the same SNPs associated with 4-year follow-up measurements of weight, when analysed longitudinally with adjustments for age, sex, study group and the time variable. Similar associations between ADIPOQ SNPs and baseline and 4-year measurements of BMI were seen (data not shown). Genotype differences were mainly seen by using additive and dominant inheritance models.
Individuals carrying the rs266729 G allele had higher baseline body weight than those homozygous for C allele. A sex-genotype interaction was found for weight and significant differences were observed in women. Likewise, during the 4-year follow-up significant differences in weight according to rs266729 were only observed in women (p = 0.007/q = 0.038 for the dominant inheritance model and p = 0.027/q = 0.309 for sex-genotype interaction).
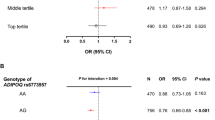
Rs16861205 G, rs1501299 G, rs3821799 C and rs6773957 G alleles associated dose-dependently with higher baseline body weight. The longitudinal data is shown for rs3821799 in figure 2. Genotype differences remained significant during the 4-year follow-up, when the additive inheritance model was used (rs16861205: p = 0.028/q = 0.049, rs1501299:p = 0.041/q = 0.049, rs3821799: p = 0.002/q = 0.049, and rs6773957: p = 0.045/q = 0.049) and were even stronger when the dominant inheritance model was used (rs16861205: p = 0.008/q = 0.049, rs1501299: p = 0.014/0.049, rs3821799: p = 0.001/q = 0.049, and rs6773957: p = 0.014/q = 0.049). For rs3821799, the genotype differences were significant also when the recessive inheritance model was used. When the longitudinal analyses were performed separately in the intervention and control group, genotype differences remained constant during the follow-up in both groups, but were statistically significant only in the control group.
Body weight from baseline to year 4 according to rs3821799. Data are predicted geometric means and 95% CI. Additive (A) and dominant (B) inheritace models; p values are adjusted for age, sex, study group and the time; Correction for multiple hypothesis testing was performed with FDR, denoted as q-value.
We also conducted multiple SNP models analysis with SNPs that associated individually with baseline body weight. In a model including all five SNPs, a trend for significance was observed for rs3821799 (p = 0.063). When the least significant SNPs were removed sequentially from the model, two SNPs remained in the final model: rs3821799 (p = 0.007) and rs16861205 (p = 0.101).
Genotype associations with Type 2 Diabetes Incidence
Altogether 184 of 507 subjects converted from IGT to T2DM during the follow-up of 0-11 (mean 6.34) years. The association of the 10 ADIPOQ SNPs with the conversion from IGT to T2DM was analysed by using Cox regression models adjusting for baseline BMI, WHR, baseline fasting plasma glucose and study group (Table 4).
T2DM was less likely developed in study participants carrying rs266729 G allele. The adjusted hazard ratio was 0.682 (95% CI 0.510-0.912, p = 0.010/q = 0.203) for the G allele carriers compared with subjects with CC genotype, 0.695 (95% CI 0.513-0.942, p = 0.019/q = 0.203) for CG genotype and 0.615 (95% CI 0.350-1.081, p = 0.091/q = 0.330) for GG genotype. In addition, subjects carrying the rare minor alleles of rs2241766 and rs2082940 (G and T, respectively) had increased risk of developing T2DM. The results were similar when a statistical model not adjusted for BMI or WHR was used (data not shown).
Since the lifestyle intervention decreased the risk of T2DM significantly in the DPS [36], the effects of each SNP were examined for the intervention and control groups separately. The genotype differences were similar in both groups according to rs266729, but statistical significance was reached only in the control group. Only two subjects were homozygous for the rare alleles of rs2241766 and rs2082940 and both were in the control group. When dominant inheritance model was used, significant differences were only seen in the control group according to rs2241766.
In a multiple SNP model with rs266729, rs2241766 and rs2082940, only rs2766729 remained significant predictor of conversion from IGT to T2DM (adjusted hazard ratio 0.693, 95% CI 0.518-0.928, p = 0.014 for the G allele carriers compared with subjects with CC genotype). A borderline significance was observed for rs2241766 when it was included in the model with rs266729 (p = 0.086).
Genotype associations with serum adiponectin levels
Baseline serum adiponectin levels differed significantly according to five ADIPOQ SNPs, when adjustments were made for age, sex and baseline WHR (Table 5): rs16861210 (p = 0.029/q = 0.032 for the additive and p = 0.008/q = 0.032 for the dominant inheritance model), rs17366568 (p = 0.003/q = 0.032 for the additive and p = 0.0007/q = 0.032 for the dominant inheritance model), rs2241766 (p = 0.056/q = 0.032 for TT vs. TG genotype), rs6773957 (p = 0.016/q = 0.032 for the additive, p = 0.021/q = 0.032 for the dominant and p = 0.022/q = 0.032 for the recessive inheritance model) and rs2082940 (p = 0.056/q = 0.032 for CC vs. CT genotype). Lower serum adiponectin concentrations were associated with rs16861210 G allele, rs17366568 A allele, rs2241766 TT genotype, rs6773957 G allele and rs2082940 CC genotype. In addition, a significant sex-genotype interaction was found for rs2241766 and rs2082940 (p = 0.020/q = 0.279 for both). When men and women were analysed separately, a significant association between rs2241766 and rs2082940 genotype and serum adiponectin levels was seen only in men (p = 0.003/q = 0.131 for both SNPs).
In multiple SNP model including rs16861210, rs17366568, rs6773957 and either rs2241766 or rs2082940, the first two SNPs remained significantly associated with baseline adiponectin levels (p = 0.012 and p = 0.003, respectively).
Baseline serum adiponectin concentrations and T2DM risk
A trend for a lower T2DM risk was seen with hazard ratio of 0.695 (0.451-1.072, p = 0.100) for those with higher than median (6.250 for men and 8.815 for women) serum adiponectin concentrations compared with those with adiponectin lower than median.
Genotype associations with 4-year change in adiponectin concentrations
Rs16861205 associated with 4-year change in serum adiponectin concentrations (p = 0.040/q = 0.798 and p = 0.014/q = 0.798 for the additive and the dominant inheritance models, respectively) (figure 3). The A allele was dose dependently associated with increase in serum adiponectin levels (AA: n = 9 and AG: n = 54) compared with the GG genotype (n = 127). When study groups were analysed separately, genotype differences were similar, but statistical significance was not reached. When individuals who lost weight (n = 144) or gained weight (n = 45) during the first year of the intervention were analysed as separate groups, statistically significant differences in the 4-year change were seen only in those who were able to lose weight (p = 0.034/q = 0.725 for the additive inheritance model, p = 0.013/q = 0.725 for the dominant inheritance model, figure 3 B). Like in the whole study population, serum adiponectin concentrations increased in subjects carrying the A allele.
Four year change in serum adiponectin levels according to rs16861205. Data are means ± SD. Dominant inheritance model for the whole DPS (A), for participants who lost weight (B), and for participants who gained weight during the first year (C). p/q value for GG genotype vs. A allele carriers, Kruskal-Wallis test; Correction for multiple hypothesis testing was performed with FDR, denoted as q-value.
Discussion
In the present study, we found novel associations between ADIPOQ variations and body weight, the risk of T2DM and serum adiponectin concentrations. The results also support the existing view that genetic variation in the ADIPOQ influences metabolic and anthropometric characteristics related to T2DM.
Body weight - SNPs in haploblocks 1 and 2
Rs266729 and rs1501299 are the most extensively studied of the SNPs associating with body weight and BMI in DPS. Earlier studies report associations to various obesity related phenotypes for rs266729 [15, 16, 29] and for rs1501299 [14, 16, 17].
We found that rs266729 G allele carriers had higher weight compared with individuals with CC genotype at baseline and a similar trend was seen during the 4-year follow-up, especially in women. Significant association that were seen more often in women than men may be explained by larger number of female subjects in the DPS. In contrast to our results, rs266729 C is a risk allele for obesity related phenotypes in most study populations [15, 16]. Genotype differences according to rs1501299 were seen in baseline weight as well as in 4-year follow-up. In agreement with this, the G allele was associated with obesity related phenotypes in some studies [14], but the T allele in others [16, 17].
To our knowledge, no previous data concerning associations of intronic SNPs rs16861205 and rs3821799, and rs6773957, located in the 3'UTR, with obesity related phenotypes exists. The strongest association was observed for rs3821799 and it remained significant also in multiple SNP analyses. During the 4-year follow-up, the genotype differences remained constant in both study groups. Thus, individuals with different ADIPOQ genotypes did not respond differently to the lifestyle modification regarding weight change.
Type 2 diabetes - rs266729, rs2241766 and rs2082940
We found significantly increased conversion rates from IGT to T2DM for subjects homozygous for the rs266729 C allele. The G allele, associated with higher body weight, associated with lower T2DM risk in statistical models with or without adjustment for obesity related covariates. It seems therefore that the effect of ADIPOQ variation on T2DM risk is independent of its effect on body weight. According to previous studies rs266729 G allele frequently associates with increased risk of T2DM [15, 23] and insulin resistance [16, 44]. On the other hand, the C allele was associated with increased risk of T2DM in a German study population [25] and with lower clamp-derived insulin sensitivity in a cohort of Europid adolescents and their parents [26].
In addition, the rare minor alleles of rs2241766 and rs2082940 associated with conversion to T2DM in DPS. Rs2241766 is one of the most extensively studied ADIPOQ SNPs. Consistent with our results, the G allele associated with T2DM in cross-sectional studies [21], and also with T2DM risk [24] and hyperglycemia [19, 20] in prospective studies. On the contrary, others have reported that the T allele associated with elevated insulin/glucose values and insulin resistance [14]. To our knowledge, no previous studies have found association between T2DM and rs2082940, which is in high LD with rs2241766 (r2 = 0.872). It is located in the 3' UTR and its functional role is currently unknown. In multiple SNP analyses rs266729 remained significant predictor of T2DM risk. Thus at least one genetic signal affecting T2DM risk exists in the ADIPOQ locus. When the study groups were analysed separately, SNPs associated with T2DM similarly in both study groups. Thus, the effect of lifestyle intervention on T2DM was not modified by the ADIPOQ SNPs studied.
Serum adiponectin levels - SNPs in intron1, exon 2 and in 3'UTR
Serum adiponectin levels are under strong genetic control [45, 46]. A genome-wide scan of plasma adiponectin levels provided evidence that variation in ADIPOQ gene, especially rs2241766, rs1501299 and +2019A/- insertion/deletion polymorphism, are responsible for linkage of adiponectin levels to 3q27 in the Old Order Amish [28]. Moreover, a recent genome wide association study found that rs17366568 explained 3.8% of variation in adiponectin levels, while 6.7% of the variation was explained by all SNPs in the ADIPOQ region [47]. In another whole genome association analysis rs6773957 and rs3774261 associated strongly with adiponectin levels [34]. Furthermore, rs6773957 and two promoter SNPs (rs17300539 and rs822387) associated with higher serum adiponectin levels and a trend for rs17366568 was also seen [33].
In our study, rs16861210, rs17366568 and rs6773957 associated strongly with baseline serum adiponectin levels in a dose dependent manner. In multiple SNP analyses the significance of rs6773957 was lost indicating LD with another, most likely rs17366568. To our knowledge, no previous studies examining the association between rs16861210 and adiponectin levels have been published. Both rs16861210 and rs17366568 are located in intron 1, but are not in strong LD with each other and represent likely independent genetic signals affecting serum adiponectin levels. Functional role of either SNP is currently unknown, but interestingly, three SNPs located immediately on either side of rs17366568 were predicted to affect transcription factor binding sites [47].
The association between rs2241766 and rs2082940 with baseline serum adiponectin remained borderline significant in multi SNP analyses and may represent genetic signal, independent of the variations in intron 1. The association of the rare minor alleles of these two SNPs with higher adiponectin levels in men, and on the other hand, higher risk of T2DM in the total population is contradictory. Especially so, since a non-significant trend for lower adiponectin levels predicting higher risk of developing T2DM was seen in our population. The number of minor allele carriers was relatively low and the results should therefore be interpreted with caution. Nevertheless, the T allele of rs2241766 associates with lower adiponectin levels [14, 28, 30, 31] and the G allele with T2DM related traits in several previous studies [19–21, 24].
While association between ADIPOQ promoter SNPs and adiponectin levels have been suggested widely [18, 23, 29, 31–33], we did not find significant association between rs266729 and adiponectin levels possibly due to lack of statistical power. The results of several functional studies suggest that rs266729 does not have influence on the transcription efficiency [16, 27, 32]. On the other hand, an important role in ADIPOQ promoter activity was recently suggested for rs266729 and two other promoter SNPs (rs16861194 and rs17300539) [48]. In addition, another study presents evidence that rs266729 alters the sequence in one of the transcriptional stimulatory protein (SP1) binding sites in the promoter region [49]. Unfortunately, in the DPS population, we did not genotype the promoter variant rs17300539 for which important functional role has been also suggested [16, 48].
Significant differences were seen in 4-year change in serum adiponectin levels according to rs16861205. Increase in adiponectin levels was greatest in A allele carriers and the most beneficial changes in serum adiponectin concentrations were seen in A allele carriers who were able to lose weight, while in a group of subjects whose weight increased, genotype differences were not seen. This may implicate that A allele carriers benefit more of weight loss in terms of change in adiponectin concentrations. Genotype differences were still significant for the dominant inheritance model when 4-year weight change was used, although the number of subjects was low.
Several ADIPOQ association studies have failed to replicate results of previous studies or have even reported opposite effects of alleles. Possible explanation is that the LD patterns differ between populations with various ethnic backgrounds. Moreover, the true functional SNPs underlying complex traits may be rare and population specific. Recently, Bowden at al. (2010) demonstrated that a rare ADIPOQ variant explained approximately 17% of the variance in plasma adiponectin in Hispanic American population. Interestingly, this variant was not observed in African American or European American populations [50].
Since ADIPOQ alleles have shown opposite effects even in populations with similar ethnic background and disease status [14, 51] other explanations, such as differences in participant inclusion criteria, diagnostic criteria and study design, are also possible. As an example, genetic effects for complex traits can vary by age and such interactions can even prevent replication of an association especially in cross-sectional studies [52]. Lastly gene-gene interactions, genetic epistasis and even epigenetic modifications may modify genetic associations. Although these phenomena are still poorly understood in human, Greene et al (2009) have shown that differences in allele frequencies and gene-gene interactions can explained results where effects of gene variations are significant, but in different directions [53]. Lastly, one obvious possibility is that these findings are simply due to chance. However, since significant associations have been replicated in many different populations, the latter alternative seems unlikely.
The two-block LD structure of ADIPOQ with the existence of two independent genetic signals corresponding to the haploblock structure reported earlier [22, 31] was also found in the DPS and different phenotypes were accounted for mostly by SNPs located in separate regions based on the LD structure of ADIPOQ. ADIPOQ is a locus of low LD and high haplotypic diversity [54] and it is therefore important to perform the association studies in genetically homogeneous population with comparatively high degree of LD. Owing to its demographic history, the Finnish population exhibits a decrease of genetic diversity and an increase in LD when compared with more admixed populations of central European background and it has been speculated that identifying genes for complex traits could be especially advantageous in this population [55]. Moreover, the participants of the DPS study were carefully selected and phenotyped, hence minimising the risk of false results due to population stratification. Moreover, the follow-up data available in DPS increases the power to find true associations compared to studies with just single measurement of a given variable. The size of the DPS population, however, is moderate for genetic association study and we were only able to measure the serum adiponectin levels from part of the study participants. This may weaken the power to find true associations or increase the risk of false positive findings.
Performing multiple statistical tests in genetic association studies is likely to lead to false positive findings and general guidelines on applying correction for multiple comparisons or threshold values do not exist. In this study, we applied FDR to control for the multiple hypothesis testing and present combination of p-value and q-value for each test performed on single SNPs. The FDR provides an estimation of the minimum false discovery rate at which the test may be called significant. FDR was low (q < 0.1) for associations between ADIPOQ SNPs and body weight/BMI and serum adiponectin levels. However, although p value was <0.05, the FDR was higher for the risk of T2DM in our analyses. Nevertheless, given that our candidate gene was selected based on solid prior information on the important role of adiponectin as a metabolic regulator, and the study design that enabled us to perform statistical analyses also on longitudinal data, we believe that our results are true associations. Again, this is further supported by the results of earlier genetic association studies on ADIPOQ.
Conclusions
This study, performed in a homogeneous group of individuals, supported the significant role of ADIPOQ variations in conditions linked to T2DM and obesity, partly by replicating earlier findings, but also by discovering novel associations. New evidence for the involvement of rs3821799 and rs6773957 in obesity was provided. Particularly, the strong association of rs6773957 with body weight and serum adiponectin levels is interesting finding as this SNP is located in the 3'UTR and could therefore have an effect on mRNA stability or translational efficiency.
References
Malecki MT, Klupa T: Type 2 diabetes mellitus: from genes to disease. Pharmacol Rep. 2005, 57 (Suppl): 20-32.
Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J, Hotta K, Shimomura I, Nakamura T, Miyaoka K, Kuriyama H, Nishida M, Yamashita S, Okubo K, Matsubara K, Muraguchi M, Ohmoto Y, Funahashi T, Matsuzawa Y: Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999, 257 (1): 79-83. 10.1006/bbrc.1999.0255.
Hotta K, Funahashi T, Arita Y, Takahashi M, Matsuda M, Okamoto Y, Iwahashi H, Kuriyama H, Ouchi N, Maeda K, Nishida M, Kihara S, Sakai N, Nakajima T, Hasegawa K, Muraguchi M, Ohmoto Y, Nakamura T, Yamashita S, Hanafusa T, Matsuzawa Y: Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol. 2000, 20 (6): 1595-1599.
Weyer C, Funahashi T, Tanaka S, Hotta K, Matsuzawa Y, Pratley RE, Tataranni PA: Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab. 2001, 86 (5): 1930-1935. 10.1210/jc.86.5.1930.
Matsubara M, Maruoka S, Katayose S: Decreased plasma adiponectin concentrations in women with dyslipidemia. J Clin Endocrinol Metab. 2002, 87 (6): 2764-2769. 10.1210/jc.87.6.2764.
Ouchi N, Kihara S, Arita Y, Maeda K, Kuriyama H, Okamoto Y, Hotta K, Nishida M, Takahashi M, Nakamura T, Yamashita S, Funahashi T, Matsuzawa Y: Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation. 1999, 100 (25): 2473-2476.
Yang WS, Lee WJ, Funahashi T, Tanaka S, Matsuzawa Y, Chao CL, Chen CL, Tai TY, Chuang LM: Weight reduction increases plasma levels of an adipose-derived anti-inflammatory protein, adiponectin. J Clin Endocrinol Metab. 2001, 86 (8): 3815-3819. 10.1210/jc.86.8.3815.
Fruebis J, Tsao TS, Javorschi S, Ebbets-Reed D, Erickson MR, Yen FT, Bihain BE, Lodish HF: Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci USA. 2001, 98 (4): 2005-2010. 10.1073/pnas.041591798.
Yamauchi T, Kamon J, Waki H, Terauchi Y, Kubota N, Hara K, Mori Y, Ide T, Murakami K, Tsuboyama-Kasaoka N, Ezaki O, Akanuma Y, Gavrilova O, Vinson C, Reitman ML, Kagechika H, Shudo K, Yoda M, Nakano Y, Tobe K, Nagai R, Kimura S, Tomita M, Froguel P, Kadowaki T: The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat Med. 2001, 7 (8): 941-946. 10.1038/90984.
Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, Komuro R, Ouchi N, Kihara S, Tochino Y, Okutomi K, Horie M, Takeda S, Aoyama T, Funahashi T, Matsuzawa Y: Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002, 8 (7): 731-737. 10.1038/nm724.
Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, Martin L, Blangero J, Comuzzie AG: Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA. 2000, 97 (26): 14478-14483. 10.1073/pnas.97.26.14478.
Vionnet N, Hani E, Dupont S, Gallina S, Francke S, Dotte S, De Matos F, Durand E, Lepretre F, Lecoeur C, Gallina P, Zekiri L, Dina C, Froguel P: Genomewide search for type 2 diabetes-susceptibility genes in French whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2-diabetes locus on chromosome 1q21-q24. Am J Hum Genet. 2000, 67 (6): 1470-1480. 10.1086/316887.
Mori Y, Otabe S, Dina C, Yasuda K, Populaire C, Lecoeur C, Vatin V, Durand E, Hara K, Okada T, Tobe K, Boutin P, Kadowaki T, Froguel P: Genome-wide search for type 2 diabetes in Japanese affected sib-pairs confirms susceptibility genes on 3q, 15q, and 20q and identifies two new candidate Loci on 7p and 11p. Diabetes. 2002, 51 (4): 1247-1255. 10.2337/diabetes.51.4.1247.
Menzaghi C, Ercolino T, Di Paola R, Berg AH, Warram JH, Scherer PE, Trischitta V, Doria A: A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes. 2002, 51 (7): 2306-2312. 10.2337/diabetes.51.7.2306.
Gu HF, Abulaiti A, Ostenson CG, Humphreys K, Wahlestedt C, Brookes AJ, Efendic S: Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish caucasians. Diabetes. 2004, 53 (Suppl 1): S31-5. 10.2337/diabetes.53.2007.S31.
Bouatia-Naji N, Meyre D, Lobbens S, Seron K, Fumeron F, Balkau B, Heude B, Jouret B, Scherer PE, Dina C, Weill J, Froguel P: ACDC/Adiponectin Polymorphisms Are Associated With Severe Childhood and Adult Obesity. Diabetes. 2006, 55 (2): 545-550. 10.2337/diabetes.55.02.06.db05-0971.
Yang WS, Yang YC, Chen CL, Wu IL, Lu JY, Lu FH, Tai TY, Chang CJ: Adiponectin SNP276 is associated with obesity, the metabolic syndrome, and diabetes in the elderly. Am J Clin Nutr. 2007, 86 (2): 509-513.
Warodomwichit D, Shen J, Arnett DK, Tsai MY, Kabagambe EK, Peacock JM, Hixson JE, Straka RJ, Province MA, An P, Lai CQ, Parnell LD, Borecki IB, Ordovas JM: ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring). 2009, 17 (3): 510-517. 10.1038/oby.2008.583.
Tso AW, Sham PC, Wat NM, Xu A, Cheung BM, Rong R, Fong CH, Xu JY, Cheng KK, Janus ED, Lam KS: Polymorphisms of the gene encoding adiponectin and glycaemic outcome of Chinese subjects with impaired glucose tolerance: a 5-year follow-up study. Diabetologia. 2006, 49 (8): 1806-1815. 10.1007/s00125-006-0324-2.
Fumeron F, Aubert R, Siddiq A, Betoulle D, Pean F, Hadjadj S, Tichet J, Wilpart E, Chesnier MC, Balkau B, Froguel P, Marre M, Epidemiologic Data on the Insulin Resistance Syndrome (DESIR) Study Group: Adiponectin gene polymorphisms and adiponectin levels are independently associated with the development of hyperglycemia during a 3-year period: the epidemiologic data on the insulin resistance syndrome prospective study. Diabetes. 2004, 53 (4): 1150-1157. 10.2337/diabetes.53.4.1150.
Hara K, Boutin P, Mori Y, Tobe K, Dina C, Yasuda K, Yamauchi T, Otabe S, Okada T, Eto K, Kadowaki H, Hagura R, Akanuma Y, Yazaki Y, Nagai R, Taniyama M, Matsubara K, Yoda M, Nakano Y, Tomita M, Kimura S, Ito C, Froguel P, Kadowaki T: Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes. 2002, 51 (2): 536-540. 10.2337/diabetes.51.2.536.
Menzaghi C, Trischitta V, Doria A: Genetic influences of adiponectin on insulin resistance, type 2 diabetes, and cardiovascular disease. Diabetes. 2007, 56 (5): 1198-1209. 10.2337/db06-0506.
Vasseur F, Helbecque N, Lobbens S, Vasseur-Delannoy V, Dina C, Clement K, Boutin P, Kadowaki T, Scherer PE, Froguel P: Hypoadiponectinaemia and high risk of type 2 diabetes are associated with adiponectin-encoding (ACDC) gene promoter variants in morbid obesity: evidence for a role of ACDC in diabesity. Diabetologia. 2005, 48 (5): 892-899. 10.1007/s00125-005-1729-z.
Zacharova J, Chiasson JL, Laakso M: The Common Polymorphisms (Single Nucleotide Polymorphism [SNP] +45 and SNP +276) of the Adiponectin Gene Predict the Conversion From Impaired Glucose Tolerance to Type 2 Diabetes: The STOP-NIDDM Trial. Diabetes. 2005, 54 (3): 893-899. 10.2337/diabetes.54.3.893.
Schwarz PE, Govindarajalu S, Towers W, Schwanebeck U, Fischer S, Vasseur F, Bornstein SR, Schulze J: Haplotypes in the promoter region of the ADIPOQ gene are associated with increased diabetes risk in a German Caucasian population. Horm Metab Res. 2006, 38 (7): 447-451. 10.1055/s-2006-947842.
Rasmussen-Torvik LJ, Pankow JS, Jacobs DR, Steinberger J, Moran A, Sinaiko AR: The association of SNPs in ADIPOQ, ADIPOR1, and ADIPOR2 with insulin sensitivity in a cohort of adolescents and their parents. Hum Genet. 2009, 125 (1): 21-28. 10.1007/s00439-008-0595-4.
Yang M, Qiu CC, Chen W, Xu LL, Yu M, Xiang HD: Identification of a regulatory single nucleotide polymorphism in the adiponectin (APM1) gene associated with type 2 diabetes in Han nationality. Biomed Environ Sci. 2008, 21 (6): 454-459. 10.1016/S0895-3988(09)60002-7.
Pollin TI, Tanner K, O'connell JR, Ott SH, Damcott CM, Shuldiner AR, McLenithan JC, Mitchell BD: Linkage of plasma adiponectin levels to 3q27 explained by association with variation in the APM1 gene. Diabetes. 2005, 54 (1): 268-274. 10.2337/diabetes.54.1.268.
Tanko LB, Siddiq A, Lecoeur C, Larsen PJ, Christiansen C, Walley A, Froguel P: ACDC/adiponectin and PPAR-gamma gene polymorphisms: implications for features of obesity. Obes Res. 2005, 13 (12): 2113-2121. 10.1038/oby.2005.262.
Jang Y, Lee JH, Chae JS, Kim OY, Koh SJ, Kim JY, Cho H, Lee JE, Ordovas JM: Association of the 276G->T polymorphism of the adiponectin gene with cardiovascular disease risk factors in nondiabetic Koreans. Am J Clin Nutr. 2005, 82 (4): 760-767.
Heid IM, Wagner SA, Gohlke H, Iglseder B, Mueller JC, Cip P, Ladurner G, Reiter R, Stadlmayr A, Mackevics V, Illig T, Kronenberg F, Paulweber B: Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes. 2006, 55 (2): 375-384. 10.2337/diabetes.55.02.06.db05-0747.
Kyriakou T, Collins LJ, Spencer-Jones NJ, Malcolm C, Wang X, Snieder H, Swaminathan R, Burling KA, Hart DJ, Spector TD, O'Dell SD: Adiponectin gene ADIPOQ SNP associations with serum adiponectin in two female populations and effects of SNPs on promoter activity. J Hum Genet. 2008, 53 (8): 718-727. 10.1007/s10038-008-0303-1.
Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J, Fox CS, O'Donnell CJ, Cupples LA, Meigs JB: Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes. 2008, 57 (12): 3353-3359. 10.2337/db08-0700.
Ling H, Waterworth DM, Stirnadel HA, Pollin TI, Barter PJ, Kesaniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP, Cohen JC, Grundy SM, Mooser VE, Mitchell BD: Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring). 2009, 17 (4): 737-744. 10.1038/oby.2008.625.
Eriksson J, Lindstrom J, Valle T, Aunola S, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Laakso M, Lauhkonen M, Lehto P, Lehtonen A, Louheranta A, Mannelin M, Martikkala V, Rastas M, Sundvall J, Turpeinen A, Viljanen T, Uusitupa M, Tuomilehto J: Prevention of Type II diabetes in subjects with impaired glucose tolerance: the Diabetes Prevention Study (DPS) in Finland. Study design and 1-year interim report on the feasibility of the lifestyle intervention programme. Diabetologia. 1999, 42 (7): 793-801. 10.1007/s001250051229.
Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, Keinanen-Kiukaanniemi S, Laakso M, Louheranta A, Rastas M, Salminen V, Uusitupa M, Finnish Diabetes Prevention Study Group: Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001, 344 (18): 1343-1350. 10.1056/NEJM200105033441801.
World Health Organization Expert Committee: Diabetes Mellitus. Technical Report Series. 1985, 742:
Lindstrom J, Louheranta A, Mannelin M, Rastas M, Salminen V, Eriksson J, Uusitupa M, Tuomilehto J, Finnish Diabetes Prevention Study Group: The Finnish Diabetes Prevention Study (DPS): Lifestyle intervention and 3-year results on diet and physical activity. Diabetes Care. 2003, 26 (12): 3230-3236. 10.2337/diacare.26.12.3230.
International HapMap Consortium: The International HapMap Project. Nature. 2003, 426 (6968): 789-796. 10.1038/nature02168.
de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D: Efficiency and power in genetic association studies. Nat Genet. 2005, 37 (11): 1217-1223. 10.1038/ng1669.
Storey JD, Tibshirani R: Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003, 100 (16): 9440-9445. 10.1073/pnas.1530509100.
Barrett JC, Fry B, Maller J, Daly MJ: Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005, 21 (2): 263-265. 10.1093/bioinformatics/bth457.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D: The structure of haplotype blocks in the human genome. Science. 2002, 296 (5576): 2225-2229. 10.1126/science.1069424.
Ferguson JF, Phillips CM, Tierney AC, Perez-Martinez P, Defoort C, Helal O, Lairon D, Planells R, Shaw DI, Lovegrove JA, Gjelstad IM, Drevon CA, Blaak EE, Saris WH, Leszczynska-Golabek I, Kiec-Wilk B, Riserus U, Karlstrom B, Miranda JL, Roche HM: Gene-nutrient interactions in the metabolic syndrome: single nucleotide polymorphisms in ADIPOQ and ADIPOR1 interact with plasma saturated fatty acids to modulate insulin resistance. Am J Clin Nutr. 2010, 91 (3): 794-801. 10.3945/ajcn.2009.28255.
Menzaghi C, Ercolino T, Salvemini L, Coco A, Kim SH, Fini G, Doria A, Trischitta V: Multigenic control of serum adiponectin levels: evidence for a role of the APM1 gene and a locus on 14q13. Physiol Genomics. 2004, 19 (2): 170-174. 10.1152/physiolgenomics.00122.2004.
Comuzzie AG, Funahashi T, Sonnenberg G, Martin LJ, Jacob HJ, Black AE, Maas D, Takahashi M, Kihara S, Tanaka S, Matsuzawa Y, Blangero J, Cohen D, Kissebah A: The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metab. 2001, 86 (9): 4321-4325. 10.1210/jc.86.9.4321.
Heid IM, Henneman P, Hicks A, Coassin S, Winkler T, Aulchenko YS, Fuchsberger C, Song K, Hivert MF, Waterworth DM, Timpson NJ, Richards JB, Perry JR, Tanaka T, Amin N, Kollerits B, Pichler I, Oostra BA, Thorand B, Frants RR, Illig T, Dupuis J, Glaser B, Spector T, Guralnik J, Egan JM, Florez JC, Evans DM, Soranzo N, Bandinelli S, Carlson OD, Frayling TM, Burling K, Smith GD, Mooser V, Ferrucci L, Meigs JB, Vollenweider P, Dijk KW, Pramstaller P, Kronenberg F, van Duijn CM: Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis. 2010, 208 (2): 412-420. 10.1016/j.atherosclerosis.2009.11.035.
Laumen H, Saningong AD, Heid IM, Hess J, Herder C, Claussnitzer M, Baumert J, Lamina C, Rathmann W, Sedlmeier EM, Klopp N, Thorand B, Wichmann HE, Illig T, Hauner H: Functional characterization of promoter variants of the adiponectin gene complemented by epidemiological data. Diabetes. 2009, 58 (4): 984-991. 10.2337/db07-1646.
Zhang D, Ma J, Brismar K, Efendic S, Gu HF: A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J Diabetes Complications. 2009, 23 (4): 265-272. 10.1016/j.jdiacomp.2008.05.004.
Bowden DW, An SS, Palmer ND, Brown WM, Norris JM, Haffner SM, Hawkins GA, Guo X, Rotter JI, Chen YD, Wagenknecht LE, Langefeld CD: Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum Mol Genet. 2010, 19 (20): 4112-4120. 10.1093/hmg/ddq327.
Filippi E, Sentinelli F, Trischitta V, Romeo S, Arca M, Leonetti F, Di Mario U, Baroni MG: Association of the human adiponectin gene and insulin resistance. Eur J Hum Genet. 2004, 12 (3): 199-205. 10.1038/sj.ejhg.5201120.
Lasky-Su J, Lyon HN, Emilsson V, Heid IM, Molony C, Raby BA, Lazarus R, Klanderman B, Soto-Quiros ME, Avila L, Silverman EK, Thorleifsson G, Thorsteinsdottir U, Kronenberg F, Vollmert C, Illig T, Fox CS, Levy D, Laird N, Ding X, McQueen MB, Butler J, Ardlie K, Papoutsakis C, Dedoussis G, O'Donnell CJ, Wichmann HE, Celedon JC, Schadt E, Hirschhorn J, Weiss ST, Stefansson K, Lange C: On the replication of genetic associations: timing can be everything!. Am J Hum Genet. 2008, 82 (4): 849-858. 10.1016/j.ajhg.2008.01.018.
Greene CS, Penrod NM, Williams SM, Moore JH: Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009, 4 (6): e5639-10.1371/journal.pone.0005639.
Gibson F, Froguel P: Genetics of the APM1 locus and its contribution to type 2 diabetes susceptibility in French Caucasians. Diabetes. 2004, 53 (11): 2977-2983. 10.2337/diabetes.53.11.2977.
Service S, DeYoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, Ospina J, Ruiz-Linares A, Macedo A, Palha JA, Heutink P, Aulchenko Y, Oostra B, van Duijn C, Jarvelin MR, Varilo T, Peddle L, Rahman P, Piras G, Monne M, Murray S, Galver L, Peltonen L, Sabatti C, Collins A, Freimer N: Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet. 2006, 38 (5): 556-560. 10.1038/ng1770.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/12/5/prepub
Acknowledgements
This study has been financially supported by grants from the Academy of Finland (117844 and 40758, 211497 and 118590 (MU); 38387 and 46558 (JT); 206310 and 73566 (SKK)), The EVO-fund of Kuopio University Hospital (5106, 5168 and 5254 (MU)), The Ministry of Education of Finland, The Finnish Diabetes Research Foundation, Yrjö Jahnsson Foundation (56358 (MK)), Sigrid Juselius Foundation, The Graduate School of Molecular Medicine (NS), Nordic Centre of Excellence (NCoE) on "Systems biology in controlled dietary interventions and cohort studies" (SYSDIET; project number 070014), TEKES 70103/06 and 40058/07 and InterAct. The technical assistance of Ms. Minna Kiuttu is acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NS participated in designing the genetic studies, performed the statistical analyses, most of the genotyping and drafted the manuscript. LP participated in designing the genetic studies and writing the manuscript. MK participated in interpretation of data and writing the manuscript. JL, JGE, MV, PIP and SKK contributed to study design and coordination and revised the manuscript. JT and MU are the principle investigators of the DPS study and participated in writing the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Siitonen, N., Pulkkinen, L., Lindström, J. et al. Association of ADIPOQ gene variants with body weight, type 2 diabetes and serum adiponectin concentrations: the Finnish Diabetes Prevention Study. BMC Med Genet 12, 5 (2011). https://doi.org/10.1186/1471-2350-12-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-12-5