Abstract
Background
Within the genus Aspergillus, A. flavus is the second most important species of clinical significance. It is predominantly associated with infections involving sinuses, eye and skin, mostly in geographic regions with hot and arid climate, including the Middle East. Recent reports on emergence of resistance to triazoles among Aspergillus spp. is a cause of concern for treatment of patients with invasive aspergillosis. In this study we present data on genetic characterization and antifungal susceptibility profile of clinical and environmental isolates of A. flavus.
Methods
Ninety-nine Aspergillus section Flavi isolates, originating from clinical (n=92) and environmental (n=7) sources, initially identified by morphological characteristics, were analyzed by partial sequencing of β-tubulin and calmodulin gene fragments and their susceptibilities to six antifungal agents was determined by Etest on RPMI1640 and Muller-Hinton agar media. Etest minimum inhibitory concentrations (MICs) of amphotericin B and voriconazole were also compared with zone of inhibition diameters obtained by disc diffusion test on RPMI agar medium.
Results
The identity of all clinical and environmental isolates was confirmed as A. flavus species by combined analysis of β-tubulin and calmodulin genes. The mean MIC90 (μg/ml) values on RPMI medium for amphotericin B, voriconazole, posaconazole, anidulafungin, micafungin and caspofungin were 3, 0.25, 0.25, 0.002, 0.002 and 0.032, respectively. No environmental isolate exhibited MIC value of >2 μg/ml for amphotericin B. For clinical isolates, the zone of inhibition diameters for amphotericin B and voriconazole ranged from 7–16 mm and 24–34 mm, respectively. Linear regression analysis between Etest MIC values and disk diffusion diameters revealed a significant inverse correlation with amphotericin B (p <0.001) and voriconazole (p<0.003).
Conclusions
The β-tubulin and calmodulin gene sequences confirmed that all 92 clinical isolates identified phenotypically belonged to A. flavus taxon, thus suggesting that the other species within Aspergillus section Flavi are of little clinical significance. Triazoles and echinocandins showed very good in vitro activity against the A. flavus, however, 10% clinical isolates showed MICs of >2 μg/ml for amphotericin B.
Similar content being viewed by others
Background
Among filamentous fungal pathogens, Aspergillus spp. account for highest rates of morbidity and mortality among severely immunocomnpromised patients [1]. Although A. fumigatus is the principal etiologic agent of invasive aspergillosis, the etiologic role of non-fumigatus Aspergillus species is being increasingly recognized [2–4]. Aspergillus flavus is the second most important species causing localized as well as systemic infections [2, 5–7]. The species is of particular significance in North Africa, India and the Middle East, where it is predominantly associated with nasal/sinus infections [8–12]. In 2008, Clinical and Laboratory Standards Institute produced standard guidelines based on broth microdilution (BMD) method for determining antifungal susceptibilities of spore-forming filamentous fungi [13]. Recently, epidemiologic cutoff values (ECVs) for triazoles and caspofungin for wild-type strains of Aspergillus spp. have been developed [14–16]. However, due to interspecies/inter-strain differences in MICs, clinical breakpoints for each Aspergillus spp. are not yet available. Occurrence of azole resistance among wild-type strains of A. fumigatus due to mutations in cyp51A gene and possibility of finding similar resistance in other Aspergillus spp [17, 18] have necessitated the need to evaluate efficacy of simple agar-based methods for antifungal susceptibility testing. Preliminary studies have suggested that disk diffusion test and Etest show comparable results with BMD method for susceptibility testing with azoles [19–22]. Here, we present molecular characterization and MIC results of six antifungal agents tested against 99 A. flavus isolates by Etest and compare amphotericin B and voriconazole MICs with zone of inhibition diameters obtained by disk diffusion test.
Methods
Aspergillus flavusisolates
Ninety-two clinical and 7 environmental isolates of A. flavus were included in the study (Table 1). The clinical isolates were recovered from a variety of specimens over an 18-year period (1993–2011). They were deposited in the culture collection of the Mycology Reference Laboratory and maintained by periodic sub-culture. Since the isolates were obtained during routine mycological investigations, there was no ethical requirement to take approval from individual patients for their subsequent use. The study (Project No. YM 03/10) was approved by the Ethical Committee of Faculty of Medicine, Health Sciences Center, Kuwait University. All isolates were identified as A. flavus by typical colony and microscopic characteristics as described by Klich [23]. The isolates were also characterized and speciated by molecular methods.
Molecular characterization
The genomic DNA from the isolates was prepared as described previously and used as template for PCR amplification [24]. The ITS region (ITS-1, 5.8 S rRNA and ITS-2) of rDNA was amplified with Aspergillus section Flavi-specific AFLF and AFLR primers as described in detail previously [10]. The species-specific identification of all isolates was studied by partial sequencing of β-tubulin and calmodulin gene fragments. The variable region of β-tubulin gene was amplified by using BTUBF (5′-TGGTAACCAAATCGGTGCTGCTT-3′) and BTUBR (5′-GCACCCTCAGTGTAGTGACCCT-3′) primers while the variable region of calmodulin gene was amplified by using Cmd5 (5′-GTCTCCGAGTACAAGGAGGC-3′) and Cmd6 (5′-TCGCCGATRGAGGTCATRACGTG-3′) primers and the amplicons were sequenced as described previously [25]. GenBank basic local alignment search tool (BLAST) searches (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi?) were performed for species identification. The DNA sequences for type strains already available in GenBank were retrieved. The gene sequences were analyzed individually or nucleotide sequences of both, β-tubulin and calmodulin gene fragments were included in the combined analysis. Multiple sequence alignments were performed with ClustalX version 2.0. The phylogenetic trees were constructed using the neighbor-joining method with pair-wise deletion of gaps option. Aspergillus parasiticus (CBS100926) was chosen as the outlying taxon and the robustness of branches was assessed by bootstrap analysis with 1,000 replicates.
Antifungal susceptibility testing
Preparation of inoculum
All isolates were freshly sub-cultured on potato dextrose agar (PDA) slants to obtain good sporulation. The culture tubes were flooded with 1 ml of 0.9% saline and vortexed for 15 seconds to dislodge the conidia. The growth suspensions were transferred to another sterile tube containing 1.5-ml saline and 0.2% Tween 80. A conidial suspension containing approximately 0.4 × 104 to 5 × 104 cells was used as inoculum [13]. Reference strains of A. flavus (CBS100927) and A. parasiticus (CBS100926) were included with each batch of susceptibility testing to ensure quality control.
Etest
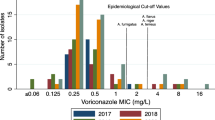
Two media, namely, RPMI 1640 supplemented with 2% glucose (pH adjusted to 7.0 with morpholinepropanesulfonic acid buffer) and Mueller-Hinton (MH) II agar (Becton, Dickinson and Company, Sparks, MD, USA) supplemented with 2% glucose and methylene blue (0.5 μg/ml) (MH-GMB) were used for Etest. Etest was performed according to the manufacturer’s instructions Briefly, each 150-mm petri plate containing 60 ml of medium was inoculated by streaking the swab over the entire surface of the medium. Etest antifungal susceptibility strips for amphotericin B, voriconazole, posaconazole, caspofungin, anidulafungin and micafungin were obtained from bioMerieux Sa, Marcy-l’Etoile, France. Before applying Etest strips, the plates were allowed to dry for about 15 min. MIC readings were taken after 24 h of incubation at 35°C. The E-test MIC was defined as the lowest drug concentration at which the border of the elliptical inhibition zone intercepted the scale on the antifungal strip, Microcolonies within the inhibition zone were ignored. Considering that newer azoles are the treatment of choice for Aspergillus infections, the European Committee on Antifungal Susceptibility Testing (EUCAST): (http://www.eucast.org/antifungal_susceptibility_testing_afst/) has recently proposed the following clinical break-points for A. fumigatus: for itraconazole and voriconazole, ≤ 1 mg/L (susceptible) and >2 mg/L (resistant) and for posaconazole, <0.12 mg/L (susceptible) and >0.25 mg/L (resistant) [26]. For A. flavus, the clinical breakpoints for itraconazole are the same as for A. fumigatus, whereas for other antifungal agents there is insufficient evidence to recommend clinical breakpoints. The epidemiologic cutoff values proposed on the basis of MIC (mg/L) distribution of wild-type strains of A. flavus are as follows: itraconazole, 1 (99.6%); posaconazole, 0.25 (95%); voriconazole, 1 (98.1%); caspofungin, 0.5 (99%) and amphotericin B, 4 (99%), which are generally one step higher than A. fumigatus[14–16].
Disk diffusion test
For the purpose of comparing zone of inhibition results, the disk diffusion test was performed on RPMI 1640 medium supplemented with 2% glucose instead of Mueller-Hinton agar medium. Disk diffusion disks for voriconazole (1-μg) and amphotericin B (100-μg) were obtained commercially (Bio-Rad Laboratories, Marnes-la-Coquette, France). After applying the discs, the plates were incubated at 35°C for 24 h. Zone diameters were measured at the point where the growth significantly decreased (80 to 100% inhibition) and were recorded to the nearest millimeter. Recently, Espinel-Ingroff et al. proposed for the first time quality control and reference zone diameter limits for A. fumigatus (ATCC MYA-3626) for amphotericin B (10-μg disk), itraconazole (10-μg disk), posaconazole (10-μg disk), and voriconazole (10-μg disk) [27]. No quality control ranges were recommended for A. flavus and A. terreus, because some results did not meet M23-A3 document requirements [28]. However, for A. flavus, the acceptable zone diameter ranges for voriconazole and posaconazole were proposed as 25–36 mm and 27–37 mm, respectively.
Statistical analysis
The Spearman correlation test was performed to determine the correlation between Etest MICs and zone of inhibition diameters. A P value of <0.05 was considered as significant. Independent samples T test was used to compare mean MIC values of A. flavus strains isolated in two different periods of 9-year each (1993–2001 and 2002–2011).
Results
Molecular characterization
All 99 A. flavus isolates included in this study were identified as belonging to Aspergillus section Flavi by specific amplification of a DNA fragment of expected size (~243 bp) in PCR with Aspergillus section Flavi-specific primers AFLF and AFLR [10]. The partial β-tubulin DNA sequences from 94 isolates were completely identical and showed no difference with sequence from reference A. flavus (CBS100927) strain while the sequences of remaining 5 isolates differed at only 1 nucleotide position. The calmodulin gene sequences of the 99 isolates were more variable (total 8 patterns) but still showed maximum identity with sequence from reference A. flavus (CBS100927) strain than with reference strains of other species belonging to Aspergillus section Flavi. The β-tubulin and calmodulin gene sequences were combined and the combined data set was compared with corresponding combined data set from reference strains of A. flavus CBS100927, A. parvisclerotigenus CBS121.62, A. minisclerotigenes CBS117635, A. parasiticus CBS100926, A. sojoe CBS100928, A. celatus CBS763.97, A. tamarii CBS104.13 and A. nomius CBS260.88 strains. The combined sequences from A. oryzae CBS100925 and other synonyms of A. flavus (A. fasciculatus CBS110.55, A. thomii CBS120.51, A. kambarensis CBS542.69 and A. subolivaceus CBS501.65) were also used [29]. The combined β-tubulin and calmodulin gene sequences of the 99 isolates also showed maximum identity with sequence from reference A. flavus (CBS100927) strain. Pair-wise sequence comparisons showed that all 99 isolates exhibited 11 different patterns with three large clusters corresponding to 3 patterns containing 78 isolates (Table 2). The neighbor-joining phylogenetic tree based on combined data set for β-tubulin and calmodulin gene sequences (using only one representative isolate from each of 11 patterns) is shown in Figure 1. All 11 representative A. flavus isolates from Kuwait clustered together with A. flavus strains on a separate branch. The unique DNA sequencing data reported here have been submitted to EMBL under accession nos. HF570030-HF570051.
Neighbor-joining phylogenetic tree based on combined β-tubulin and calmodulin gene sequence data for selected A. flavus isolates, each representing the 11 unique patterns, from Kuwait together with reference strains of several species belonging to Aspergillus section Flavi . Total number of isolates in various clusters are indicated within brackets. Numbers on the nodes branches are bootstrap frequencies. Only values above 50% are indicated.
Antifungal susceptibility
The results of Etest antifungal susceptibility of clinical isolates of A. flavus are presented in Table 3. In general, all tested antifungal agents showed good in vitro activity against A. flavus. The MIC range and MIC90 values on RPMI 1640 medium were as follows: amphotericin B, 0.064 – 4 and 3 μg/ml; voriconazole, 0.064 – 0.25 and 0.25 μg/ml; posaconazole, 0.016 – 0.38 and 0.25 μg/ml; anidulafungin, 0.002 – 0.006 and 0.002 μg/ml; micafungin, 0.002 – 0.008 and 0.002 μg/ml and caspofungin, 0.002 – 0.125 and 0.032 μg/ml, respectively. The MIC90 values obtained on MH-GMB were similar to RPMI values for all the tested antifungal agents except amphotericin B where it was 4 μg/ml.
The cumulative percentage of A. flavus isolates inhibited at different MIC values are presented in Table 4. Of the 92 clinical isolates tested, 74.2% (n=69) and 59.1% (n=55) isolates were inhibited by amphotericin B at MIC of ≤1.024 μg/ml on RPMI and MH-GMB medium, respectively. About 11% (n=10) of the isolates showed MICs of >2 μg/ml on RPMI medium. Among azoles, voriconazole inhibited 100% of the isolates at a concentration of 0.256 μg/ml on both test media. At this concentration, posaconazole inhibited 100% of the isolates on RPMI and 93.5% of the isolates on MH-GMB. For three echinocandins, micafungin and anidulafungin demonstrated comparable in vitro activity inhibiting 100% of the isolates at a MIC of ≤0.008 μg/ml on RPMI medium, whereas caspofungin inhibited 63.4% of the isolates on RPMI and 33.3% of the isolates on MH-GMB. A comparison of the mean MIC values of A. flavus isolates recovered in two 9-year periods (1993–2001 and 2002–2011) did not show any significant difference for the antifungal drugs tested.
The mean and range of MIC values of seven environmental A. flavus isolates on RPMI medium for the six antifungal agents were as follows: amphotericin B, 1.25 μg/ml ± 0.577 (range 0.75-2 μg/ml); voriconazole, 0.138 μg/ml ± 0.050 (range 0.094-0.25 μg/ml); posaconazole, 0.079 μg/ml ± 0.087 (range 0.008-0.25 μg/ml); anidulafungin, 0.002 μg/ml ± 0.00 (range 0.002-0.002 μg/ml); micafungin, 0.002 ± 0.00 (range 0.002-0.002 μg/ml) and caspofungin, 0.071 μg/ml ± 0.082 (range 0.008-0.19 μg/ml), respectively. These mean MIC values were comparable with those obtained on MH-GMB medium. None of the environmental isolates exhibited MIC value of >2 μg/ml for amphotericin B.
Disk diffusion test
Using amphotericin B (100-μg) and voriconazole (1-μg) disks, the mean values for zone of inhibition for clinical isolates were 10.38 ± 1.655 mm (range 7–16 mm) and 28.88 ± 2.321 mm (24–34 mm), respectively (Table 5). For environmental isolates, the mean values for zone of inhibition were 10.71 ± 1.38 mm (range 8–12 mm) and 27.71 ± 1.603 mm (26–30 mm) for amphotericin B and voriconazole, respectively. The differences in inhibition zone diameters between clinical and environmental isolates for voriconazole and amphotericin B were not statistically significant. Linear regression analysis between Etest MIC values and disk diffusion diameters obtained with clinical isolates revealed a significant inverse correlation, both with amphotericin B (R2 = 0.2536, p <0.001) and voriconazole (R2 = 0.4043, p<0.003).
Discussion
This study provides molecular characterization and in vitro antifungal susceptibility data on clinical and environmental isolates of A. flavus from Kuwait. All clinical and environmental isolates were identified as A. flavus strains based on the partial β-tubulin and calmodulin sequences. The phylogenetic tree derived from combined β-tubulin and calmodulin gene sequences also showed that all 99 isolates analyzed in this study clustered with reference strains of A. flavus or synonyms of A. flavus[29] clearly distinct from A. parvisclerotigenus, A. minisclerotigenes and A. parasiticus, the latter being used as an out-group. These studies established species-specific identity of all clinical and environmental isolates used in this study.
The in vitro drug susceptibility testing data showed that all the tested drugs exhibited good activity except amphotericin B, where ~10% isolates showed MIC of >2 μg/ml on RPMI medium. Although no antifungal susceptibility breakpoints are available for A. flavus, there is a consensus that isolates demonstrating MIC values of >1 μg/ml may be considered resistant to amphotericin B [26, 30, 31]. Badiee et al. [32] determined antifungal susceptibility of 66 A. flavus isolates from Iranian patients by Etest and CLSI methods. Thirty-six of the isolates were resistant to amphotericin B by both the methods. In another study from Tunesia, 31 of 37 isolates of A. flavus obtained from 14 patients with hematological malignancies were resistant (≥2 μg/ml) to amphotericin B [33]. Notably, survival rate was much lower among patients who yielded resistant strains (22% versus 67%) and were treated with amphotericn B. Similar results were obtained in a previous study [31]. However, with the introduction of voriconazole as a primary therapy for invasive aspergillosis, the concerns about amphotericin B resistance have been largely addressed in resource-rich countries.
All our A. flavus isolates were inhibited at a concentration of ≤0.256 μg/ml, which is lower or equal to the recently proposed clinical breakpoints for resistance to voriconazole (>2 μg/ml) and posaconazole (>0.25μg/ml) for A. fumigatus complex isolates [26]. Consistent with our observations, none of the 98 A. flavus clinical isolates tested in the Netherlands exhibited resistance to itraconazole or voriconazole [17]. So far, acquired resistance to azoles in A. flavus is extremely rare [34]. However, recent reports of voriconazole resistance in an A. flavus isolate cultured from lung specimen of a patient in China and a case of voriconazole-refractory eye infection in a patient from India have posed new therapeutic challenges [35, 36]. It is unclear why itraconazole/voriconazole-resistance among A. flavus isolates is so rare despite their wide-spread environmental prevalence and exposure to same azole fungicides that are apparently related to development of resistance in A. fumigatus[17], more so, when voriconazole-resistant strains of A. flavus can be readily obtained in the laboratory [37]. Although we tested only seven environmental isolates of A. flavus, their mean MIC values were marginally lower than clinical isolates for voriconazole and posaconazole but higher for amphotericin B. In one previous study, MIC values of environmental isolates (n=59) for amphotericin B and itraconazole were found to be significantly lower than the clinical isolates (n=29) (p < 0.05) [38].
All three echinocandins showed good in vitro activity against A. flavus by Etest. Consistent with several previous reports, anidulafungin and micafungin appeared more potent than caspofungin [21, 22, 39, 40]. Pfaller et al. reported that, caspofungin, micafungin and anidulafungin inhibited all A. flavus isolates (n=64) at a concentration of ≤0.06 μg/ml [39]. Recently, Espinel-Ingroff et al. determined epidemiological cut-off values (ECVs) for triazoles, caspofungin and amphotericin B using wild-type isolates of Aspergillus spp. employing CLSI microdilution methodology [14–16]. Our Etest MICs obtained with A. flavus isolates compared well with ECVs described in the above studies, involving 100% of the isolates with voriconazole, 98.9% of the isolates with posaconazole, and 100% of the isolates with caspofungin and amphotericin B (Table 6) [14–16]. Although we used Etest, the data suggest that there are no notable differences in the MICs of clinical and wild-type isolates of A. flavus. Furthermore, there was no significant difference in the mean MIC values of 14 A. flavus isolates recovered in the first 9-year period (1993–2001) compared to 78 A. flavus isolates recovered in the second 9-year period (2002–2011) for all the antifungal drugs tested.
A limitation of our study is the non-availability of BMD data for comparison with Etest MICs. Similar to some other studies, we also ignored the presence of microcolonies within the Etest inhibition zone [21, 22]. A good concordance has been reported between MICs obtained by Etest and BMD test for posaconazole (84% to 98%) [41, 42], itraconazole (100%) [43, 44], and voriconazole (85%) [44–46] against Aspergillus spp. at both, 24 and 48 h readings in studies that have used these two methods. Recently, Colozza et al. compared CLSI and Etest MICs for determining Aspergillus spp. susceptibility to amphotericin B and AmBisome [30]. A high percentage of A. flavus complex isolates were resistant (>1 μg/ml) to AmBisome (43.7%) than amphotericin B (16.7%) by BMD. This was in contrast with the amphotericin B susceptibility results obtained by BMD method for the same isolates, but not with those obtained by the Etest. The authors inferred that Etest may be a superior method than BMD for determining amphotericin B-resistant Aspergillus strains, perhaps because of the wider range of MIC distribution available on the Etest strip.
There is paucity of data on correlation between inhibition zone diameters and Etest MICs against Aspergillus spp. A few studies available in the literature suggest that agar diffusion methods have potential value for evaluating susceptibility to antifungal agents [47, 48]. In a multicenter study, Espinel-Ingroff et al. evaluated agar diffusion method for susceptibility testing of filamentous fungi on plain Muller-Hinton agar medium [47]. The investigators suggested that using 5-μg disk of posaconazole and caspofungin, 1-μg disk of voriconazole, and 10-μg disk for itraconazole, and an incubation time of 24 h are optimal for determining susceptibility of three Aspergillus spp. (A. fumigatus, A. flavus, and A. niger). Recently, Espinel-Ingroff et al. [27], following CLSI described guidelines [49], established zone diameter limits for disk diffusion susceptibility testing for A. fumigatus. No quality control ranges were recommended for A. flavus and A. terreus as some of the results did not satisfy the M23-A3 document requirements [28]. However, the zone diameter ranges of 27 to 37 mm for posaconazole and 25 to 36 mm for voriconazole obtained by the authors for A. flavus isolates were considered acceptable. We obtained a comparable zone diameter range of 24 to 34 mm with 1-μg voriconazol disk, whereas zone diameters for amphotericin B were much narrower (7 to 16 mm) despite the fact that we used 100-μg disks. More recently, Martos et al. evaluated disk diffusion method for determining susceptibility to echinocandins [21]. For all Aspergillus spp. (including 18 A. flavus isolates), caspofungin disk provided narrower zone of inhibition (14–29 mm) than micafungin (14–40 mm) and anidulafungin (22–45 mm) at 24 h incubation, which is in concordance with relatively higher Etest MICs obtained with caspofungin than micafungin and anidulafungin obtained in the present study.
Conclusion
All 92 clinical isolates collected over an 18-year period in Kuwait and phenotypically identified as A. flavus, were also characterized as A. flavus strains by partial sequencing of β-tubulin and calmodulin gene fragments. The triazoles and echinocandins showed good activity against clinical as well as environmental A. flavus isolates, however, nearly 11% and 18% isolates showed MIC of >2 μg/ml against amphotericin B on RPMI agar medium and Mueller-Hinton agar medium, respectively. There was a significant inverse correlation between Etest MICs and inhibition zone diameter values with voriconazole and amphotericin B. To the best of our knowledge, this is the first study reporting results of molecular characterization of A. flavus isolates from the Middle East.
References
Dasbach EJ, Davies GM, Teutsch SM: Burden of aspergillosis-related hospitalizations in the United States. Clin Infect Dis. 2000, 31: 1524-1528. 10.1086/317487.
Krishnan S, Manavathu EK, Chandrasekar PH: Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses. 2009, 52: 206-222. 10.1111/j.1439-0507.2008.01642.x.
Hageskal G, Kristensen R, Fristad RF, Skaar I: Emerging pathogen Aspergillus calidoustus colonizes water distribution systems. Med Mycol. 2011, 49: 588-593.
Coelho D, Silva S, Vale-Silva LS, Gomes H, Pinto EN, Sarmento AN, Pinheiro M: Aspergillus viridinutans: an agent of adult chronic invasive aspergillosis. Med Mycol. 2011, 49: 755-759.
Fadl FA, Hassan KM, Faizuddin M: Allergic fungal rhinosinusitis: report of 4 cases from Saudi Arabia. Saudi Med J. 2000, 21: 581-584.
Al-Dousary SH: Allergic fungal sinusitis: radiological and microbiological features of 59 cases. Am J Otolaryngol. 2004, 25: 213-218. 10.1016/j.amjoto.2003.12.003.
Taj-Aldeen SJ, Hilal AA, Chong-Lopez A: Allergic Aspergillus flavus rhinosinusitis: a case report from Qatar. Eur Arch Otorhinolaryngol. 2003, 260: 331-335. 10.1007/s00405-002-0547-x.
Taj-Aldeen SJ, Hilal AA, Schell WA: Allergic fungal rhinosinusitis: a report of 8 cases. Ann Saudi Med. 2008, 28: 17-21. 10.4103/0256-4947.51772.
Hedayati MT, Bahoosh M, Kasiri A, Ghasemi M, Motahhari SJ, Poormosa R: Prevalence of fungal rhinosinusitis among patients with chronic rhinosinusitis from Iran. J Med Mycol. 2010, 20: 298-303. 10.1016/j.mycmed.2010.09.002.
Khan ZU, Ahmad S, Mokaddas E, Said T, Nair MP, Halim MA, Nampoory MR, McGinnis MR: Cerebral aspergillosis diagnosed by detection of Aspergillus flavus-specific DNA, galactomannan and (1@@3)-beta-D-glucan in clinical specimens. J Med Microbiol. 2007, 56: 129-132. 10.1099/jmm.0.46549-0.
Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW: Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007, 153: 1677-692. 10.1099/mic.0.2007/007641-0.
Khan ZU, Sanyal SC, Mokaddas E, Vislocky I, Anim JT, Salama AL, Shuhaiber H: Endocarditis due to Aspergillus flavus. Mycoses. 1997, 40: 213-217. 10.1111/j.1439-0507.1997.tb00217.x.
Clinical and Laboratory Standards Institute: Reference method for broth dilution antifungal susceptibility testing for filamentous fungi; approved standard. Document M38-A2. 2008, Wayne, PA: Clinical and Laboratory Standards Institute
Espinel-Ingroff A, Cuenca-Estrella M, Fothergill A, Fuller J, Ghannoum M, Johnson E, Pelaez T, Pfaller MA, Turnidge J: Wild-type MIC distributions and epidemiological cutoff values for amphotericin B and Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). Antimicrob Agents Chemother. 2011, 55: 5150-5154. 10.1128/AAC.00686-11.
Espinel-Ingroff A, Fothergill A, Fuller J, Johnson E, Pelaez T, Turnidge J: Wild-type MIC distributions and epidemiological cutoff values for caspofungin and Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). Antimicrob Agents Chemother. 2011, 55: 2855-2859. 10.1128/AAC.01730-10.
Espinel-Ingroff A, Diekema DJ, Fothergill A, Johnson E, Pelaez T, Pfaller MA, Rinaldi MG, Canton E, Turnidge J: Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J Clin Microbiol. 2010, 48: 3251-7. 10.1128/JCM.00536-10.
van der Linden JW, Snelders E, Kampinga GA, Rijnders BJ, Mattsson E, Debets-Ossenkopp YJ, Kuijper EJ, Van Tiel FH, Melchers WJ, Verweij PE: Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerg Infect Dis. 2011, 17: 1846-1854. 10.3201/eid1710.110226.
Chowdhary A, Kathuria S, Randhawa HS, Gaur SN, Klaassen CH, Meis JF: Isolation of multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR/L98H mutations in the cyp51A gene in India. J Antimicrob Chemother. 2012, 67: 362-366. 10.1093/jac/dkr443.
Espinel-Ingroff A: Evaluation of broth microdilution testing parameters and agar diffusion Etest procedure for testing susceptibilities of Aspergillus spp. to caspofungin acetate (MK-0991). J Clin Microbiol. 2003, 41: 403-409. 10.1128/JCM.41.1.403-409.2003.
Espinel-Ingroff A, Rezusta A: E-test method for testing susceptibilities of Aspergillus spp. to the new triazoles voriconazole and posaconazole and to established antifungal agents: comparison with NCCLS broth microdilution method. J Clin Microbiol. 2002, 40: 2101-2107. 10.1128/JCM.40.6.2101-2107.2002.
Martos AI, Martín-Mazuelos E, Romero A, Serrano C, González T, Almeida C, Puche B, Cantón E, Pemán J, Espinel-Ingroff A: Evaluation of disk diffusion method compared to broth microdilution for antifungal susceptibility testing of 3 echinocandins against Aspergillus spp. Diagn Microbiol Infect Dis. 2012, 73: 53-56. 10.1016/j.diagmicrobio.2012.01.006.
Martos AI, Romero A, González MT, González A, Serrano C, Castro C, Pemán J, Cantón E, Martín-Mazuelos E: Evaluation of the Etest method for susceptibility testing of Aspergillus spp. and Fusarium spp. to three echinocandins. Med Mycol. 2010, 48: 858-861. 10.3109/13693781003586943.
Klich MA: Identification of common Aspergillus species. 2002, Utrecht, The Netherlands: Centraalbureau voor Schimmelcultures Publication
Ahmad S, Khan ZU, Theyyathel AM: Diagnostic value of DNA, (1–3)-beta-d-glucan, and galactomannan detection in serum and bronchoalveolar lavage of mice experimentally infected with Aspergillus terreus. Diagn Microbiol Infect Dis. 2007, 59: 165-171. 10.1016/j.diagmicrobio.2007.04.009.
Khan Z, Ahmad S, Al-Ghimlas F, Al-Mutairi S, Joseph L, Chandy R, Sutton DA, Guarro J: Purpureocillium lilacinum as a cause of cavitary pulmonary disease: a new clinical presentation and observations on atypical morphologic characteristics of the isolate. J Clin Microbiol. 2012, 50: 1800-1804. 10.1128/JCM.00150-12.
European Committee on Antimicrobial Susceptibility Testing: 2013, (http://www.eucast.org/antifungal_susceptibility_testing_afst/): Antifungal agents - Breakpoint tables for interpretation of MICs, Version 5.0
Espinel-Ingroff A, Canton E, Fothergill A, Ghannoum M, Johnson E, Jones RN, Ostrosky-Zeichner L, Schell W, Gibbs DL, Wang A, Turnidge J: Quality control guidelines for amphotericin B, itraconazole, posaconazole, and voriconazole disk diffusion susceptibility tests with non-supplemented Mueller-Hinton Agar (CLSI M51-A document) for nondermatophyte filamentous fungi. J Clin Microbiol. 2011, 49: 2568-2571. 10.1128/JCM.00393-11.
Clinical Laboratory Standards Institute: Development of in vitro susceptibility testing criteria and quality control parameters; approved guideline, 3rd ed. 2008, Villanova, PA: CLSI document M23-A3: Clinical and Laboratory Standards Institute
Varga J, Frisvad JC, Samson RA: Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Studies Mycol. 2011, 69: 57-80. 10.3114/sim.2011.69.05.
Colozza C, Posteraro B, Santilli S, De Carolis E, Sanguinetti M, Girmenia C: In vitro activities of amphotericin B and Am Bisome against Aspergillus isolates recovered from Italian patients treated for haematological malignancies. Int J Antimicrob Agents. 2012, 39: 440-443. 10.1016/j.ijantimicag.2012.01.013.
Lass-Flörl C, Kofler G, Kropshofer G, Hermans J, Kreczy A, Dierich MP, Niederwieser D: In-vitro testing of susceptibility to amphotericin B is a reliable predictor of clinical outcome in invasive aspergillosis. J Antimicrob Chemother. 1998, 42: 497-502. 10.1093/jac/42.4.497.
Badiee P, Alborzi A, Moeini M, Haddadi P, Farshad S, Japoni A, Ziyaeyan M: Antiungal susceptibility of the Aspergillus species by Etest and CLSI reference methods. Arch Iran Med. 2012, 15: 429-432.
Hadrich I, Makni F, Neji S, Cheikhrouhou F, Bellaaj H, Elloumi M, Ayadi A, Ranque S: Amphotericin B in vitro resistance is associated with fatal Aspergillus flavus infection. Med Mycol. 2012, 50: 829-834. 10.3109/13693786.2012.684154.
Pfaller MA, Messer SA, Boyken L, Rice C, Tendolkar S, Hollis RJ, Diekema DJ: In vitro survey of triazole cross-resistance among more than 700 clinical isolates of Aspergillus species. J Clin Microbiol. 2008, 46: 2568-2572. 10.1128/JCM.00535-08.
Mittal V, Mittal R, Sharma PC: Voriconazole-refractory fungal infection of phacoemulsification tunnel. Indian J Ophthalmol. 2010, 58: 434-437. 10.4103/0301-4738.67072.
Liu W, Sun Y, Chen W, Liu W, Wan Z, Bu D, Li R: T778G mutation in cyp51C gene confers voriconazole-resistance in Aspergillus flavus causing aspergillosis. Antimicrob Agents Chemother. 2012, 56: 2598-2603. 10.1128/AAC.05477-11.
Krishnan-Natesan S, Chandrasekar PH, Alangaden GJ, Manavathu EK: Molecular characterisation of cyp51A and cyp51B genes coding for P450 14alpha-lanosterol demethylases A (CYP51Ap) and B (CYP51Bp) from voriconazole-resistant laboratory isolates of Aspergillus flavus. Int J Antimicrob Agents. 2008, 32: 519-524. 10.1016/j.ijantimicag.2008.06.018.
Araujo R, Pina-Vaz C, Rodrigues AG: Susceptibility of environmental versus clinical strains of pathogenic Aspergillus. Int J Antimicrob Agents. 2007, 29: 108-111. 10.1016/j.ijantimicag.2006.09.019.
Pfaller MA, Boyken L, Hollis RJ, Kroeger J, Messer SA, Tendolkar S, Diekema DJ: In vitro susceptibility of clinical isolates of Aspergillus spp. to anidulafungin, caspofungin, and micafungin: a head-to-head comparison using the CLSI M38-A2 broth microdilution method. J Clin Microbiol. 2009, 47: 3323-3325. 10.1128/JCM.01155-09.
Shivaprakash MR, Geertsen E, Chakrabarti A, Mouton JW, Meis JF: In vitro susceptibility of 188 clinical and environmental isolates of Aspergillus flavus for the new triazole isavuconazole and seven other antifungal drugs. Mycoses. 2011, 54: e583-589. 10.1111/j.1439-0507.2010.01996.x.
Pfaller MA, Messer SA, Boyken L, Hollis RJ, Diekema DJ: In vitro susceptibility testing of filamentous fungi: comparison of Etest and reference M38-A microdilution methods for determining posaconazole MICs. Diagn Microbiol Infect Dis. 2003, 45: 241-244. 10.1016/S0732-8893(02)00544-8.
Messer SA, Diekema DJ, Hollis RJ, Boyken LB, Tendolkar S, Kroeger J, Pfaller MA: Evaluation of disk diffusion and Etest compared to broth microdilution for antifungal susceptibility testing of posaconazole against clinical isolates of filamentous fungi. J Clin Microbiol. 2007, 45: 1322-1324. 10.1128/JCM.02450-06.
Pfaller MA, Messer SA, Mills K, Bolmström A: In vitro susceptibility testing of filamentous fungi: comparison of Etest and reference microdilution methods for determining itraconazole MICs. J Clin Microbiol. 2000, 38: 3359-3361.
Ozkutuk A, Ergon C, Metin DY, Yucesoy M, Polat SH: Comparison of disk diffusion, E-test and broth microdilution test in determination of susceptibility of Aspergillus species to amphotericin B, itraconazole and voriconazole. J Chemother. 2008, 20: 87-92.
Pfaller JB, Messer SA, Hollis RJ, Diekema DJ, Pfaller MA: In vitro susceptibility testing of Aspergillus spp.: comparison of Etest and reference microdilution methods for determining voriconazole and itraconazole MICs. J Clin Microbiol. 2003, 41: 1126-1129. 10.1128/JCM.41.3.1126-1129.2003.
Guinea J, Peláez T, Alcalá L, Bouza E: Correlation between the Etest and the CLSI M-38 A microdilution method to determine the activity of amphotericin B, voriconazole, and itraconazole against clinical isolates of Aspergillus fumigatus. Diagn Microbiol Infect Dis. 2007, 57: 273-276. 10.1016/j.diagmicrobio.2006.09.003.
Espinel-Ingroff A, Arthington-Skaggs B, Iqbal N, Ellis D, Pfaller MA, Messer S, Rinaldi M, Fothergill A, Gibbs DL, Wang A: Multicenter evaluation of a new disk agar diffusion method for susceptibility testing of filamentous fungi with voriconazole, posaconazole, itraconazole, amphotericin B, and caspofungin. J Clin Microbiol. 2007, 45: 1811-1820. 10.1128/JCM.00134-07.
Espinel-Ingroff A, Canton E: Comparison of Neo-Sensitabs tablet diffusion assay with CLSI broth microdilution M38-A and disk diffusion methods for testing susceptibility of filamentous fungi with amphotericin B, caspofungin, itraconazole, posaconazole, and voriconazole. J Clin Microbiol. 2008, 46: 1793-1803. 10.1128/JCM.01883-07.
Laboratory Standards Institute: Performance standards for antifungal disk diffusion susceptibility testing of non-dermatophyte filamentous fungi; Informational supplement, 1st ed. CLSI document M51-A. 2010, Wayne, PA: Clinical and Laboratory Standards Institute
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/13/126/prepub
Acknowledgments
This work was supported by Research Administration grant YM 03/10 and the College of Graduate Studies, Kuwait University. The authors are thankful to Dr. Wassim Chehadeh for helping with statistical analyses.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZUK and SA conceived the study, supervised it and finalized the manuscript. FAW did the work which formed part of her Master thesis and contributed to writing of the manuscript. All authors have read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Al-Wathiqi, F., Ahmad, S. & Khan, Z. Molecular identification and antifungal susceptibility profile of Aspergillus flavus isolates recovered from clinical specimens in Kuwait. BMC Infect Dis 13, 126 (2013). https://doi.org/10.1186/1471-2334-13-126
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2334-13-126