Abstract
Background
Pectins are acidic sugar-containing polysaccharides that are universally conserved components of the primary cell walls of plants and modulate both tip and diffuse cell growth. However, many of their specific functions and the evolution of the genes responsible for producing and modifying them are incompletely understood. The moss Physcomitrella patens is emerging as a powerful model system for the study of plant cell walls. To identify deeply conserved pectin-related genes in Physcomitrella, we generated phylogenetic trees for 16 pectin-related gene families using sequences from ten plant genomes and analyzed the evolutionary relationships within these families.
Results
Contrary to our initial hypothesis that a single ancestral gene was present for each pectin-related gene family in the common ancestor of land plants, five of the 16 gene families, including homogalacturonan galacturonosyltransferases, polygalacturonases, pectin methylesterases, homogalacturonan methyltransferases, and pectate lyase-like proteins, show evidence of multiple members in the early land plant that gave rise to the mosses and vascular plants. Seven of the gene families, the UDP-rhamnose synthases, UDP-glucuronic acid epimerases, homogalacturonan galacturonosyltransferase-like proteins, β-1,4-galactan β-1,4-galactosyltransferases, rhamnogalacturonan II xylosyltransferases, and pectin acetylesterases appear to have had a single member in the common ancestor of land plants. We detected no Physcomitrella members in the xylogalacturonan xylosyltransferase, rhamnogalacturonan I arabinosyltransferase, pectin methylesterase inhibitor, or polygalacturonase inhibitor protein families.
Conclusions
Several gene families related to the production and modification of pectins in plants appear to have multiple members that are conserved as far back as the common ancestor of mosses and vascular plants. The presence of multiple members of these families even before the divergence of other important cell wall-related genes, such as cellulose synthases, suggests a more complex role than previously suspected for pectins in the evolution of land plants. The presence of relatively small pectin-related gene families in Physcomitrella as compared to Arabidopsis makes it an attractive target for analysis of the functions of pectins in cell walls. In contrast, the absence of genes in Physcomitrella for some families suggests that certain pectin modifications, such as homogalacturonan xylosylation, arose later during land plant evolution.
Similar content being viewed by others
Background
Pectins make up approximately one third of the dry mass of primary cell walls in eudicots, affecting both water dynamics and the mechanical behavior of the wall [1]. Pectins consist of four domains: homogalacturonan (HG), xylogalacturonan (XGA), rhamnogalacturonan I (RG-I), and rhamnogalacturonan II (RG-II) [2]. Homogalacturonan makes up the majority of the pectic component of the cell wall and also serves as the backbone of XGA and RG-II. Xylogalacturonan is made up of HG with attached xylose side-groups, whereas RG-II has four complex and distinct side-chains [3]. Rhamnogalacturonan I has side-chains containing galactose and arabinose, but its backbone consists of alternating rhamnose and galacturonic acid. These complex polysaccharides are almost universally conserved in land plants and are also present in some algae [4], although structural diversity in pectins is present between some species. For instance, there is evidence for RG-II in all land plant species analyzed to date [3, 5] but its side chains are not perfectly conserved [6], and the side chains of RG-I vary among species [1]. Additionally, XGA has not been detected in Physcomitrella patens[7].
Pectins are important determinants of wall remodeling during cellular growth [8]. Pairs of HG molecules can be bound together by Ca2+ bridges, stiffening the wall [9], and RG-II side-chains dimerize via borate diol ester bonds [10]. A decreased ability to form RG-II dimers leads to dwarfism [11]. Modifications to pectin can enhance or prevent these interactions and thus affect the properties of the wall as a whole: for example, alterations in wall stiffness mediated by pectin methylation have been implicated in organ primordium initiation and cell elongation [8, 12]. Pectins also appear to be essential for normal cell-cell adhesion, since some pectin methylation-defective mutants lack tissue cohesion [13, 14].
The complex structures of pectins require a large suite of biosynthetic genes, many of which are inferred only by the biochemical reactions required to synthesize the many linkages in pectins [15, 16]. Nevertheless, many pectin-related genes have been identified, and modification of their expression can have serious effects on the development and growth of mutant plants [17–20]. Pectins play an especially important role in the tip growth of pollen tubes, with methylation status regulating the yielding properties of the tip and side walls [21, 22], but this system does not allow for easy genetic manipulation. Physcomitrella patens, the model moss [23], represents an attractive experimental system for the genetic and molecular analysis of pectins in the walls of tip-growing cells. Its primary growth form is a mass of protonemal filaments that extend exclusively via tip growth and might therefore rely heavily on pectins for normal development [24, 25]. Genes in the Physcomitrella genome [26] can be modified directly using high-efficiency homologous recombination [27], which, combined with the dominant haploid generation of this moss, makes it ideal for genetic modification and analysis. As a moss, Physcomitrella is also likely to resemble an early stage in the transition of plants from aquatic to terrestrial life, giving us a clearer view of the cell wall architectures and physiology that made this transition possible.
As diverse plant genomes are sequenced, there are new opportunities to study gene families in an evolutionary context. The PlantTribes 2.0 database [28] is an objective gene family classification that can be used to investigate gene family composition and phylogeny on a global scale. By using the complete inferred protein sequences from ten diverse plant genomes (seven angiosperms plus the lycophyte Selaginella moellendorffii, the moss Physcomitrella, and the chlorophyte Chlamydomonas reinhardtii; see Figure 1), orthologous gene clusters (orthogroups) were identified that represent deeply conserved, but often narrowly defined gene families. Orthogroups were constructed using OrthoMCL [29], resulting in gene clusters that typically align well across their length and have a conserved domain structure [30]. Leveraging the PlantTribes 2.0 classification is a conservative approach to identify gene family members from sequenced genomes, avoiding false positive hits that may be identified using less structured search algorithms (e.g. BLAST). To assess the complexity of the pectin biosynthetic and modification machinery in Physcomitrella and to investigate the evolutionary history of pectin-related gene families in land plants, we performed an orthogroup-based phylogenetic study of 16 gene families associated with pectin production and modification and mapped the relationships of these genes among terrestrial plant species with sequenced genomes. These analyses reveal that the Physcomitrella genome contains at least one member in most of the families analyzed and that the total number of pectin-related gene family members in Physcomitrella is much lower than that in Arabidopsis. Analysis of these families not only identified members in Physcomitrella, it also reveals that several pectin-related gene families likely had multiple members in the land-plant common ancestor.
Summary of land plant phylogeny. The evolutionary relationships of the ten PlantTribes species used in this study (land plants and Chlamydomonas) and the charophycean algae used as additional outgroups. Note that only one moss and one lycophyte genome has been sequenced to represent early-diverging lineages of land plants, compared with many genomes representing angiosperms.
Results
Identification of pectin-related genes using PlantTribes 2.0
We used a set of genes in Arabidopsis belonging to 16 pectin-related gene families identified in the literature (Additional file 1) to select orthogroups in the PlantTribes 2.0 database for in-depth phylogenetic analysis (Additional file 2) [28]. The number of genes from each species in each family is displayed in Additional file 3. We found at least one Physcomitrella gene in 12 of the 16 families examined (Table 1). Notably, no Physcomitrella members of the xylogalacturonan xylosyltransferase (Additional file 4), rhamnogalacturonan-I arabinosyltransferases (Additional file 5), pectin methylesterase inhibitor (Additional file 6), or polygalacturonase inhibitor protein (Additional file 7) families were detected. There were fewer Physcomitrella members in most of the pectin-related gene families than in Arabidopsis, with the exception of the UDP-rhamnose synthase (four Arabidopsis, six Physcomitrella), β-1,4-galactan β-1,4-galactosyltransferase (three Arabidopsis, four Physcomitrella), and UDP-glucuronic acid (UDP-GlcA) epimerase (five Arabidopsis, nine Physcomitrella) families.
Phylogenetic analysis of pectin-related gene families
Our identification of pectin-related genes in ten diverse plant species (Figure 1) provided an opportunity to examine their phylogenetic patterns [31]. To analyze the evolutionary relationships between gene family members, we aligned the sequences from the PlantTribes 2.0 search results for each family using the MUSCLE algorithm [32] followed by manual curation, and constructed maximum likelihood trees from these alignments using RAxML [33]. Where possible, we also included a homologous gene from a green alga to root the trees. We tested the hypothesis that each pectin-related gene family would trace back to a single ancestral gene in the common ancestor of land plants, with any Physcomitrella genes forming a clade sister to all other land plants. Surprisingly, this was the case for only seven of the 16 families examined (Table 1). Five of the trees have multiple well-supported land plant-wide clades (Figures 2, 3, 4, Additional file 8 and Additional file 9). Each clade is evidence for a separate ancestral gene in the early land plant ancestor of the terrestrial species examined. These trees and their implications are explored below.
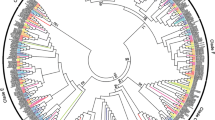
GAUT family tree. Three well-supported clades that suggest ancestral GAUTs are highlighted (blue, pink, and green clouds), and an unresolved polytomy near the root of the tree is indicated in light grey. The green and pink clades, as well as the polytomy, contain monocot, eudicot, Selaginella, and Physcomitrella members, whereas the blue clade does not have any Physcomitrella members. The algal root gene from Spirogyra pratensis falls within the polytomy.
Polygalacturonase family tree. Four monophyletic clades (blue, pink, green, and yellow clouds) contain monocot, eudicot, Selaginella, and Physcomitrella genes. The tree contains two large polytomies, indicated in light grey and labeled “A” and “B”. Polytomy B contains unresolved Physcomitrella and Selaginella members. The algal root gene is from C. reinhardtii, a chlorophytic alga.
Pectin-methylesterase family tree. Two large polytomies, labeled “A” and “B” and shown in light grey, indicate poor resolution of some of this family’s lineages. Four monophyletic clades contain members from the monocots, eudicots, Selaginella, and Physcomitrella. One of these clades (blue cloud) consists of polytomy B and a smaller clade of Physcomitrella and Selaginella genes. Additional moss and tracheophyte genes remain poorly resolved in polytomy A. The algal root (from P. margaritaceum) is within one of the polytomies.
The GAUTsuperfamily contains at least five ancestral land plant genes
The GAUT superfamily consists of the GAUT and the distantly-related GAUT-like (GATL) families [34, 35]. Some galacturonosyltransferases (GAUTs) are responsible for constructing HG and use UDP-galacturonic acid (UDP-GalA) as a substrate [34]. In Arabidopsis, mutations in GAUTs cause phenotypes ranging from changes in sugar composition of the wall to severe dwarfism to apparent lethality [34, 36–38]. In our analysis, the GAUT family tree contains three large well-resolved clades, as well as an unresolved polytomy (Figure 2). Genes from Physcomitrella and tracheophytes are present in two of these clades and within the polytomy from which the root algal gene is not resolved. The third of these clades includes genes from Selaginella, monocots, and eudicots but no Physcomitrella genes. This tree suggests a minimum of four ancestral GAUTs in the earliest land plant.
The roles of the GATL proteins are not all clearly established: some of them have been implicated in pectin production, while at least one seems to be involved in xylan synthesis [38, 39]. When we generated an alignment and phylogenetic tree of the entire superfamily (Figure 5), the GATL family (yellow cloud) appeared as a well-resolved but distant clade derived from within the GAUT family that also contains representatives from all of the land plant species queried.
GAUT superfamily tree. In this tree, phylogenetic distance is indicated by branch length. The GATL gene family (yellow cloud) is well-supported as being derived from within the GAUTs; due to a polytomy in the GATL family, clade relationships within this family are not well resolved. The distance of the GATLs from the GAUTs suggests an ancient divergence, but the position of the algal root supports the hypothesis that the GATLs descended from the GAUTs rather than diverging from a common ancestor. Scale bar, 0.7 substitutions/site.
Polygalacturonase and pectin methylesterase families are large and deeply conserved
Whereas GAUTs build the HG backbone of pectins, polygalacturonases (PGs) hydrolyze it, weakening the pectin matrix and potentially loosening the wall [40]. In eudicots, PGs are important in cell expansion and also in abscission and fruit softening [41]. The PG family is very large in Arabidopsis, with over 65 known members. Our phylogenetic analysis for these genes resulted in two large unresolved polytomies, each containing several monophyletic groups, four of which contain representatives from mosses, lycophytes, monocots, and eudicots (Figure 3). Although the placement of several of the Physcomitrella genes is unresolved, the gene tree suggests a minimum of five genes in the common ancestor.
Like the PGs, the pectin methylesterase (PME) family is very large in Arabidopsis[42]. Galacturonic acid residues in the HG backbones of pectins often have attached methyl ester groups at the C6 position that can prevent pectin-modifying enzymes as well as interactions with other HG chains. Thus, the amount and pattern of methylation can affect wall dynamics in several ways. PMEs remove methyl groups from pectin, rendering it more prone to degradation by hydrolytic enzymes as well as to calcium cross-linking, potentially either weakening or stiffening the wall. This is complicated by the tendency of different PMEs to remove methyl groups in random or block-wise patterns: lone de-methylated GalAs make the polymer prone to enzyme degradation, whereas consecutive exposed carboxylate groups favor calcium-bridging [43]. Like the PGs, the PME gene tree we generated has two large polytomies and two smaller resolved clades (Figure 4). Unlike the PG tree, the algal root is a member of one of the polytomies. Within this polytomy are two well-supported land plant-wide monophyletic clades. Resolved from this polytomy is a third land plant-wide clade. Several Physcomitrella and Selaginella genes are in a clade that is sister to the second polytomy, which consists entirely of angiosperm genes. This tree suggests that a minimum of five PMEs existed in the common ancestor of the species examined.
Many pectin-related gene families appear to have had only one or two members in the common ancestor of land plants
Like the polygalacturonases, pectate lyase-like proteins cleave the HG backbone of pectins (Additional file 8) [44]. Homogalacturonan methyltransferases are responsible for methylating newly synthesized HG (Additional file 9) [13]. Both of these family trees indicate the existence of multiple members in the common ancestor by having multiple supported clades with members from every division of the plant lineage. The final seven of the family trees have Physcomitrella genes grouped sister to the other land plants, indicating a single ancestral gene prior to the divergence of Physcomitrella and the tracheophytes: the UDP-GlcA epimerases, the UDP-rhamnose synthases, the pectin acetylesterases, the pectin acetyltransferases, the RG-II xylosyltransferases, the β-1,4-galactan β-1,4-galactosyltransferases, and the GATLs (Additional files 10, 11, 12, 13, 14, 15 and 16). These families are listed as having one supported common ancestral gene in Table 1. The UDP-GlcA epimerase, UDP-rhamnose synthase, β-1,4-galactan β-1,4-galactosyltransferase, and GATL families all likely expanded in Physcomitrella after its divergence from the tracheophytes.
Discussion
Search and tree-building criteria for pectin-related genes
We adopted a relatively stringent set of criteria to identify putative orthologs of Arabidopsis pectin-related genes in Physcomitrella and other plant species, and used these genes to build phylogenetic trees of pectin-related gene families. Rather than simply using database searches and overall sequence similarity to identify homologous genes, we leveraged the network of global gene relationships in the PlantTribes 2.0 database to identify clusters of orthologous genes (orthogroups) from the other species for analysis. Using BLAST to identify putative gene orthologs is a common practice, but increases the number of false positive sequences obtained because hits may only share high similarity in a small portion of the gene (i.e. a conserved domain), but may not be closely related and align poorly across the full length of the sequence. In contrast to BLAST-based methods, the use of PlantTribes 2.0 orthogroups increases the probability of identifying genes within the same evolutionary lineage, thus reflecting the history of these gene families more accurately. In some cases our search method detected fewer Physcomitrella members than other analyses of these families [40, 45, 46]. In all of these cases the researchers used shared protein domains or sequence homology to identify their genes of interest. The search method we used was intended to identify high-confidence candidate genes for further experimental analysis that are more likely to share conserved functions within other model systems. We therefore employed a higher-stringency approach at the cost of missing more distantly related homologs.
Although our trees largely agree with previously published phylogenies for some pectin-related gene families [35, 36, 40, 45–49], the larger number of species we used improved our ability to resolve gene family topologies and to detect basal branchpoints that have been obscured in analyses using genome data from fewer species [36, 40, 46–49]. An exception to this is the work of Wang et al., which identified PMEs and PMEIs in the same land plant species we examined, as well as Amborella trichopoda[45]. Wang et al. searched for conserved PME and PMEI protein domains and identified 35 putative Physcomitrella PMEs as compared with our ten. They also produced a large PMEI tree that included a putative Physcomitrella member. In contrast to our approach, their domain-based approach likely resulted in the detection of distantly related genes not included in our results.
Several pectin-related gene families likely had multiple members in the common ancestor of mosses and tracheophytes
The topologies of the trees we generated provide clues to the evolutionary relationships between known pectin-related genes and their orthologs in other species. This allows us to hypothesize about the state of the gene families in the last common ancestor of Physcomitrella and vascular plants. In seven of the families we analyzed, the paralogs in Physcomitrella are sister to all other genes in vascular plants. On the other hand, several of the families (GAUTs, HG methyltransferases, PMEs, PGs, pectate lyase-like proteins) each appear to have had multiple members in the common ancestor of land plants. Our analyses suggest that the suite of genes for the production, modification, and degradation of pectins had already diversified prior to the radiation of land plants. This contrasts with the cellulose synthase gene family (CESA), which likely contained a single gene in the ancestor of land plants and subsequently diversified after the divergence of mosses and vascular plants [50]. Multiple members of a gene family often have different expression patterns, allowing for tissue-specific regulation of the associated activity; for example, PpCESA5 is required only for gametophore development, implying that other PpCESAs produce cellulose in protonemal tissue [51]. Intriguingly, others have hypothesized that pectin synthesis and modification might originally have been central in wall production and modulation, with the importance of cellulose arising later [52]. There is also evidence for further diversification of these families before the flowering plant divergence in the form of angiosperm-wide clades in the GAUTs, PMEs, PGs, pectate lyase-like proteins, UDP-glucuronic acid epimerases, UDP-rhamnose synthases, and pectin acetylesterases.
Some pectin-related gene families were not detected in Physcomitrella
Since orthogroups in the PlantTribes 2.0 database generally represent narrowly defined gene lineages that typically align well across the whole length of the gene, we are confident that distantly related genes have been excluded from our analyses. However, it is possible that we failed to detect highly divergent members of some of these gene families. Nevertheless, most of the searches yielded at least one Physcomitrella gene per family. This was not true of the XGA xylosyltransferases, the RG I arabinosyltransferases, the PGIPs, and the PMEIs. It is not surprising that XGA xylosyltransferases were not detected in Physcomitrella given that a previous study using comprehensive microarray polymer profiling (COMPP) did not detect XGA in Physcomitrella cell walls [7]. On the other hand, α(1–5)-arabinans characteristic of RG I were detected in the pectic fraction of Physcomitrella walls, which combined with the failure to detect Physcomitrella orthologs of AtARAD genes in this study and others [49] raises the possibility of the existence of other arabinan-arabinosyltransferases that are only distantly related to the currently known genes.
Although there are not any studies indicating that PGIPs are absent in Physcomitrella, we also did not detect any PGIP genes in Selaginella, suggesting that this gene family may have evolved after the divergence of lycophytes and euphyllophytes. PGIPs are thought to play a role in pathogen defense by preventing foreign PGs from degrading the plant cell wall [53], and it is interesting that none were detected in either our representative moss or lycophyte, given that Physcomitrella and other mosses are susceptible to fungal pathogens [54]. The PMEI tree we generated only contains genes from Arabidopsis and Medicago truncatula, and might not adequately represent the diversity in this gene family. This might be due to insufficient numbers of query genes to allow for the detection of all the family members, or because coding sequence information for some of the species might have been incomplete. Importantly, the Arabidopsis query genes were both contained within one orthogroup. Genome data for additional plant species and/or future improvements in genome annotations could potentially overcome this limitation.
Arabidopsishas an abundance of pectin-related genes, whereas grasses appear to have fewer pectin-related genes in some families
In nine of the 16 families analyzed, Arabidopsis had more members than any of the other species (Additional file 3). This might be the result of the more extensive annotation of the Arabidopsis genome as compared to other species in the database, or the unique genome duplication histories of the species analyzed [30]. We see a general trend of more pectin-related genes in the eudicots than in the monocots and more in the monocots than in the more basal species such as Physcomitrella and Selaginella. This may reflect the lower levels of pectin in the walls of grasses compared to other flowering plants [55], as well as the relatively high abundance of other acidic polymers such as glucuronoarabinoxylans in grasses [56]. Further phylogenetic analyses of non-commelinid monocots, which have Type I cell walls [57], might be informative in determining the relationship between the elaboration of pectin-related gene families and the abundance of pectins in the cell wall.
Conclusions
Pectins play a key role in the cell walls of plants. We analyzed 16 gene families involved in the production, modification, and degradation of pectins in nine land plant species. Our analysis indicates that although many of these families appear to trace back to a single gene in the last common ancestor to the mosses and the vascular plants, several of the major families involved in pectin regulation likely contained multiple genes. We did not detect Physcomitrella or Selaginella genes in four of the studied families, providing some evidence that they might have evolved after the divergence of seed plants from the lycophytes. This study has allowed us to identify Physcomitrella orthologs related to known pectin-related genes in Arabidopsis for in-depth experimental analysis. Our results also shed light on the evolutionary history of pectin biosynthesis and modification, suggesting that pectins may have played an important role in the transition from an aquatic to a terrestrial environment.
Methods
Identification of pectin-related gene families
We compiled a list of Arabidopsis genes with known and predicted pectin-related functions using TAIR and Uniprot annotations, as well as relevant literature (Additional file 1) [1, 34, 42, 53, 58–64]. In total, we used 108 genes from Arabidopsis to identify putative pectin-related gene families in the PlantTribes 2.0 database [65]. PlantTribes 2.0 is an objective gene family classification of protein coding genes from ten sequenced green plant genomes that have been clustered into orthogroups (putatively monophyletic gene lineages) using OrthoMCL [28]. Orthogroups containing pectin-related genes from Arabidopsis were extracted for phylogenetic analysis. This approach enabled us to include additional homologous genes from Arabidopsis not annotated with pectin-related gene functions. In some cases, the pectin-related query genes from Arabidopsis did not belong to an orthogroup (i.e., they were singletons). The closest Physcomitrella gene to each singleton Arabidopsis gene was identified via TBLASTX and added to the family alignment. Because PlantTribes 2.0 includes the Physcomitrella patens version 1.1 gene annotations from Phytozome [66], we used a nucleotide BLAST+ search of a local database of Physcomitrella patens version 1.6 annotated coding sequences to identify the current gene annotations for ease of reference (Additional file 2, which includes all of the genes used in this paper). Although PlantTribes 2.0 does include the chlorophyte alga Chlamydomonas reinhardtii, many of the gene families still lacked a non-land plant outgroup. To enhance the possibility of rooting our trees using an outgroup, we also included homologous transcript sequences from three additional green algae (Nitella hyalina, Penium margaritaceum, and Spirogyra pratensis) where possible [67]. We searched each transcriptome separately with coding sequences from Physcomitrella using TBLASTX with an E-value cutoff of 10-10. Full-length coding sequences were identified for the GAUT, pectin methylesterase, UDP-rhamnose synthase, rhamnogalacturonan I arabinosyltransferase, and rhamnogalacturonan II xylosyltransferase families.
Phylogenetic analysis
Sequences for each family were aligned by translation in Geneious using MUSCLE (default parameters) [32], manually curated, and saved as relaxed Phylip files (Additional files 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32 and 33). In some cases this required removing non-homologous genes and gene fragments from poorly annotated genomes. To generate trees (Additional files 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49 and 50), maximum likelihood phylogenetic analysis was performed using RAxML [33] with the following parameters: rapid bootstrap analysis and search for best-scoring maximum likelihood tree in one run, GTRGAMMA model of nucleotide evolution, random seed 12345, 1000 bootstrap replicates. Nodes with less than 50% bootstrap support were collapsed using TreeCollapserCL4 [68] and were visualized using FigTree [69]. Figures were manually edited for readability using Adobe Illustrator.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
References
Atmodjo MA, Hao Z, Mohnen D: Evolving views of pectin biosynthesis. Annu Rev Plant Biol. 2013, 64 (April): 747-779.
Mohnen D: Pectin structure and biosynthesis. Curr Opin Plant Biol. 2008, 11: 266-277. 10.1016/j.pbi.2008.03.006.
Matsunaga T, Ishii T, Matsumoto S, Higuchi M, Darvill A, Albersheim P, O’Neill MA: Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes, and bryophytes. Implications for the evolution of vascular plants. Plant Physiol. 2004, 134: 339-351. 10.1104/pp.103.030072.
Domozych DS, Serfis A, Kiemle SN, Gretz MR: The structure and biochemistry of charophycean cell walls: I. Pectins of Penium margaritaceum. Protoplasma. 2007, 230: 99-115. 10.1007/s00709-006-0197-8.
Pérez S, Rodríguez-Carvajal MA, Doco T: A complex plant cell wall polysaccharide: rhamnogalacturonan II. A structure in quest of a function. Biochimie. 2003, 85: 109-121. 10.1016/S0300-9084(03)00053-1.
Pabst M, Fischl RM, Brecker L, Morelle W, Fauland A, Köfeler H, Altmann F, Léonard R: Rhamnogalacturonan II structure shows variation in the side chains monosaccharide composition and methylation status within and across different plant species. Plant J. 2013, 76: 61-72.
Moller I, Sørensen I, Bernal AJ, Blaukopf C, Lee K, Øbro J, Pettolino F, Roberts A, Mikkelsen JD, Knox JP, Bacic A, Willats WGT: High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J. 2007, 50: 1118-1128. 10.1111/j.1365-313X.2007.03114.x.
Derbyshire P, McCann MC, Roberts K: Restricted cell elongation in Arabidopsis hypocotyls is associated with a reduced average pectin esterification level. BMC Plant Biol. 2007, 7: 31-10.1186/1471-2229-7-31.
Braccini I, Pérez S: Molecular basis of Ca2+-induced gelation in alginates and pectins: the egg-box model revisited. Biomacromolecules. 2001, 2: 1089-1096. 10.1021/bm010008g.
O’Neill MA, Warrenfeltz D, Kates K, Pellerin P, Doco T, Darvill AG, Albersheim P: Rhamnogalacturonan-II, a pectic polysaccharide in the walls of growing plant cell, forms a dimer that is covalently cross-linked by a borate ester. J Biol Chem. 1996, 271: 22923-22930. 10.1074/jbc.271.37.22923.
O’Neill MA, Eberhard S, Albersheim P, Darvill AG: Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science. 2001, 294: 846-849. 10.1126/science.1062319.
Peaucelle A, Braybrook S, Le Guillou L, Bron E, Kuhlemeier C, Höfte H: Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr Biol. 2011, 21: 1720-1726. 10.1016/j.cub.2011.08.057.
Mouille G, Ralet M-C, Cavelier C, Eland C, Effroy D, Hématy K, McCartney L, Truong HN, Gaudon V, Thibault J-F, Marchant A, Höfte H: Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 2007, 50: 605-614. 10.1111/j.1365-313X.2007.03086.x.
Krupková E, Immerzeel P, Pauly M, Schmülling T: The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 2007, 50: 735-750. 10.1111/j.1365-313X.2007.03123.x.
Harholt J, Suttangkakul A, Vibe Scheller H: Biosynthesis of pectin. Plant Physiol. 2010, 153: 384-395. 10.1104/pp.110.156588.
Mohnen D, Bar-Peled M, Somerville C: Cell wall polysaccharide synthesis. Biomass Recalcitrance: Deconstructing Plant Cell Wall Bioenergy. Edited by: Himmel M. 2008, Oxford: Blackwell Publishing, 94-187.
Atkinson RG, Schröder R, Hallett IC, Cohen D, MacRae EA: Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002, 129: 122-133. 10.1104/pp.010986.
Iwai H, Masaoka N, Ishii T, Satoh S: A pectin glucuronyltransferase gene is essential for intercellular attachment in the plant meristem. Proc Natl Acad Sci U S A. 2002, 99: 16319-16324. 10.1073/pnas.252530499.
Orfila C, Seymour GB, Willats WG, Huxham IM, Jarvis MC, Dover CJ, Thompson AJ, Knox JP: Altered middle lamella homogalacturonan and disrupted deposition of (1- > 5)-alpha-ʟ-arabinan in the pericarp of Cnr, a ripening mutant of tomato. Plant Physiol. 2001, 126: 210-221. 10.1104/pp.126.1.210.
Hongo S, Sato K, Yokoyama R, Nishitani K: Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell. 2012, 24: 2624-2634. 10.1105/tpc.112.099325.
Röckel N, Wolf S, Kost B, Rausch T, Greiner S: Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 2008, 53: 133-143. 10.1111/j.1365-313X.2007.03325.x.
Parre E, Geitmann A: Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta. 2005, 220: 582-592. 10.1007/s00425-004-1368-5.
Quatrano RS, McDaniel SF, Khandelwal A, Perroud P-F, Cove DJ: Physcomitrella patens: mosses enter the genomic age. Curr Opin Plant Biol. 2007, 10: 182-189. 10.1016/j.pbi.2007.01.005.
Lee KJD, Sakata Y, Mau S-L, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP: Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell. 2005, 17: 3051-3065. 10.1105/tpc.105.034413.
Menand B, Calder G, Dolan L: Both chloronemal and caulonemal cells expand by tip growth in the moss Physcomitrella patens. J Exp Bot. 2007, 58: 1843-1849. 10.1093/jxb/erm047.
Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y, Tanahashi T, Sakakibara K, Fujita T, Oishi K, Shin-I T, Kuroki Y, Toyoda A, Suzuki Y, Hashimoto S-I, Yamaguchi K, Sugano S, Kohara Y, Fujiyama A, Anterola A, Aoki S, Ashton N, Barbazuk WB, Barker E, Bennetzen JL, Blankenship R, et al: The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008, 319: 64-69. 10.1126/science.1150646.
Kamisugi Y, Schlink K, Rensing S, Schween G, von Stackelberg M, Cuming AC, Reski R, Cove DJ: The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 2006, 34: 6205-6214. 10.1093/nar/gkl832.
Wall PK, Leebens-Mack J, Müller KF, Field D, Altman NS, DePamphilis CW: PlantTribes: a gene and gene family resource for comparative genomics in plants. Nucleic Acids Res. 2008, 36 (Database issue): D970-D976.
Li L, Stoeckert CJ, Roos DS: OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13: 2178-2189. 10.1101/gr.1224503.
Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, Soltis DE, Clifton SW, Schlarbaum SE, Schuster SC, Ma H, Leebens-Mack J, de Pamphilis CW: Ancestral polyploidy in seed plants and angiosperms. Nature. 2011, 473: 97-100. 10.1038/nature09916.
The Tree of Life Web Project. http://tolweb.org,
Edgar RC: MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32: 1792-1797. 10.1093/nar/gkh340.
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Sterling JD, Atmodjo MA, Inwood SE, Kumar Kolli VS, Quigley HF, Hahn MG, Mohnen D: Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc Natl Acad Sci U S A. 2006, 103: 5236-5241. 10.1073/pnas.0600120103.
Yin Y, Chen H, Hahn MG, Mohnen D, Xu Y: Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol. 2010, 153: 1729-1746. 10.1104/pp.110.154229.
Caffall KH, Pattathil S, Phillips SE, Hahn MG, Mohnen D: Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol Plant. 2009, 2: 1000-1014. 10.1093/mp/ssp062.
Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, Orlando R, Scheller HV, Mohnen D: Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc Natl Acad Sci U S A. 2011, 108: 20225-20230. 10.1073/pnas.1112816108.
Kong Y, Zhou G, Yin Y, Xu Y, Pattathil S, Hahn MG: Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol. 2011, 155: 1791-1805. 10.1104/pp.110.163220.
Lee C, Zhong R, Richardson E, Himmelsbach DS, McPhail BT, Ye Z-H: The PARVUS gene is expressed in cells undergoing secondary wall thickening and is essential for glucuronoxylan biosynthesis. Plant Cell Physiol. 2007, 48: 1659-1672. 10.1093/pcp/pcm155.
Yang Z-L, Liu H-J, Wang X-R, Zeng Q-Y: Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 2013, 197: 1353-1365. 10.1111/nph.12107.
Hadfield KA, Bennett AB: Polygalacturonases: many genes in search of a function. Plant Physiol. 1998, 117: 337-343. 10.1104/pp.117.2.337.
Pelloux J, Rustérucci C, Mellerowicz EJ: New insights into pectin methylesterase structure and function. Trends Plant Sci. 2007, 12: 267-277. 10.1016/j.tplants.2007.04.001.
Wolf S, Mouille G, Pelloux J: Homogalacturonan methyl-esterification and plant development. Mol Plant. 2009, 2: 851-860. 10.1093/mp/ssp066.
Palusa SG, Golovkin M, Shin S-B, Richardson DN, Reddy ASN: Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 2007, 174: 537-550. 10.1111/j.1469-8137.2007.02033.x.
Wang M, Yuan D, Gao W, Li Y, Tan J, Zhang X: A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS One. 2013, 8: e72082-10.1371/journal.pone.0072082.
Yin Y, Huang J, Gu X, Bar-Peled M, Xu Y: Evolution of plant nucleotide-sugar interconversion enzymes. PLoS One. 2011, 6: e27995-10.1371/journal.pone.0027995.
Kim B-G, Jung WD, Ahn J-H: Cloning and characterization of a putative UDP-rhamnose synthase 1 from Populus euramericana Guinier. J Plant Biol. 2013, 56: 7-12. 10.1007/s12374-012-0333-2.
Egelund J, Damager I, Faber K, Olsen C-E, Ulvskov P, Petersen BL: Functional characterisation of a putative rhamnogalacturonan II specific xylosyltransferase. FEBS Lett. 2008, 582: 3217-3222. 10.1016/j.febslet.2008.08.015.
Harholt J, Sørensen I, Fangel J, Roberts A, Willats WGT, Scheller HV, Petersen BL, Banks JA, Ulvskov P: The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of its cell wall. PLoS One. 2012, 7: e35846-10.1371/journal.pone.0035846.
Roberts AW, Bushoven JT: The cellulose synthase (CESA) gene superfamily of the moss Physcomitrella patens. Plant Mol Biol. 2007, 63: 207-219.
Goss CA, Brockmann DJ, Bushoven JT, Roberts AW: A CELLULOSE SYNTHASE (CESA) gene essential for gametophore morphogenesis in the moss Physcomitrella patens. Planta. 2012, 235: 1355-1367. 10.1007/s00425-011-1579-5.
Peaucelle A, Braybrook S, Höfte H: Cell wall mechanics and growth control in plants: the role of pectins revisited. Front Plant Sci. 2012, 3 (June): 121-
Di C-X, Zhang H, Sun Z-L, Jia H-L, Yang L-N, Si J, An L-Z: Spatial distribution of polygalacturonase-inhibiting proteins in Arabidopsis and their expression induced by Stemphylium solani infection. Gene. 2012, 506: 150-155. 10.1016/j.gene.2012.06.085.
Akita M, Lehtonen MT, Koponen H, Marttinen EM, Valkonen JPT: Infection of the Sunagoke moss panels with fungal pathogens hampers sustainable greening in urban environments. Sci Total Environ. 2011, 409: 3166-3173. 10.1016/j.scitotenv.2011.05.009.
Carpita NC: Structure and Biogenesis of the Cell Walls of Grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996, 47: 445-476. 10.1146/annurev.arplant.47.1.445.
Anders N, Wilkinson MD, Lovegrove A, Freeman J, Tryfona T, Pellny TK, Weimar T, Mortimer JC, Stott K, Baker JM, Defoin-Platel M, Shewry PR, Dupree P, Mitchell RAC: Glycosyl transferases in family 61 mediate arabinofuranosyl transfer onto xylan in grasses. Proc Natl Acad Sci U S A. 2012, 109: 989-993. 10.1073/pnas.1115858109.
Carpita NC, Gibeaut DM: Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993, 3: 1-30. 10.1111/j.1365-313X.1993.tb00007.x.
Gu X, Bar-Peled M: The biosynthesis of UDP-galacturonic acid in plants. Functional cloning and characterization of Arabidopsis UDP-D-glucuronic acid 4-epimerase. Plant Physiol. 2004, 136: 4256-4264. 10.1104/pp.104.052365.
Liwanag AJM, Ebert B, Verhertbruggen Y, Rennie EA, Rautengarten C, Oikawa A, Andersen MCF, Clausen MH, Scheller HV: Pectin biosynthesis: GALS1 in Arabidopsis thaliana is a β-1,4-galactan β-1,4-galactosyltransferase. Plant Cell. 2012, 24: 5024-5036. 10.1105/tpc.112.106625.
Jensen JK, Sørensen SO, Harholt J, Geshi N, Sakuragi Y, Møller I, Zandleven J, Bernal AJ, Jensen NB, Sørensen C, Pauly M, Beldman G, Willats WGT, Scheller HV: Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell. 2008, 20: 1289-1302. 10.1105/tpc.107.050906.
Raiola A, Camardella L, Giovane A, Mattei B, De Lorenzo G, Cervone F, Bellincampi D: Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett. 2004, 557: 199-203. 10.1016/S0014-5793(03)01491-1.
González-Carranza ZH, Elliott KA, Roberts JA: Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J Exp Bot. 2007, 58: 3719-3730. 10.1093/jxb/erm222.
Sun L, van Nocker S: Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 2010, 10: 152-10.1186/1471-2229-10-152.
Manabe Y, Nafisi M, Verhertbruggen Y, Orfila C, Gille S, Rautengarten C, Cherk C, Marcus SE, Somerville S, Pauly M, Knox JP, Sakuragi Y, Scheller HV: Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011, 155: 1068-1078. 10.1104/pp.110.168989.
PlantTribes 2.0 Database. http://fgp.bio.psu.edu/tribedb/10_genomes/index.pl,
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS: Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40 (Database issue): D1178-D1186.
Timme RE, Bachvaroff TR, Delwiche CF: Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012, 7: e29696-10.1371/journal.pone.0029696.
TreeCollapserCL4. http://emmahodcroft.com/TreeCollapseCL.html,
Acknowledgements
Thanks to William Ehlhardt, William Murphy, and John Doyle for help in building scripts to streamline database searches, and to Eric Wafula for bioinformatic assistance. Phylogenetic analysis was supported as part of The Center for LignoCellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences under Award # DE-SC0001090 (TWM and CTA), and development of the PlantTribes 2.0 database was supported by NSF Plant Genome grant #0922742 (JPD, LAH, and CWD).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TWM contributed to experimental design, collected query sequences, performed the database searches, identified algal roots, performed the sequence alignments, ran the phylogenetic analyses, prepared the figures, and participated in drafting the manuscript. JPD contributed to experimental design, assisted in sequence alignment, and participated in drafting the manuscript. LAH contributed to experimental design, assisted in sequence alignment, and participated in drafting the manuscript. CWD contributed to experimental design and participated in drafting the manuscript. CTA contributed to experimental design and participated in drafting the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12870_2013_1768_MOESM1_ESM.xls
Additional file 1: Table S1: Query Arabidopsis genes. A list of all the Arabidopsis genes used as queries to the PlantTribes 2.0 database and the sources for collecting them. (XLS 39 KB)
12870_2013_1768_MOESM2_ESM.zip
Additional file 2: Table S2: Pectin-related genes. This table contains all of the genes examined in this study. (ZIP 455 KB)
12870_2013_1768_MOESM3_ESM.zip
Additional file 3: Table S3: Species distribution by family. Plant Tribes 2.0 species list, with the number of pectin-related genes found in each. (ZIP 715 bytes)
12870_2013_1768_MOESM4_ESM.pdf
Additional file 4: Figure S1: Xylogalacturonan xylosyltransferase family tree. Physcomitrella and Selaginella genes were not detected in this family. (PDF 61 KB)
12870_2013_1768_MOESM5_ESM.pdf
Additional file 5: Figure S2: Rhamnogalacturonan I arabinosyltransferase family tree. This tree contains no Physcomitrella members and two algal members, one from Penium margaritaceum and one from Nitella hyalina. (PDF 101 KB)
12870_2013_1768_MOESM6_ESM.pdf
Additional file 6: Figure S3: Pectinmethylesterase inhibitor (PMEI) family tree. This tree contains only Arabidopsis and Medicago trunculata members and likely does not represent the whole family. (PDF 56 KB)
12870_2013_1768_MOESM7_ESM.pdf
Additional file 7: Figure S4: Polygalacturonase inhibitor protein family tree. Physcomitrella and Selaginella genes were not detected in this family. Monocot and eudicot family members are contained in separate clades that are well-resolved from each other. (PDF 106 KB)
12870_2013_1768_MOESM8_ESM.pdf
Additional file 8: Figure S5: Pectate lyase-like (PLL) family tree. A small land plant-wide clade is resolved from the rest of the tree (pink cloud), indicating at least two genes in the common ancestor of land plants. (PDF 466 KB)
12870_2013_1768_MOESM9_ESM.pdf
Additional file 9: Figure S6: Homogalacturonan methyltransferase family tree. This tree consists of three monophyletic clades, two of which are land plant-wide. An algal root with reasonably homology was not detected for this gene family, preventing the determination of whether two or three ancestral genes were present in the common ancestor of land plants. (PDF 175 KB)
12870_2013_1768_MOESM10_ESM.pdf
Additional file 10: Figure S7: UDP-Glucuronic acid epimerase family tree. This family appears to be land plant-wide and is rooted by a gene from C. reinhardtii. However, the grouping of all the Physcomitrella genes into one monophyletic clade implies that there was only one family member in the common ancestor. (PDF 183 KB)
12870_2013_1768_MOESM11_ESM.pdf
Additional file 11: Figure S8: UDP-Rhamnose synthase family tree. Not only is this family land plant-wide, it includes members from the algae C. reinhardtii, Spirogyra pratensis, and Penium margaritaceum, but the grouping of all the Physcomitrella genes into one monophyletic clade implies that there was only one family member in the common ancestor. (PDF 178 KB)
12870_2013_1768_MOESM12_ESM.pdf
Additional file 12: Figure S9: Pectin acetyltransferase family tree. This family appears to be land plant-wide and is rooted by a gene from C. reinhardtii. The grouping of all the Physcomitrella genes into one monophyletic clade implies that there was only one family member in the common ancestor. (PDF 130 KB)
12870_2013_1768_MOESM13_ESM.pdf
Additional file 13: Figure S10: Pectin acetylesterase family tree. This family contains only one Physcomitrella and no Selaginella members. (PDF 219 KB)
12870_2013_1768_MOESM14_ESM.pdf
Additional file 14: Figure S11: Rhamnogalacturonan II xylosyltransferase family tree. This family appears to be land plant-wide, with one member in the common ancestor of land plants. The algal root gene is from Nitella hyalina. (PDF 89 KB)
12870_2013_1768_MOESM15_ESM.pdf
Additional file 15: Figure S12: β-1,4-Galactan β-1,4-Galactosyltransferase family tree. This tree has no algal root. The Physcomitrella genes are grouped together in a well-supported clade separate from other species. There is no evidence for more than one gene in the common ancestor. (PDF 133 KB)
12870_2013_1768_MOESM16_ESM.pdf
Additional file 16: Figure S13: GATL family tree. This tree is poorly resolved, with no root and large polytomies. The Physcomitrella genes group together in one well-supported clade. (PDF 260 KB)
12870_2013_1768_MOESM17_ESM.zip
Additional file 17: galactangalactosyltransferasefamilyalignment.phy. Phylip gene alignment, .phy. Raw β-1,4-galactan β-1,4-galactosyltransferase alignment. β-1,4-galactan β-1,4-galactosyltransferase family alignment file. (ZIP 9 KB)
12870_2013_1768_MOESM18_ESM.zip
Additional file 18: GATLfamilyalignment.phy. Phylip gene alignment, .phy. Raw GATL alignment. GATL family alignment file. (ZIP 17 KB)
12870_2013_1768_MOESM19_ESM.zip
Additional file 19: GAUTfamilyalignment.phy. Phylip gene alignment, .phy. Raw GAUT alignment. GAUT family alignment file. (ZIP 59 KB)
12870_2013_1768_MOESM20_ESM.zip
Additional file 20: GAUTsuperfamilyalignment.phy.Phylip gene alignment, .phy. Raw GAUT superfamily alignment. GAUT superfamily alignment file. (ZIP 80 KB)
12870_2013_1768_MOESM21_ESM.zip
Additional file 21: homogalacturonanmethyltransferasefamilyalignment.phy. Phylip gene alignment, .phy. Raw homogalacturonan methyltransferase alignment. Homogalacturonan methyltransferase family alignment file. (ZIP 17 KB)
12870_2013_1768_MOESM22_ESM.zip
Additional file 22: pectatelyaselikefamilyalignment.phy. Phylip gene alignment, .phy. Raw pectate lyase-like alignment. Pectate lyase-like family alignment file. (ZIP 46 KB)
12870_2013_1768_MOESM23_ESM.zip
Additional file 23: pectinacetylesterasefamilyalignment.phy. Phylip gene alignment, .phy. Raw pectin acetylesterase alignment. Pectin acetylesterase family alignment file. (ZIP 17 KB)
12870_2013_1768_MOESM24_ESM.zip
Additional file 24: pectinacetyltransferasefamilyalignment.phy. Phylip gene alignment, .phy. Raw pectin acetyltransferase alignment. Pectin acetyltransferase family alignment file. (ZIP 10 KB)
12870_2013_1768_MOESM25_ESM.zip
Additional file 25: PGIPfamilyalignment.phy. Phylip gene alignment, .phy. Raw polygalacturonase inhibitor protein alignment. Polygalacturonase inhibitor protein family alignment file. (ZIP 5 KB)
12870_2013_1768_MOESM26_ESM.zip
Additional file 26: PMEfamilyalignment.phy. Phylip gene alignment, .phy. Raw pectin methylesterase alignment. Pectin methylesterase family alignment file. (ZIP 171 KB)
12870_2013_1768_MOESM27_ESM.zip
Additional file 27: PMEIfamilyalignment.phy. Phylip gene alignment, .phy. Raw pectin methylesterase inhibitor alignment. Pectin methylesterase inhibitor family alignment file. (ZIP 1 KB)
12870_2013_1768_MOESM28_ESM.zip
Additional file 28: polygalacturonasefamilyalignment.phy. Phylip gene alignment, .phy. Raw polygalacturonase alignment. Polygalacturonase family alignment file. (ZIP 145 KB)
12870_2013_1768_MOESM29_ESM.zip
Additional file 29: RGIarabinosyltransferasefamilyalignment.phy. Phylip gene alignment, .phy. Raw rhamnogalacturonan I arabinosyltransferase alignment. Rhamnogalacturonan I arabinosyltransferase family alignment file. (ZIP 5 KB)
12870_2013_1768_MOESM30_ESM.zip
Additional file 30: RGIIxylosyltransferasefamilyalignment.phy. Phylip gene alignment, .phy. Raw rhamnogalacturonan II xylosyltransferase alignment. Rhamnogalacturonan II xylosyltransferase family alignment file. (ZIP 4 KB)
12870_2013_1768_MOESM31_ESM.zip
Additional file 31: UDPGlcAepimerasefamilyalignment.phy. Phylip gene alignment, .phy. Raw UDP-glucuronic acid epimerase alignment. UDP-glucuronic acid epimerase family alignment file. (ZIP 14 KB)
12870_2013_1768_MOESM32_ESM.zip
Additional file 32: UDPrhamnosesynthasefamilyalignment.phy. Phylip gene alignment, .phy. Raw UDP-rhamnose synthase alignment. UDP-Rhamnose synthase family alignment file. (ZIP 15 KB)
12870_2013_1768_MOESM33_ESM.zip
Additional file 33: xylogalacturonanxylosyltransferasefamilyalignment.phy. Phylip gene alignment, .phy. Raw xylogalacturonan xylosyltransferase alignment. Xylogalacturonan xylosyltransferase family alignment file. (ZIP 4 KB)
12870_2013_1768_MOESM34_ESM.zip
Additional file 34: galactangalactosyltransferase.tree. Newick tree, .tree. Raw β-1,4-galactan β-1,4-galactosyltransferase tree. β-1,4-galactan β-1,4-galactosyltransferase family tree file with bootstrap values. (ZIP 879 bytes)
12870_2013_1768_MOESM35_ESM.zip
Additional file 35: GATL.tree. Newick tree, .tree. Raw GATL tree. GATL family tree file with bootstrap values. (ZIP 2 KB)
12870_2013_1768_MOESM36_ESM.zip
Additional file 36: GAUT_superfamily.tree. Newick tree, .tree. Raw GAUT superfamily tree. GAUT superfamily tree file with bootstrap values. (ZIP 5 KB)
12870_2013_1768_MOESM37_ESM.zip
Additional file 37: GAUT.tree. Newick tree, .tree. Raw GAUT tree. GAUT family tree file with bootstrap values. (ZIP 3 KB)
12870_2013_1768_MOESM38_ESM.zip
Additional file 38: homogalacturonanmethyltransferase.tree. Newick tree, .tree. Raw homogalacturonan methyltransferase tree. Homogalacturonan methyltransferase family tree file with bootstrap values. (ZIP 1 KB)
12870_2013_1768_MOESM39_ESM.zip
Additional file 39: pectatelyaselike.tree. Newick tree, .tree. Raw pectate lyase-like tree. Pectate lyase-like family tree file with bootstrap values. (ZIP 4 KB)
12870_2013_1768_MOESM40_ESM.zip
Additional file 40: pectinacetylesterase.tree. Newick tree, .tree. Raw pectin acetylesterase tree. Pectin acetylesterase family tree file with bootstrap values. (ZIP 2 KB)
12870_2013_1768_MOESM41_ESM.zip
Additional file 41: pectinacetyltransferase.tree. Newick tree, .tree. Raw pectin acetyltransferase tree. Pectin acetyltransferase family tree file with bootstrap values. (ZIP 865 bytes)
12870_2013_1768_MOESM42_ESM.zip
Additional file 42: PGIP.tree. Newick tree, .tree. Raw polygalacturonase inhibitor protein tree. Polygalacturonase inhibitor protein family tree file with bootstrap values. (ZIP 681 bytes)
12870_2013_1768_MOESM43_ESM.zip
Additional file 43: PME.tree. Newick tree, .tree. Raw pectin methylesterase tree. Pectin methylesterase family tree file with bootstrap values. (ZIP 10 KB)
12870_2013_1768_MOESM44_ESM.zip
Additional file 44: PMEI.tree. Newick tree, .tree. Raw pectin methylesterase inhibitor tree. Pectin methylesterase inhibitor family tree file with bootstrap values. (ZIP 363 bytes)
12870_2013_1768_MOESM45_ESM.zip
Additional file 45: polygalacturonase.tree. Newick tree, .tree. Raw polygalacturonase tree. Polygalacturonase family tree file with bootstrap values. (ZIP 9 KB)
12870_2013_1768_MOESM46_ESM.zip
Additional file 46: RGIarabinosyltransferase.tree. Newick tree, .tree. Raw rhamnogalacturonan I arabinosyltransferase tree. Rhamnogalacturonan I arabinosyltransferase family tree file with bootstrap values. (ZIP 678 bytes)
12870_2013_1768_MOESM47_ESM.zip
Additional file 47: RGIIxylosyltransferase.tree. Newick tree, .tree. Raw rhamnogalacturonan II xylosyltransferase tree.Rhamnogalacturonan II xylosyltransferase family tree file with bootstrap values. (ZIP 618 bytes)
12870_2013_1768_MOESM48_ESM.zip
Additional file 48: UDPGlcAepimerase.tree. Newick tree, .tree. Raw UDP-glucuronic acid epimerase tree. UDP-glucuronic acid epimerase family tree file with bootstrap values. (ZIP 1 KB)
12870_2013_1768_MOESM49_ESM.zip
Additional file 49: UDPrhamnosesynthase.tree. Newick tree, .tree. Raw UDP-rhamnose synthase tree. UDP-Rhamnose synthase family tree file with bootstrap values. (ZIP 1 KB)
12870_2013_1768_MOESM50_ESM.zip
Additional file 50: xylogalacturonanxylosyltransferase.tree. Newick tree, .tree. Raw xylogalacturonan xylosyltransferase tree. Xylogalacturonan xylosyltransferase family tree file with bootstrap values. (ZIP 471 bytes)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
McCarthy, T.W., Der, J.P., Honaas, L.A. et al. Phylogenetic analysis of pectin-related gene families in Physcomitrella patensand nine other plant species yields evolutionary insights into cell walls. BMC Plant Biol 14, 79 (2014). https://doi.org/10.1186/1471-2229-14-79
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2229-14-79