Abstract
Background
Neuron-derived orphan receptor (Nor) 1, nuclear receptor (Nur) 77, and nuclear receptor-related protein (Nurr) 1 constitute the NR4A family of orphan nuclear receptors which were recently found to modulate hepatic glucose production, insulin signalling in adipocytes, and oxidative metabolism in skeletal muscle. In this study, we assessed whether common genetic variation within the NR4A3 locus, encoding Nor-1, contributes to the development of prediabetic phenotypes, such as glucose intolerance, insulin resistance, or β-cell dysfunction.
Methods
We genotyped 1495 non-diabetic subjects from Southern Germany for the five tagging single nucleotide polymorphisms (SNPs) rs7047636, rs1526267, rs2416879, rs12686676, and rs10819699 (minor allele frequencies ≥ 0.05) covering 100% of genetic variation within the NR4A3 locus (with D' = 1.0, r2 ≥ 0.9) and assessed their association with metabolic data derived from the fasting state, an oral glucose tolerance test (OGTT), and a hyperinsulinemic-euglycemic clamp (subgroup, N = 506). SNPs that revealed consistent associations with prediabetic phenotypes were subsequently genotyped in a second cohort (METSIM Study; Finland; N = 5265) for replication.
Results
All five SNPs were in Hardy-Weinberg equilibrium (p ≥ 0.7, all). The minor alleles of three SNPs, i.e., rs1526267, rs12686676, and rs10819699, consistently tended to associate with higher insulin release as derived from plasma insulin at 30 min(OGTT), AUCC-peptide-to-AUCGluc ratio and the AUCIns30-to-AUCGluc30 ratio with rs12686676 reaching the level of significance (p ≤ 0.03, all; additive model). The association of the SNP rs12686676 with insulin secretion was replicated in the METSIM cohort (p ≤ 0.03, additive model). There was no consistent association with glucose tolerance or insulin resistance in both study cohorts.
Conclusion
We conclude that common genetic variation within the NR4A3 locus determines insulin secretion. Thus, NR4A3 represents a novel candidate gene for β-cell function which was not covered by the SNP arrays of recent genome-wide association studies for type 2 diabetes mellitus.
Similar content being viewed by others
Background
The NR4A family of orphan nuclear receptors comprises nuclear receptor (Nur) 77 (gene: NR4A1), nuclear receptor-related protein (Nurr) 1 (gene: NR4A2), and neuron-derived orphan receptor (Nor) 1 (gene: NR4A3). These ligand-independent constitutively active transcription factors are co-expressed in many metabolically relevant tissues, such as skeletal muscle, adipose tissue, liver, heart, and brain, and are thought to be predominantly regulated at the transcriptional level (for review, see [1]). In this context, a plethora of stimuli was identified, including fatty acids [2], growth factors [3], inflammatory cytokines [4, 5], peptide hormones [6, 7], membrane depolarisation [8], and stress [9], regulating NR4A family member expression in a tissue-specific manner. Recently, important metabolic functions of NR4A transcription factors were revealed. All three NR4A family members are induced in the liver upon fasting and glucagon stimulation, and their hepatic expression was found to be increased in both type 1 and type 2 diabetic mouse models [10].
Moreover, adenoviral over-expression of these receptors induced the expression of gluconeogenetic enzymes and enhanced hepatic glucose production in vitro as well as in vivo [10]. In 3T3-L1 adipocytes, NR4A1 (Nur-77) and NR4A3 (Nor-1) expression was reported to be induced upon insulin and thiazolidinedione treatment.
Furthermore, the expression of both receptors was found to be decreased in skeletal muscle and adipose tissue of multiple rodent models of insulin resistance and diabetes [11]. As lentiviral Nor-1 over-expression additionally was shown to enhance adipocyte insulin signalling and glucose transporter-4 translocation [11] and siRNA knock-down showed that Nor-1 regulates gene expression for oxidative metabolism in skeletal muscle [12], we asked whether common genetic variation within the genetic locus harbouring the Nor-1 gene NR4A3 (OMIM ID 600542, Entrez Gene ID 8013) contributes to the development of prediabetic phenotypes, especially such as glucose intolerance and insulin resistance.
To this end, we genotyped 1495 non-diabetic subjects from Southern Germany for the five tagging single nucleotide polymorphisms (SNPs) rs7047636, rs1526267, rs2416879, rs12686676, and rs10819699 (minor allele frequencies (MAFs) ≥ 0.05) covering 100% of genetic variation within the NR4A3 locus (with D' = 1.0 and r2 ≥ 0.9) and assessed their association with metabolic data derived from the fasting state, an oral glucose tolerance test (OGTT), and, in a subgroup of 506 subjects, from a hyperinsulinemic-euglycemic clamp. Found SNP-genotype associations were subjected to further analysis in the METSIM Study cohort from Finland (N = 5265) for replication purposes.
Methods
Study participants
TÜF/TULIP cohort
Data from 1495 non-diabetic participants of the Tübingen Family Study and the Tuebingen Lifestyle Intervention Program [13] were analyzed for this study. The participants (characteristics given in Table 1) did not take any medication known to affect glucose tolerance or insulin secretion, and were requested not to smoke 24 h before and during the test period. Informed written consent was obtained from all study participants, and the local ethics committee has approved the protocol.
METSIM cohort
The METabolic Syndrome In Men (METSIM) Study addresses a random sample of 6147 Finnish men aged from 45 to 70 years in Eastern Finland (Kuopio, see Table 1). All study participants underwent an oral glucose tolerance test (OGTT, see below), and the study protocol was approved by the local ethics committee. For the study of prediabetic traits, only non-diabetic METSIM participants were analyzed (N = 5265, aged 58 ± 6 years). METSIM participants with newly diagnosed diabetes (N = 882) according to the WHO criteria [14] allowed estimation for diabetes risk using logistic regression analysis.
Anthropometrics
Body mass index (BMI) was calculated as weight divided by the square of height (kg/m2). Body fat was determined by bioelectrical impedance (RJL, Detroit, MI, USA).
Analytical procedures
Blood glucose was measured with a bedside glucose analyzer (Yellow Springs Instruments, Yellow Springs, OH, USA). Plasma insulin and C-peptide concentrations were measured by commercial chemiluminescence assays for ADVIA Centaur (Siemens Medical Solutions, Fernwald, Germany) according to the manufacturer's instructions. In the METSIM Study, plasma glucose was measured by enzymatic hexokinase photometric assay (Konelab Systems Reagents, Thermo Fischer Scientific, Vantaa, Finland), and insulin was determined by immunoassay (ADVIA Centaur Insulin IRI, no 02230141, Siemens Medical Solutions Diagnostics, Tarrytown, NY).
Oral glucose tolerance test
The oral glucose tolerance test was performed according to the WHO recommendations after a 12-h fasting period [14]. In addition to plasma glucose, C-peptide levels and plasma insulin were measured at 0, 30, 60, 90 and 120 min in the TÜF/TULIP cohort. In the METSIM Study, plasma glucose and insulin levels were measured at 0, 30 and 120 min.
Calculations on insulin sensitivity and insulin secretion
Insulin sensitivity was estimated from insulin and glucose values obtained during the OGTT according to the (HOMA) homeostasis model assessment [15] or the method proposed by Matsuda and DeFronzo [16]. In 506 TÜF/TULIP Study participants, insulin sensitivity was additionally measured by a euglycemic hyperinsulinemic clamp, as described elsewhere [17]. In TÜF/TULIP, insulin secretion was estimated from insulin plasma levels measured at 30 min of the OGTT and the area under the curve (AUC) of C-peptide levels divided by the corresponding AUC of plasma glucose (AUCC-peptide-to-AUCGluc ratio) levels [18]. In both study cohorts, insulin secretion was estimated from insulin plasma levels measured at 30 min during the OGTT and from the AUCIns30-to-AUCGluc30 ratio [(insulin0 min + insulin30 min)/glucose0 min + glucose30 min)] that recently has been shown to be the best surrogate parameter for first phase insulin secretion in an intravenous glucose tolerance test [19].
Genotyping and selection of tagging SNPs
Genomic DNA was isolated from EDTA blood samples by blood cell lysis, protein precipitation and a washing protocol as previously described [17]. Genotyping was accomplished by use of the TaqMan Assay (Eurogentec, Liege, Belgium) and an ABI Prism 7500 sequence detection system (Applied Biosystems, Foster City, CA, USA) for selected tagging SNPs. Selection criteria for tagging SNPs of the NR4A3 locus on chromosome 9q22 were a minor allele frequency (MAF) ≥ 0.05 and a linkage disequilibrium measure r2 ≤ 0.8 in the CEU population of the HapMap project (Utah residents with ancestry from Northern and Western Europe).
Gene expression analyses in human islet and adipose tissue
Pancreata were obtained from brain-dead multiorgan donors and islet isolations were performed as previously described according to the Ricordi method with local (Geneva) adaptations [20, 21]. Briefly, the pancreas was distended by intraductal infusion of a cold collagenase solution. After digestion at 37°C in a modified Ricordi chamber, separated exocrine and endocrine tissues were washed and purified in a continuous Biocoll gradient. After several washings, islets were incubated in CMRL 1066 medium containing 5.6 mM glucose. RNA was extracted from cultivated (2d) human islets and adipose tissue specimens with PeqGOLD Tri Fast™ (PEQLAB Biotechnologie, Erlangen, Germany) according to the protocol supplied by the manufacturer, and quantitative real-time PCR was performed with Roche's LightCycler® 480 (Roche, Basel, Switzerland).
Statistical analyses
The Hardy-Weinberg equilibrium was tested with the χ2-test. Three-group comparisons without adjustments were performed using ANOVA. In addition, multivariate linear regression models with adjustments to relevant covariates were undertaken for genotype-phenotype association analyses. Non-normally distributed parameters were log-transformed. All data are presented as means ± SD. The software package JMP 7.0 (SAS Institute, Cary, NC, USA) and SPSS 14.0 (SPSS, Chicago, IL, USA) was used for statistical analysis. A p-value < 0.05 was considered to be statistically significant. Linkage disequilibrium was analyzed with the JLIN software [22].
Results
Study population
The TÜF/TULIP Study cohort of 1495 (506 male/989 female) non-diabetic subjects had a mean age of 39 ± 13 years. 10% had an impaired fasting glucose (IFG), 9.5% showed an impaired glucose tolerance (IGT) and 7.6% presented with both IFG and IGT in the OGTT. METSIM non-diabetic participants (N = 5265 men) had an average age of 58 ± 6 years, and 16.6 (9.5; 6.5)% were diagnosed with IFG (IGT; IFG+IGT; see Table 1). A total of 882 METSIM participants (59 ± 6 years) genotyped for NR4A3 were diagnosed with diabetes. Further details on basal anthropometric and metabolic traits of the investigated populations are provided in Table 1.
Genetic analyses
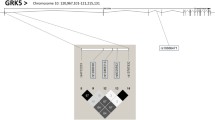
HapMap analysis of the NR4A3 locus revealed five tagging SNPs (rs7047636, rs1526267, rs2416879, rs12686676 and rs10819699) to cover 100% of variation in the NR4A3 gene locus (55 kb) including 5 kb of its 5'-flanking region and 5 kb of its 3'-flanking region (see Figure 1). Genotype call rates were > 98.4% for all investigated SNPs, and minor allele frequencies of the tagging SNPs ranged from 0.10 (rs2416879) to 0.46 (rs12686676; see Table 2) in the TÜF/TULIP cohort. All SNPs were in Hardy-Weinberg equilibrium (p ≥ 0.7, all; χ2-test), and linkage disequilibrium of the five selected tagging SNPs was rather low (r2 < 0.6, all; see Table 2).
Fifty-five-kb genomic region of human chromosome 9q22 harbouring the NR4A3 gene and HapMap LD data of the 17 common (minor allele frequency ≥ 0.05) informative SNPs within this region. The NR4A3 gene consists of eight exons and seven introns and spans 45 kb from nucleotide 101,623,958 to nucleotide 101,668,994. The locations of the five tagging SNPs rs7047636, rs1526267, rs2416879, rs12686676, and rs10819699 are indicated by black arrows. The Haploview LD colour scheme 'R-squared' was chosen to visualize regions of high linkage disequilibrium (black diamonds: complete LD; dark grey diamonds: strong LD; light grey diamonds: weak LD). Within the diamonds, the r2 values are given (black diamonds: r2 = 1.0). LD – linkage disequilibrium; SNP – single nucleotide polymorphism.
Association of NR4A3SNPs with anthropometric and metabolic traits in TÜF/TULIP
We found a significant association of the minor allele of rs7047636 with increased plasma glucose at 120 min of the OGTT (p = 0.03, additive model, adjusted for age, gender and BMI). The minor allele of rs1526267 was associated with a higher BMI (p = 0.02, additive model, adjusted for age and gender) and with decreased insulin sensitivity measured by clamp (p = 0.03, additive model, adjusted for age, gender and BMI). We did not detect any significant association of the four other tagging SNPs with BMI, body fat, blood glucose levels (fasting state and 120 min OGTT) and insulin sensitivity (see Table 3 and [Additional file 1]). The most consistent finding in the TÜF/TULIP population was a trend towards an association of the minor alleles of rs1526267, rs12686676 and rs10819699 with increased insulin secretion, with rs12686676 reaching the level of significance (p ≤ 0.03 after adjustment for age, gender, BMI and insulin sensitivity; Table 3). This finding was independent of the insulin secretion estimation model used for the analysis (insulin at 30 min OGTT, AUCC-peptide-to-AUCGluc ratio, AUCIns30-to-AUCGluc30 ratio; Table 3). All these findings could be reproduced if the Matsuda model [16] instead of the HOMA model [15] was used as an estimate of insulin sensitivity. In the dominant model of e.g. AUCC-peptide-to-AUCGluc ratio, the increase in insulin secretion amounted to 6.2% for rs1526267, 6.0% for rs12686676 and 3.7% for rs10819699. The intronic SNP rs1526267 is in a very low LD with the NR4A3-3'-flanking SNPs rs12686676 and rs10819699 in both the CEU population of the HapMap project (Figure 1) and our cohort (Table 2), and the fact that three rather independent SNPs associate with insulin secretion further underscores the role of NR4A3 in beta-cell function. The SNP rs2416879 also showed a significant association with two insulin secretion estimation models (insulin at 30 min OGTT, AUCIns30-to-AUCGluc30 ratio; p < 0.001 for adjusted models, [see Additional file 1]). However, no clear allele dose effect could be detected. Therefore, we doubted about the reliability of this finding and did not replicate this SNP in the METSIM Study.
Replication in the METSIM Study
Next, we tried to replicate the most reliable associations between SNPs (rs1526267, rs12686676, rs10819699) and insulin secretion in the METSIM cohort. Adjustment for age, BMI and insulin sensitivity (HOMA) revealed a significant association with both insulin secretion estimation models (insulin at 30 min OGTT, AUCIns30-to-AUCGluc30 ratio) in rs12686676, which yet showed the most convincing association in the TÜF/TULIP Study. In contrast to TÜF/TULIP, rs1526267 had no effect on BMI or insulin sensitivity in METSIM participants, and both rs1526267 and rs10819699 did not associate with insulin secretion (Table 4). Since the METSIM Study design also allows the analysis of NR4A3 genetic variants for the endpoint diabetes in a population-based cohort, we tested whether rs1526267, rs12686676 or rs10819699 associated with the prevalence of diabetes. However, comparing diabetic with non-diabetic study participants by logistic regression analysis did not reveal a significantly altered diabetes prevalence in minor allele carriers of the three investigated SNPs (p > 0.11; all, [see Additional file 2]). There were also no significant differences in genotype distribution of the investigated NR4A3 SNPs according to the glucose tolerance status (normal glucose tolerance, impaired fasting glucose, impaired glucose tolerance, diabetes) of both TUEF/TULIP and METSIM participants [see Additional file 3].
NR4A3 expression in human islets
NR4A3 gene expression was determined in four independent, freshly isolated, human islet preparations. Signal intensities of NR4A3 were comparable for all four islet preparations. Interestingly, NR4A3 signals were ~1.5 fold stronger in human islets as in two samples of adipose tissue included as a positive control, and remarkably high in comparison to two other positive controls, namely the house-keeping gene RPS13 (40S ribosomal protein S13) and IRS2 (insulin-receptor-substrate 2; Figure 2).
NR4A3 gene expression in human islets. RNA was isolated from four independent (islet1-4) freshly isolated human islet preparations, and NR4A3 gene expression was determined with quantitative real-time PCR in comparison with the house-keeping gene RPS13 (40S ribosomal protein S13) and insulin-receptor-substrate 2 (IRS2). RNA preparations of two adipose tissue samples were included as a positive control for NR4A3 expression analysis.
Discussion
This study investigated the role of NR4A3 for metabolic traits in a prediabetic population. Our primary hypothesis was that NR4A3 variants may associate with glucose intolerance or insulin resistance, based on published data on Nor-1 functions in liver and skeletal muscle [10–12, 23]. However, we did not detect any convincing associations between the selected NR4A3 tagging SNPs and glucose intolerance/insulin resistance in the two investigated study cohorts. Contrary to our expectation, the principal finding was that three out of the five investigated tagging SNPs, namely rs1526267, rs12686676, and rs10819699, showed a consistent trend towards an association with insulin secretion in the TÜF/TULIP cohort, with rs12686676 reaching a significant level after adjustment for confounding variables. This finding could be replicated in a second independent cohort of Finnish male subjects (METSIM Study). Summarizing our data obtained from both cohorts, we assume a rather dominant effect of NR4A3 on insulin secretion. However, further research is needed to verify this hypothesis in other cohorts, as all other detected diabetes risk genes show an additive allele effect [24]. It is of note that the tested SNPs were not covered by the SNP arrays used in recent genome-wide association studies (GWA), according to currently available statistic data http://www.wtccc.org.uk. Interestingly, most diabetes genes detected by the GWA studies are also involved in insulin secretory function [25–28], as it is the case for NR4A3.
As the effect of NR4A3 on insulin secretion was not dependent on the levels of non-esterified fatty acids as a read-out of adipose tissue metabolism determined in our study cohort (data not shown), we assume that NR4A3 genetic variability is an independent determinant for insulin secretion. Our finding of a remarkably high NR4A3 gene expression in freshly isolated human islets also supports a physiological role of NR4A3 in beta cells. Further evidence for this hypothesis arises from a study on target genes of a fusion protein encoded by a t(9;22) chromosomal translocation. The resulting EWS/NOR-1 fusion induces serum- and glucocorticoid-regulated kinase 1 (SGK1) [29], which mediates inhibition of insulin release in insulin-secreting cells [30].
As mentioned above, there was no replicable significant effect of NR4A3 genetic variability on insulin sensitivity in the two study populations, despite that both NR4A1 and NR4A3 expression is reduced in skeletal muscle of various diabetic animal models [11] and increased in L6 skeletal muscle cells and muscle biopsies upon insulin stimulation [31] or dietary restriction [32]. We also were not able to show a replicable association with BMI in our two study cohorts. This finding may reflect the fact that the NR4A family of nuclear orphan receptors was shown to be dispensable for adipogenesis, at least in the 3T3L1 adipocyte cellular model [33].
The following limitations of our study should be addressed: we did not a priori correct our data for multiple comparisons. Bonferroni correction for 5 independently analyzed SNPs (corrected α-level: 0.0102) would render all reported findings nominal. However, association with insulin secretion was consistently seen in three tagging SNPs in the NR4A3 gene and independent of the model used for estimation of insulin secretion (plasma insulin at 30 min OGTT, AUCC-peptide-to-AUCGluc ratio, AUCIns30-to-AUCGluc30 ratio) in the TÜF/TULIP Study cohort, and the most significant association of rs12686676 could be replicated in a second larger population (METSIM Study). As recent GWA studies mainly detected genes involved in insulin secretion as diabetes risk genes [24], one may assume that NR4A3 genetic variants have an impact on diabetes prevalence. Surprisingly, this was not the case in the METSIM cohort. As the METSIM study (diabetes prevalence: 13.8%) provides a power of 80% to detect a genotype relative risk of 1.15 assuming a SNP minor allele frequency of ~0.35 [see Additional file 3], we conclude that NR4A3 genetic variability may confer a rather modest effect on diabetes risk, and that analysis of this locus necessitates even larger replication samples.
Conclusion
We are confident that the described association between NR4A3 genetic variants and insulin secretion is not a by-chance finding, and that further research on the role of orphan nuclear receptors in beta cell function and diabetes pathogenesis is warranted. In addition, it would be interesting to investigate the role of NR4A3 genetic variants in large studies employing a case-control design.
References
Maxwell MA, Muscat GE: The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006, 4: e002-10.1621/nrs.04002.
Roche E, Buteau J, Aniento I, Reig JA, Soria B, Prentki M: Palmitate and oleate induce the immediate-early response genes c-fos and nur-77 in the pancreatic beta-cell line INS-1. Diabetes. 1999, 48 (10): 2007-2014. 10.2337/diabetes.48.10.2007.
Hazel TG, Nathans D, Lau LF: A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci USA. 1988, 85 (22): 8444-8448. 10.1073/pnas.85.22.8444.
Borghaei RC, Sinai RS, Mochan E, Pease EA: Induction of mitogen-inducible nuclear orphan receptor by interleukin 1 in human synovial and gingival fibroblasts. Biochem Biophys Res Commun. 1998, 251 (1): 334-338. 10.1006/bbrc.1998.9477.
Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P: Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005, 280 (32): 29256-29262. 10.1074/jbc.M502606200.
Tetradis S, Bezouglaia O, Tsingotjidou A, Vila A: Regulation of the nuclear orphan receptor Nur77 in bone by parathyroid hormone. Biochem Biophys Res Commun. 2001, 281 (4): 913-916. 10.1006/bbrc.2001.4459.
Tetradis S, Bezouglaia O, Tsingotjidou A: Parathyroid hormone induces expression of the nuclear orphan receptor Nurr1 in bone cells. Endocrinology. 2001, 142 (2): 663-670. 10.1210/en.142.2.663.
Katagiri Y, Hirata Y, Milbrandt J, Guroff G: Differential regulation of the transcriptional activity of the orphan nuclear receptor NGFI-B by membrane depolarization and nerve growth factor. J Biol Chem. 1997, 272 (50): 31278-31284. 10.1074/jbc.272.50.31278.
Honkaniemi J, Kononen J, Kainu T, Pyykonen I, Pelto-Huikko M: Induction of multiple immediate early genes in rat hypothalamic paraventricular nucleus after stress. Brain Res Mol Brain Res. 1994, 25 (3–4): 234-241. 10.1016/0169-328X(94)90158-9.
Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P: NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat Med. 2006, 12 (9): 1048-1055. 10.1038/nm1471.
Fu Y, Luo L, Luo N, Zhu X, Garvey WT: NR4A orphan nuclear receptors modulate insulin action and the glucose transport system: potential role in insulin resistance. J Biol Chem. 2007, 282 (43): 31525-31533. 10.1074/jbc.M701132200.
Pearen MA, Myers SA, Raichur S, Ryall JG, Lynch GS, Muscat GE: The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008, 149 (6): 2853-2865. 10.1210/en.2007-1202.
Schafer S, Kantartzis K, Machann J, Venter C, Niess A, Schick F, Machicao F, Haring HU, Fritsche A, Stefan N: Lifestyle intervention in individuals with normal versus impaired glucose tolerance. European Journal of Clinical Investigation. 2007, 37 (7): 535-543. 10.1111/j.1365-2362.2007.01820.x.
Alberti KG, Zimmet PZ: Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998, 15 (7): 539-553. 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S.
Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC: Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985, 28 (7): 412-419. 10.1007/BF00280883.
Matsuda M, DeFronzo RA: Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999, 22 (9): 1462-1470. 10.2337/diacare.22.9.1462.
Weyrich P, Machicao F, Staiger H, Simon P, Thamer C, Machann J, Schick F, Guirguis A, Fritsche A, Stefan N, et al: Role of AMP-activated protein kinase gamma 3 genetic variability in glucose and lipid metabolism in non-diabetic whites. Diabetologia. 2007, 50 (10): 2097-2106. 10.1007/s00125-007-0788-8.
Utzschneider KM, Prigeon RL, Tong J, Gerchman F, Carr DB, Zraika S, Udayasankar J, Montgomery B, Mari A, Kahn SE: Within-subject variability of measures of beta cell function derived from a 2 h OGTT: implications for research studies. Diabetologia. 2007, 50 (12): 2516-2525. 10.1007/s00125-007-0819-5.
Stancakova A, Javorsky M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M: Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009, 58 (5): 1212-1221. 10.2337/db08-1607.
Ricordi C, Lacy PE, Finke EH, Olack BJ, Scharp DW: Automated method for isolation of human pancreatic islets. Diabetes. 1988, 37 (4): 413-420. 10.2337/diabetes.37.4.413.
Bucher P, Mathe Z, Morel P, Bosco D, Andres A, Kurfuest M, Friedrich O, Raemsch-Guenther N, Buhler LH, Berney T: Assessment of a novel two-component enzyme preparation for human islet isolation and transplantation. Transplantation. 2005, 79 (1): 91-97. 10.1097/01.TP.0000147344.73915.C8.
Carter KW, McCaskie PA, Palmer LJ: JLIN: a java based linkage disequilibrium plotter. BMC Bioinformatics. 2006, 7: 60-10.1186/1471-2105-7-60.
Kumar N, Liu D, Wang H, Robidoux J, Collins S: Orphan nuclear receptor NOR-1 enhances 3',5'-cyclic adenosine 5'-monophosphate-dependent uncoupling protein-1 gene transcription. Mol Endocrinol. 2008, 22 (5): 1057-1064. 10.1210/me.2007-0464.
Doria A, Patti ME, Kahn CR: The emerging genetic architecture of type 2 diabetes. Cell Metab. 2008, 8 (3): 186-200. 10.1016/j.cmet.2008.08.006.
Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al: Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007, 316 (5829): 1336-1341. 10.1126/science.1142364.
Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, et al: Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007, 316 (5829): 1331-1336. 10.1126/science.1142358.
Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al: A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007, 316 (5829): 1341-1345. 10.1126/science.1142382.
Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007, 445 (7130): 881-885. 10.1038/nature05616.
Poulin H, Filion C, Ladanyi M, Labelle Y: Serum- and glucocorticoid-regulated kinase 1 (SGK1) induction by the EWS/NOR1(NR4A3) fusion protein. Biochem Biophys Res Commun. 2006, 346 (1): 306-313. 10.1016/j.bbrc.2006.05.134.
Ullrich S, Berchtold S, Ranta F, Seebohm G, Henke G, Lupescu A, Mack AF, Chao CM, Su J, Nitschke R, et al: Serum- and glucocorticoid-inducible kinase 1 (SGK1) mediates glucocorticoid-induced inhibition of insulin secretion. Diabetes. 2005, 54 (4): 1090-1099. 10.2337/diabetes.54.4.1090.
Wu X, Wang J, Cui X, Maianu L, Rhees B, Rosinski J, So WV, Willi SM, Osier MV, Hill HS, et al: The effect of insulin on expression of genes and biochemical pathways in human skeletal muscle. Endocrine. 2007, 31 (1): 5-17. 10.1007/s12020-007-0007-x.
Oita RC, Mazzatti DJ, Lim FL, Powell JR, Merry BJ: Whole-genome microarray analysis identifies up-regulation of Nr4a nuclear receptors in muscle and liver from diet-restricted rats. Mech Ageing Dev. 2009, 130 (4): 240-247. 10.1016/j.mad.2008.12.004.
Au WS, Payne VA, O'Rahilly S, Rochford JJ: The NR4A family of orphan nuclear receptors are not required for adipogenesis. Int J Obes (Lond). 2008, 32 (2): 388-392. 10.1038/sj.ijo.0803763.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2350/10/77/prepub
Acknowledgements
We thank all study participants for their cooperation. We thank the International HapMap Consortium for the public allocation of genotype data. We gratefully acknowledge the excellent technical assistance of Alke Guirguis, Melanie Weisser, Anna Bury, Heike Luz and Roman-Georg Werner. The study was supported in part by grants from the German Research Foundation (KFO 114/2 and GA386/9-1) and the European Community's frame program FP6 EUGENE 2 (LSHM-CT-2004-512013). Human islets were obtained from Geneva through the European Consortium for Islet Transplantation (ECIT), supported by JDRF, grant number 31-2008-416.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PW and HS were responsible for the complete statistical analysis and prepared all tables, figures and the manuscript. AS did the complete statistical analysis for the METSIM cohort and prepared tables and the manuscript. SAS and KK made contributions to phenotyping of TÜF/TULIP participants. FM is responsible of the genotyping facility at the University of Tübingen and additionally performed the real-time PCR studies. SU, FR and BG provided freshly isolated human islet preparations. ML and JK are the principal investigators of the METSIM Study and responsible for METSIM patient and data management. NS, AF and HUH acted as principal investigators for the TÜF/TULIP study, and were responsible for patient and data management. All authors read and approved the final manuscript.
Peter Weyrich, Harald Staiger contributed equally to this work.
Electronic supplementary material
12881_2009_491_MOESM1_ESM.xls
Additional file 1: Associations of NR4A3 SNPs rs7047636 and rs2416879 with anthropometric and metabolic data (TÜF/TULIP cohort, N = 1495). Data are given as means ± SD. For statistical analysis, data were log-transformed. p1 – non-adjusted additive model; p2 – adjusted additive model: age was adjusted for gender; BMI and body fat were adjusted for gender and age; glucose (Gluc) levels as well as insulin sensitivity (HOMA model, clamp) and insulin (Ins) secretion (AUCC-peptide-to-AUCGluc ratio, AUCIns30-to-AUCGluc30 ratio, plasma insulin at 30 min OGTT) were adjusted for gender, age, and BMI; p3 – Ins secretion parameters with adjustment for age, BMI, gender and insulin sensitivity. *clamped subgroup (N = 506). AUC – area under the curve; BMI – body mass index; OGTT – oral glucose tolerance test; SNP – single nucleotide polymorphism. (XLS 20 KB)
12881_2009_491_MOESM2_ESM.doc
Additional file 2: Logistic regression analysis for differences of NR4A3 SNP's minor allele prevalences in individuals with normal glucose tolerance (NGT) and overt diabetes mellitus (DM) in the METSIM Study. The [95%] confidence interval of the data provided is indicated for all different statistical models (additive, dominant, recessive) for all investigated SNPs. (DOC 70 KB)
12881_2009_491_MOESM3_ESM.doc
Additional file 3: Distribution of NR4A3 SNP minor allele frequencies according to glucose tolerance status in the TÜF/TULIP (N = 1495) and the METSIM (N = 6147) cohort. Minor allele frequencies for each investigated NR4A3 SNP are presented for study participants (TUEF-TULIP/METSIM trial) with normal glucose tolerance (NGT), impaired fasting glucose and/or impaired glucose tolerance (IFG/IGT) and with manifest diabetes (METSIM participants only). p1 – NGT vs. IFG/IGT in TUEF-TULIP/METSIM(χ2-test); p2 – NGTvs.IFG/IGTvs.DIABETESinMETSIM(χ2-test). SNPs screened in TUEF/TULIP only and not replicated in METSIM are marked with an asterisk. (DOC 36 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Weyrich, P., Staiger, H., Stančáková, A. et al. Common polymorphisms within the NR4A3 locus, encoding the orphan nuclear receptor Nor-1, are associated with enhanced β-cell function in non-diabetic subjects. BMC Med Genet 10, 77 (2009). https://doi.org/10.1186/1471-2350-10-77
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2350-10-77