Abstract
Background
Recent genome-wide association studies have identified several genetic loci linked to coronary artery disease (CAD) and myocardial infarction (MI). The 9p21.3 locus was verified by numerous replication studies to be the first common locus for CAD and MI. In the present study, we investigated whether six single nucleotide polymorphisms (SNP) rs1333049, rs1333040, rs10757274, rs2383206, rs10757278, and rs2383207 representing the 9p21.3 locus were associated with the incidence of an acute MI in patients with the main focus on the familial aggregation of the disease.
Methods
The overall cohort consisted of 976 unrelated male patients presenting with an acute coronary syndrome (ACS) with ST-elevated (STEMI) as well as non-ST-elevated myocardial infarction (NSTEMI). Genotyping data of the investigated SNPs were generated and statistically analyzed in comparison to previously published findings of matchable control cohorts.
Results
Statistical evaluation confirmed a highly significant association of all analyzed SNP's with the occurrence of MI (p < 0.0001; OR: 1.621-2.039). When only MI patients with a positive family disposition were comprised in the analysis a much stronger association of the accordant risk alleles with incident disease was found with odds ratios up to 2.769.
Conclusions
The findings in the present study confirmed a strong association of the 9p21.3 locus with MI particularly in patients with a positive family history thereby, emphasizing the pathogenic relevance of this locus as a common genetic cardiovascular risk factor.
Similar content being viewed by others
Background
In recent years, genome-wide association studies (GWAS) have displayed an effective approach to localize genomic regions predisposing to common, polygenetic disorders, including cardiovascular disorders[1, 2]. By means of independent genome-wide association studies using single nucleotide polymorphism (SNP) arrays the first common chromosomal locus has been identified which confers susceptibility for coronary artery disease (CAD) and myocardial infarction (MI)[3–5]. Initially, the two SNPs rs10757274 and rs2383206 consistently associated with CAD were identified in a large scale study population of Caucasian origin[3]. Both polymorphisms are located within a locus spanning a 58-kilobase region on chromosome 9p21.3. Three additional SNPs, rs1333040, rs2383207, and rs10757278 on the 9p21.3 locus and in genetic disequilibrium were identified independently as being associated with MI[4]. Moreover, large-scale genome-wide association scans identified further polymorphisms, amongst others rs1333049, within the same genomic region to be associated with MI and CAD and therefore verified chromosome 9p21.3 as a susceptibility locus for the incidence of the disease[5, 6]. Based on these findings, replication studies using case-control designs proved that the identified SNPs were associated with CAD and MI in a large variety of study population[7–17]. In all studies, the association of the 9p21.3 locus with CAD and MI was shown to be independent of conventional risk factors. In a subsequent study the association of the rs1333049 risk variant with the extend of CAD could be established[18]. Thus, even though the responsible gene in this region is still unknown, the 9p21.3 locus was verified to be the first common genetic factor affecting the risk of CAD and MI. Further studies may provide new insights into the impact of chromosome 9p21.3 on the pathogenesis of CAD and the occurrence of a MI.
In the present study the influence of six SNPs (rs1333049, rs1333040, rs10757274, rs2383206, rs10757278, and rs2383207) representing the 9p21.3 region on the occurrence of MI was investigated. The main focus of our investigation was on an association of the 9p21.3 locus with a positive family history of MI in a large cohort of patients presenting with an acute coronary syndrome (ACS).
Methods
Study subjects
Over a three year span (2004 to 2006), a total of 976 unrelated male patients younger than 65 years were enrolled into a study due to a symptomatic ACS with STEMI as well as NSTEMI. This prospective multi-centre registry involved four heart centres and six cardiologic departments in Germany. The study was approved by the ethical committee of the University Witten/Herdecke, Germany, and conformed to the declaration of Helsinki. All participants gave written informed consent to participation.
MI was defined as follows: ischemic type chest pain lasting for more than 20 minutes, at least 0.1 mV of ST-segment elevation in the limb leads and/or at least 0.2 mV elevation in the precordial leads and one of the following criteria: left bundle branch block, new Q wave (at least 0.03 s), elevated creatine kinase or positive troponin T or I. All cases underwent cardiac catheterization and interventional or surgical revascularization. Patients < 18 and > 65 years, with unstable angina or without central European origin were excluded from further studies.
Blood samples and DNA preparation
EDTA-blood samples were drawn from each participant at baseline ward round. Genomic DNA was extracted from 350 μl of these samples using the BioRobot EZ1 and the EZ1 blood extraction kit according to the manufacturer's instructions (QIAGEN; Hilden, Germany). DNA was quantified using the BioPhotometer (Eppendorf; Hamburg, Germany) and each sample was diluted to a final concentration of 25 ng/μl.
PCR amplification and genotyping
Genotyping of the investigated SNPs (rs1333049, rs1333040, rs10757274, rs2383206, rs10757276, and rs2383207) was carried out by real time PCR and subsequent melting curve analysis on a LightCycler 480 instrument (Roche Applied Science; Mannheim, Germany). The primers and hybridization probes were designed and synthesized by Tib MolBiol GmbH (Berlin, Germany). PCR was carried out in 96-well plates (Roche Applied Science; Mannheim, Germany) using 12.5 ng of genomic DNA as template in a final reaction volume of 5 μl. Reaction mixtures contained 0.5 μM of each primer, 0.15 μM of SNP-specific hybridization probes and 1 μl of LightCycler 480 Genotyping Master (Taq DNA polymerase, reaction buffer, 15 mM MgCl2, and a dNTP mixture with UTP instead of dTTP) (Roche Diagnostics GmbH, Mannheim, Germany).
The cycling program consisted of 10 minutes of initial denaturation at 95°C, followed by 35 cycles (rs1333049, rs2383206, rs2383207, rs10757274, rs10757278) or 40 cycles (rs1333040) of denaturation at 95°C for 5 seconds, annealing at 53°C (rs1333049, rs2383206, rs2383207), at 59°C (rs1333040) or at 62°C (rs10757274, rs10757278) for 10 seconds, and extension at 72°C for 10 seconds. After PCR melting curves were generated by holding the reaction mixture at 95°C for 1 minute, stepwise lowering of the temperature to 65°C, 55°C and 45°C, holding for 30 seconds at each temperature, lowering to 40°C for 2 minutes, followed by continuously heating to 75°C. Melting curve analyses were conducted using the LightCycler 480 software according to the manufacturer's instructions (Roche Diagnostics GmbH; Mannheim, Germany).
Statistical analysis
The distribution of genotypes and the allelic frequencies were investigated and analyzed by alignment to previously published data of applicable controls. Genotypes were tested for Hardy-Weinberg equilibrium among MI cases and controls using a chi-square test with one degree of freedom. Differences in the genotype distribution were tested for statistically significance and odds ratios (ORs) were determined using the 2-way contingency table chi-square test using a freely accessible program (http://statpages.org/ctab2x2.html). For all data, the association was considered to be significant for p < 0.05.
Results
Patients
A total of 976 patients, aged 24 to 65, were included into the present study to investigate the association between the 9p21.3 locus polymorphisms and acute MI. 72.8% of the ACS patients hospitalised with MI were diagnosed with ST-segment elevation (STEMI), 27.2% of the patients with non ST-segment elevation (NSTEMI). Of 976 patients 361 patients (37%) declared a positive family history of MI, defined as at least one first-degree relative affected before the age of 60 years, 404 patients (41.4%) had a negative family history, whereas 211 patients (21.6%) were not aware of another MI case in their family.
Controls were selected from previously published genotyping data. These data comprised subjects from Europe which were enrolled for independent studies as previously described elsewhere[3, 4, 11]. The baseline clinical characteristics and procedural parameters of the study population are presented in Table 1.
Replicated association of six variants on 9p21.3 with acute MI
Six SNPs on chromosome 9p21.3 were selected for genotyping based on strength of association in previous studies[3, 4, 11]. In order to confirm the reported association of the investigated SNPs with MI, genotyping data of the overall study cohort were compared to previously published data of control subjects (Table 2). All controls were without known history of CAD. Furthermore, only control cohorts were selected, which were comparable to the MI cases of the present study concerning geographic origin (northern and central Europe), age and wherever applicable, gender.
The rs1333049 genotype distribution and allelic frequencies were compared to the data of the PopGen controls[11]. For analysing the data of rs1333040, rs10757278, and rs2383207 we used the controls of the Iceland B collectives [4] and for the analysis of rs2383206 and rs10757274 the Copenhagen City Heart Study (CCHS) controls[3]. The genotype distribution in MI cases and controls did not significantly (P > 0.1) deviate from the Hardy-Weinberg equilibrium.
As shown in table 2, a statistically significant difference in the distribution of genotypes of MI cases compared to controls was observed for all analyzed SNPs (p < 0.0001). Odds ratios determined for the respective high-risk homozygous alleles were in the range of 1.621 to 2.039. These findings verified the strong association of the 9p21.3 locus with MI in this large scale study population of male patients presented with ACS.
Variants on 9p21.3 are associated with a positive family history of MI
To investigate the association between the sequence variants and a positive family history of MI, patients with at least one first degree-relative who had suffered from a premature MI (n = 361) were selected and the genotype distribution and allelic frequencies for all variants were analyzed and compared with the data of the corresponding control cohorts (Table 3) and with the data of the patients without a positive family history (Table 4).
Statistical analyses revealed odds ratios in the range of 1.892 to 2.769 for MI patients with a positive family history demonstrating a much stronger association than for the overall cohort of MI patients and therefore a considerable increased risk for MI patients with a family burden of the disease (Figure 1).
These findings could be confirmed comparing MI patients with a positive family history with patients without a positive family history for all SNP variants with the exception of the rs1333040 SNP (Table 4).
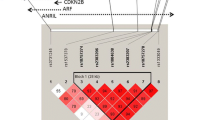
These results could also be confirmed in an additional haplotype analysis (Figure 2). For the haplotypes GGGGC and AAAAG spanning the rs10757274, rs2383206, rs2383207, rs10757278, and rs1333049 SNPs a significant association of a positive vs. a negative family history could be shown (p = 0.0051 and p = 0.0087 respectively). Adjustment for other cardiovascular risk factors (smoking, body mass index, hypertension, diabetes mellitus and hyperlipidemia) revealed no higher frequencies of the risk variants within the 9p21.3 region in MI patients examined by means of subgroup analyses (data not shown). These findings emphasize this genetic region as a genetic risk factor for MI independent of other risk factors.
Haplotype analysis for the investigated SNPs. As can be seen, all SNPs with the exception of the rs1333040 show a positive association of the phenotype MI in the presents of a positive family history of the disease. The haplotypes GGGGC and AAAAG revealed the strongest association of MI with a positive family history.
Discussion
In recent years, genome wide association studies focused on genomic factors involved in development of CAD and MI. In these studies and subsequent replication studies the 9p21.3 locus was found to be the most common locus associated with CAD and MI. Moreover, in a subsequent study, a strong association of the SNP rs1333049 representing the 9p21.3 risk variant with the extent of CAD could be shown[18].
In the present study, we investigated the association of six previously reported sequence variants representing the 9p21.3 locus in a large scale study population of male patients who had suffered from an acute MI. In this well defined study cohort, the exclusive inclusion of patients aged ≤ 65 years increased the probability that genetic factors are involved to a significant degree in the onset of MI. Moreover, only men were enrolled which excludes gender specific differences having an influence on the development of CHD. It is well known that women develop CHD about 10 years later than men, probably due to the protective effects of female sex hormones but also due to the different frequency of classical risk factors such as diabetes, hypertension and smoking habits[19, 20].
Genotyping of the study population showed similar or even higher high-risk allele frequencies compared to those in previous studies which reported the association of 9p21.3 sequence variants with CHD and MI[3, 4, 11]. Comparison was carried out using previously cited control subjects from central or northern Europe and without a known history of CAD. Moreover, in the German PopGen study only men were used as control subjects[11]. Thus, the selected controls were comparable to the present MI cohort and therefore justifiably applied in the presented association analyses. Comparison of the data with control subjects revealed a statistical significant difference in the genotype distribution and allelic frequencies of all analyzed polymorphisms within the 9p21.3 region. The observed higher odds ratios are probably due to the much more homogeneity of the study cohort investigated in the present study. No significant difference was observed in the genotype distribution of our MI cases and MI patients from previous studies. Hence, we confirmed the association of this locus with MI in the present study cohort. This emphasises once more the relevance of this genetic factor in the pathogenesis of CAD and MI.
Having replicated the association of all investigated SNPs with MI, patients having definitively presented with positive family history of MI were selected and compared to the corresponding control cohorts or a subpopulation of patients with a definitively negative family history. For the accuracy of this analysis, patients who provided no information on a family history of MI were excluded. This analysis revealed a significantly stronger association in patients having presented with a positive family history of MI than in the overall cohort of MI patients with calculated odds ratios within the range of 1.892 to 2.769.
Classification according to a positive family history provides an improved risk prediction for the patients. In fact, the increased risk of carriers of the homozygous risk alleles and a positive family history is comparable to the increased risk of hypertensive patients (OR 1.91), patients with diabetes mellitus (OR 2.37) or current smokers (OR 2.87) calculated in a large case-control study of acute MI in 52 countries[21].
Conclusions
To our knowledge the present study is the first one to analyse the association of all six 9p21.3 SNPs with acute MI in relation to a positive family history. Indeed, few other studies reported the association with CAD after classification of patients according to their family history. However, our study differs from the others in the origin of the patients, in the strict inclusion criteria and in the larger number of patients. Moreover, the quantity of analyzed sequence variants in the present study was higher.
Our findings made evident the correlation between high-risk variants on the 9p21.3 locus and male patients with acute MI and a positive family history. Over the past few years, risk evaluation has exclusively been performed on the prevalence of classical risk factors such as age and gender, LDL-cholesterol, hypertension, diabetes mellitus, smoking habits, diet, a family history and physical activity[22, 23]. The findings of the present study may help to improve risk assessment and early prevention particularly for patients with a positive family history of CAD when 9p21.3 sequence variants are taken into account.
Limitations of the study
One limitation of the study is the lack of an own control group. Using previous reported data about the allelic distribution of SNP's will be of a limited bias due to the fact, that thousands of chromosomes where analysed in previous reported studies. Another limitation is that not a consistent control group could be used for comparison.
References
Hirschhorn JN, Daly MJ: Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005, 6 (2): 95-108. 10.1038/nrg1521.
Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007, 447 (7145): 661-678. 10.1038/nature05911.
McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al: A common allele on chromosome 9 associated with coronary heart disease. Science. 2007, 316 (5830): 1488-1491. 10.1126/science.1142447.
Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, et al: A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007, 316 (5830): 1491-1493. 10.1126/science.1142842.
Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, et al: Genomewide association analysis of coronary artery disease. N Engl J Med. 2007, 357 (5): 443-453. 10.1056/NEJMoa072366.
Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, et al: Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009, 41 (3): 334-341. 10.1038/ng.327.
Abdullah KG, Li L, Shen GQ, Hu Y, Yang Y, MacKinlay KG, Topol EJ, Wang QK: Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest). Ann Hum Genet. 2008, 72 (Pt 5): 654-657. 10.1111/j.1469-1809.2008.00454.x.
Assimes TL, Knowles JW, Basu A, Iribarren C, Southwick A, Tang H, Absher D, Li J, Fair JM, Rubin GD, et al: Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum Mol Genet. 2008, 17 (15): 2320-2328. 10.1093/hmg/ddn132.
Chen Z, Qian Q, Ma G, Wang J, Zhang X, Feng Y, Shen C, Yao Y: A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Mol Biol Rep. 2009, 36 (5): 889-893. 10.1007/s11033-008-9259-7.
Hamsten A, Eriksson P: Identifying the susceptibility genes for coronary artery disease: from hyperbole through doubt to cautious optimism. J Intern Med. 2008, 263 (5): 538-552. 10.1111/j.1365-2796.2008.01958.x.
Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, et al: Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008, 117 (13): 1675-1684. 10.1161/CIRCULATIONAHA.107.730614.
Matarin M, Brown WM, Singleton A, Hardy JA, Meschia JF: Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke. 2008, 39 (5): 1586-1589. 10.1161/STROKEAHA.107.502963.
Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, Chida K, Sawabe M, Arimura T, Sato A, et al: Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008, 53 (4): 357-359. 10.1007/s10038-008-0248-4.
Talmud PJ, Cooper JA, Palmen J, Lovering R, Drenos F, Hingorani AD, Humphries SE: Chromosome 9p21.3 coronary heart disease locus genotype and prospective risk of CHD in healthy middle-aged men. Clin Chem. 2008, 54 (3): 467-474. 10.1373/clinchem.2007.095489.
Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, et al: The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008, 40 (2): 217-224. 10.1038/ng.72.
Shen GQ, Rao S, Martinelli N, Li L, Olivieri O, Corrocher R, Abdullah KG, Hazen SL, Smith J, Barnard J, et al: Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J Hum Genet. 2008, 53 (2): 144-150. 10.1007/s10038-007-0230-6.
Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D'Agostino RB, Fox CS, Govindaraju DR, Guo CY, Heard-Costa NL, Hwang SJ, et al: Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007, 8 (Suppl 1): S5-10.1186/1471-2350-8-S1-S5.
Dandona S, Stewart AF, Chen L, Williams K, So D, O'Brien E, Glover C, Lemay M, Assogba O, Vo L, et al: Gene dosage of the common variant 9p21 predicts severity of coronary artery disease. J Am Coll Cardiol. 2010, 56 (6): 479-486. 10.1016/j.jacc.2009.10.092.
Anand SS, Islam S, Rosengren A, Franzosi MG, Steyn K, Yusufali AH, Keltai M, Diaz R, Rangarajan S, Yusuf S: Risk factors for myocardial infarction in women and men: insights from the INTERHEART study. Eur Heart J. 2008, 29 (7): 932-940. 10.1093/eurheartj/ehn018.
Kannel WB, Hjortland MC, McNamara PM, Gordon T: Menopause and risk of cardiovascular disease: the Framingham study. Ann Intern Med. 1976, 85 (4): 447-452.
Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais P, Varigos J, et al: Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004, 364 (9438): 937-952. 10.1016/S0140-6736(04)17018-9.
Assmann G, Schulte H: Diabetes mellitus and hypertension in the elderly: concomitant hyperlipidemia and coronary heart disease risk. Am J Cardiol. 1989, 63 (16): 33H-37H. 10.1016/0002-9149(89)90113-6.
Wilson PW, D'Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB: Prediction of coronary heart disease using risk factor categories. Circulation. 1998, 97 (18): 1837-1847.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2261/11/9/prepub
Acknowledgements
We would like to thank Kirsten Rackebrandt for technical assistance and Joanne Davies for technical assistance and linguistic corrections, and all co-workers in the participating centres of the "Forschungsverbund Herz-Kreislauf in NRW".
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TS, MS, AH and GH have made substantial contributions to conception and design as well as analysis and interpretation of the data. KS and PB have made the genetic analyses. HRO, WS, JT, WM, F-JH, ChS, ThD, HG, HH, WD acquired patients and clinical data, TS and KS wrote the manuscript. All authors have read and approved the final version of the manuscript.
Thomas Scheffold, Silke Kullmann contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Scheffold, T., Kullmann, S., Huge, A. et al. Six sequence variants on chromosome 9p21.3 are associated with a positive family history of myocardial infarction: a multicenter registry. BMC Cardiovasc Disord 11, 9 (2011). https://doi.org/10.1186/1471-2261-11-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2261-11-9