Abstract
Background
Phosphorus is an essential macronutrient for the growth of plants. However, in most soils a large portion of phosphorus becomes insoluble and therefore, unavailable to plants. Knowledge on biodiversity of phosphate-solubilizing fluorescent pseudomonads is essential to understand their ecological role and their utilization in sustainable agriculture.
Results
Of 443 fluorescent pseudomonad strains tested, 80 strains (18%) showed positive for the solubilization of tri-calcium phosphate (Ca3(PO4)2) by the formation of visible dissolution halos on Pikovskaya's agar. These phosphate solubilizing strains showed high variability in utilizing various carbon sources. Numerical taxonomy of the phosphate solubilizing strains based on their carbon source utilization profiles resulted into three major phenons at a 0.76 similarity coefficient level. Genotypic analyses of strains by BOX (bacterial repetitive BOX element)-polymerase chain reaction (PCR) resulted into three distinct genomic clusters and 26 distinct BOX profiles at a 80% similarity level. On the basis of phenotypic characterization and 16S rRNA gene phylogenetic analyses strains were identified as Pseudomonas aeruginosa, P. mosselii, P. monteilii, P. plecoglossicida, P. putida, P. fulva and P. fluorescens. These phosphate solubilizing strains also showed the production of plant growth promoting enzymes, hormones and exhibited antagonism against phytopathogenic fungi that attack on various crops. Gene specific primers have identified the putative antibiotic producing strains. These putative strains were grown in fermentation media and production of antibiotics was confirmed by thin layer chromatography (TLC) and high performance liquid chromatography (HPLC).
Conclusion
Present study revealed a high degree of functional and genetic diversity among the phosphate solubilizing fluorescent pseudomonad bacteria. Due to their innate potential of producing an array of plant growth promoting enzymes, hormones and antifungal metabolites these phosphate solubilizing strains are considered to play a vital role in plant growth promotion, disease suppression and subsequent enhancement of yield.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
Phosphorous deficiency is a major constraint to crop production. Plants absorb only inorganic form of phosphorous and the level of inorganic phosphorus is very low in the soil because most of the phosphorous is present as insoluble forms. Agricultural soils possess considerable accumulation of phosphorous due to the regular excessive applications of chemical fertilizers. The overuse of chemicals and chemical fertilizers has led to the lethal consequences to useful arthropods and other beneficial microbes as well as led to soil pollution. A large proportion of these fertilizers added to the soil are also converted to insoluble form and become unavailable to plants [1].
Microorganisms have the ability to solubilize the insoluble phosphates and maintain the soil health and quality [2]. Bacteria use several direct and indirect mechanisms of action to improve plant growth and health. Mechanisms such as phosphate solubilization [3], aminocyclopropane-1-carboxylate (ACC) deaminase [4], nitrogen cycle [5] and phytohormone production [6] are considered as popular mechanisms. Beneficial effects of phosphate solubilizing bacteria to crops have been documented well [1, 7, 8]. It is believed that microbial solubilization of phosphate in soil was correlated with the ability of microbes in producing selected organic acids and or extracellular polysaccharides [3, 9]. This hypothesis has been corroborated by cloning pyrroloquinoline (PQQ) synthase [10–12] and gab Y genes involved in gluconic acid production. Gluconic acid is the principal organic acid produced due to direct oxidation of glucose by Pseudomonas which was found to be involved in phosphate solubilization [13].
Among phosphate solubilizing bacteria, fluorescent pseudomonads that colonize aggressively the plant roots have been considered as an important group of bacteria due to their biofertilizing and biocontrol properties. In many occasions, plant growth promoting rhizobacteria often exhibit the production of antimicrobial metabolites, which take part in suppression of diseases caused by soil-borne phytopathogens [6, 14, 15] as well as involve in the induction of systemic resistance against insects and nematodes [16–18]. Selective strains of fluorescent pseudomonad bacteria have also been reported for biodegradation of agricultural pollutants [15, 19] as well as for weed control in agricultural fields [20–22]. These bacteria have been considered as an important bioinoculants due to their innate potential to produce plant growth promoting hormones and enzymes [6, 16, 23–25]. Though the bacteria belonging to Mesorhizobium, Rhizobium, Klebsiella, Acinetobacter, Enterobacter, Erwinia, Achrobacter, Micrococcus, Aerobacter and Bacillus have been reported as phosphate solubilizers, strains belong to pseudomonads are considered as efficient phosphate solubilizers [26].
Rice and banana are most important food crops of the world and are widely grown in developing countries. Fungal pathogens are important production constraints of rice, banana and other crops. Indiscriminate use of fungicides in agriculture is known to be hazardous to the environment and lethal toward other beneficial organisms; it can lead to the development of resistance against target organisms. Microbial solubilization of phosphorus, biodegradaton of chemical pollutants and biocontrol of plant pathogens are cost-effective, novel biological restoration and ecofriendly techniques. Considering the multiple applications of phosphate solubilizing fluorescent pseudomonads it is essential to study their diversity, which will be useful in designing strategies to use these native strains as bioinoculants for sustainable and organic agriculture without causing harm to the environment and farmers. The objective of the present investigation was to study the genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads associated with rhizospheric soils of rice and banana by an array of in vitro assays, gene amplification techniques, fermentation methods and chromatographic analyses. Taxonomic affiliation of bacteria was done on the basis of 16S rRNA gene similarity and molecular phylogenetic analyses.
Methods
Isolation of fluorescent pseudomonad strains
Rhizospheric soil samples of rice (Oryza sativa L.) and banana (Musa spp. AAA) were collected from agricultural fields located at Puducherry, India. Samples were stored at 4°C before being processed within 24 h of collection. The soil was sand clay loam and its characteristics were as follows; pH 7.3, 74 μg g-1 nitrogen, 2.9 μg g-1 phosphorus, 5.29 μg g-1 potassium, 0.2 mOhms cm-1 of electrical conductivity (EC). Isolation of fluorescent pseudomonad bacteria from rhizospheric soil was performed as described earlier [25]. Briefly, soil suspension was obtained by shaking 10 g of soil sample having roots with tightly adhering soil in 90 ml of 0.1 M MgSO4.7H2O buffer for 10 min at 180 rpm on a rotary shaker. Resulting suspensions were serially diluted and 0.1 ml aliquots of each dilution were spread onto King's medium B (KB) agar in triplicates. After incubation at 28°C for 2 days, fluorescent pseudomonad colonies from replicate plates were identified under UV light (366 nm). Purified single colonies were further streaked onto KB agar plates to obtain pure cultures. Stock cultures were made in Luria Bertani (LB) broth containing 50% (w/v) glycerol and stored at -86°C.
Screening of phosphate solubilizers and estimation of phosphate solubilization
To detect the phosphate solubilizing bacteria, strains were streaked onto Pikovskaya's agar medium, which contains (per liter): 0.5 g yeast extract, 10 g dextrose, 5 g Ca3(PO4)2, 0.5 g (NH4)2SO4, 0.2 g KCl, 0.1 g MgSO4.7H2O, 0.0001 g MnSO4.H2O, 0.0001 g FeSO4.7H2O and 15 g agar. After 3 days of incubation at 28°C, strains that induced clear zone around the colonies were considered as positive [27]. Determination of phosphate solubilizing activity by the strains was carried out following standard method [28]. Briefly, cells were grown in liquid medium (pH 7.0) at 28°C up to 10 days and on 1, 3, 5, 7 and 10 day an aliquot of 5 ml was collected and cells were removed by centrifugation at 9,000 g for 20 min. Soluble free phosphate in culture supernatant was estimated from the absorbance values obtained using the calibration curve with KH2PO4 at 600 nm for each strain. Also, pH variation in Pikovskaya's medium during the growth of each strain was also observed.
Phenotypic characterization of phosphate solubilizing bacteria
In order to determine the phenotypic diversity of 80 phosphate solubilizing bacteria characterization was done on the basis of fluorescence on King's B (KB) medium, levan formation, gelatin liquefaction, nitrate reduction and growth at 4°C and 42°C [29, 30]. In order to identify fluorescence of the strains, overnight grown cells on KB agar medium were visualized under UV light (366 nm). Gram's reaction was determined by the KOH technique [31]. Briefly, visible amount of overnight grown cells from agar plate was smeared (1–2 cm2 area) onto glass slide containing loopful (3 mm) of 3% aqueous KOH solution. Gram negative strains were identified as viscous gel that string out along with the loop. To test the presence of cytochrome oxidase, bacterial culture grown for 24 h on nutrient agar (NA) supplemented with 1% glucose was used. A loopful cells were rubbed onto a filter paper impregnated with 1% (wt/vol) aqueous N,N,N',N'-tetra-methyl-p-phenylenediamine dihydrochloride solution. A change in the color of the cultures to deep purple within 10 s was registered as a positive result. Presence of arginine dihydrolase and levan sucrase was tested on Thornley's medium and NA supplemented with arginine and sucrose (20 g l-1), respectively [29].
Carbon utilization profiles were tested using Hicarbohydrate™ kit as descried by the manufacturer (Himedia Laboratories, Mumbai, India). Cells were grown in nutrient broth to reach density of 0.5 O.D. at 600 nm. An aliquot of 50 μl of this suspension was inoculated to each well of Hicarbohydrate™ kit, incubated at 30°C for 24 h and the results were registered according to the instructions of the manufacturer. The experiment was done with three replicates. On the basis of data derived from the carbon source utilization profiles, a matrix with binary code composing positive (1) and negative (0) values was made. SIMQUAL program was used to compute the symmetric matrix in the form of average taxonomic distances. Sequential, agglomerative, hierarchical and nested (SAHN) clustering was used for the cluster analyses [32]. Dendrogram was constructed from the similarity matrix by the unweighted pair group with mathematical averages (UPGMA) using NTSYS-pc2.02a (Exeter software, New York, USA) numerical taxonomy and multivariate analysis system.
BOX-PCR based genotypic analysis
Bacteria were cultured in LB broth at 28°C for 18 h and the total genomic DNA was extracted as described [25]. The BOX-A1R (5'-CTACGGCAAGGCGACGCTGACG-3') primer for genotypic analysis [33] was synthesized by Integrated DNA Technologies Inc. (Coralville, IA, USA). PCR reaction (50 μl) contained 50 pM of primer, 50 ng of genomic DNA, 1× Taq DNA polymerase buffer, 1 U of Taq DNA polymerase (Promega, Madison, WI), 0.2 mM of each deoxynucleotide triphosphate (dNTP), and 1.5 mM MgCl2. Amplification was performed in a DNA thermal cycler (2400 cycler, Perkin Elmer International, Rotkreuz, Switzerland) programmed with an initial denaturation at 95°C for 7 min, 30 cycles at 94°C for 1 min, 53°C for 1 min, and 65°C for 8 min, with an extension at 65°C for 15 min. A 10 μl of PCR product was separated using 1.5% agarose gel stained with ethidium bromide in 1× tris acetate ethylenediaminetetraacetic acid (TAE). The image of the gel was digitized by using BIO-CAPT system (Vilber Lourmat, France) and stored as TIFF files. BOX-PCR band profiles were detected by QuantityOne program (Bio-Rad Laboratories, CA, USA). Computer assisted analysis of genomic fingerprints was performed by using the BIO-GENE software program v11.02 (Vilber Lourmat, France). Similarity matrices of whole densitometric curves of the gels tracks were calculated by using the Dice coefficient. Cluster analysis of similarity matrices was performed by the un-weighted pair group with mathematical average (UPGMA) algorithm.
16S rRNA gene amplification, sequencing and phylogenetic tree analysis
Amplification of 16S rRNA gene was performed from the genomic DNA of strains using universal primers fD1 (5'-GAGTTTGATCCTGGCTCA-3') and rP2 (5'-ACGGCTACCTTGTTACGACTT-3') [34]. PCR cocktails (50 μl) contained 50 pM of primer, 50 ng of genomic DNA, 1× Taq DNA polymerase buffer, 1 U of Taq DNA polymerase (Promega, Madison, WI, USA), 0.2 mM of each dNTP, and 1.5 mM MgCl2. Amplification was performed in a DNA thermo cycler (2400 cycler, Perkin Elmer International, Rotkreuz, Switzerland) at 94°C for 3 min, followed by 30 cycles of 10 s at 94°C, 1 min at 56°C and 30 s at 72°C with an extension of 72°C for 5 min. A 5 μl aliquot of each amplification product was electrophoresed on a 0.7% agarose gel in 1× TAE buffer at 50 V for 45 min, stained with ethidium bromide and the PCR products were visualised with a UV transilluminator. PCR products were purified using Quick PCR purification column (Promega, Madison, USA). Purified PCR products were sequenced with automated DNA sequencer with specific primers using the facility at Macrogen Inc. (Seoul, Korea). To perform molecular phylogenetic analyses, reference sequences required for comparison were downloaded from the EMBL database using the site http://www.ncbi.nlm.nih.gov/Genbank. All the sequences of 16S rRNA were aligned using the multiple sequence alignment program CLUSTAL W [35]. The aligned sequences were then checked for gaps manually, arranged in a block of 600 bp in each row [36] and saved as molecular evolutionary genetics analysis (MEGA) format in software MEGA v3.0. The pair wise evolutionary distances were computed using the Kimura 2-parameter model [37]. To obtain the confidence values, the original data set was resampled 1000 times using the bootstrap analysis method. The bootstrapped data set was used directly for constructing the phylogenetic tree using the MEGA v3.0 program for calculating the multiple distance matrixes [38]. The multiple distance matrix obtained was then used to construct phylogenetic trees using neighbor-joining (NJ) method [39].
Nucleotide sequence accession numbers
The nucleotide sequences of 16S rRNA were deposited in GenBank. The accession numbers of the 16S rRNA nucleotide sequences of the strains are presented in Additional file 1.
Production of other plant growth promoting enzymes and hormones
Indole-3-acetic acid (IAA)
Production of IAA was determined following the standard method [40]. Briefly, overnight grown single colony was streaked onto LB medium agar, which contains (per liter): 10 g tryptone, 5 g yeast extract, 5 g NaCl, 15 g agar amended with 5 mM L-tryptophan, 0.06% sodium dodecyl sulphate and 1% glycerol. Plates were overlaid with sterile Whatman no. 1 filter paper (82 mm diameter) and bacterial strain was allowed to grow for 3 days at 28°C. After incubation, the paper was removed and treated with Salkowski's reagent [41] having the formulation of 2% of 0.5 M ferric chloride in 35% perchloric acid at room temperature for 60 min. In a Petri dish, the filter papers were saturated by soaking in Salkowski's reagent and the production of IAA was identified by the formation of a red halo on the paper immediately surrounding the colony.
Aminocyclopropane-1-carboxylate (ACC) deaminase
The ACC deaminase activity was determined as described earlier [16] on Dworkin and Foster (DF) minimal salts medium, which contains (per liter): 4 g KH2PO4, 6 g Na2HPO4, 0.2 g MgSO4.7H2O, 2 g glucose, 0.2 g; 2 g gluconic acid and 2 mg citric acid with trace element solution (1 mg FeSO4.7H2O, 10 μg H3BO3, 11.19 μg MnSO4.H2O, 124.6 μg ZnSO4.7H2O, 78.22 μg CuSO4.5H2O and 10 μg MoO3). Filter sterilized ACC solution (3 mM) was spread over the agar plates, allowed to dry for 10 min and inoculated with bacterial strains. Observation of the growth made after 2 days incubation at 28°C as described earlier [4].
Siderophore
Production of siderophore by the strains was determined using the FeCl3 test and the chrome azurol S agar assay. Briefly, inoculum (10 μl) of bacterial strains dropped onto the center of a CAS plate. After incubation at 25°C for 3 days, siderophore production was assessed on the basis of change in color of the medium from blue to orange [42–44].
Production of fungal cell wall degrading enzymes
Protease
The protease activity was determined using skim milk agar medium, which contains (per liter): 5 g pancreatic digest of casein, 2.5 g yeast extract, 1 g glucose, 7% skim milk solution and 15 g of agar. Bacterial cells were spot inoculated and after 2 days incubation at 28°C proteolytic activity was identified by clear zone around the cells [45].
Chitinase
The chitinase activity of strains was tested on chitin agar medium, which contains (per liter): 1.62 g nutrient broth, 0.5 g NaCl, 6 g M9 salts, 8 g colloidal chitin and 15 g agar. Bacterial cells were spot inoculated and after 5 days incubation at 30°C, chitinase activity was identified by clear zone around the cells [17].
Production of other beneficial enzymes
Cellulase
Strains were screened for cellulase production by plating onto M9 medium agar amended with 10 g of cellulose and 1.2 g of yeast extract per liter. After 8 days of incubation at 28°C, colonies surrounded by clear halos were considered positive for cellulase production [23].
Pectinase
Pectinase production was determined using M9 medium amended with 4.8 g of pectin per liter. After 2 days of incubation at 28°C, plates were flooded with 2 mol l-1 HCl and strains surrounded by clear halos were considered positive for pectinase production [23].
Test for antagonism
Strains were tested for in vitro antagonism towards fungal pathogens by following standard co-inoculation technique on potato dextrose agar (PDA) [36, 46]. Fungal phytopathogens, Fusarium oxysporum f. sp. cubense FOC (discoloration of banana), Cylindrocladium floridanum ATCC42971 (root necrosis of banana), Cylindrocladium scoparium ATCC46300 (root necrosis of banana), were kindly provided by J. M. Risede, UMR de pathologie végétale INRA-INH-Universite d'Angers, Beaucouze Cedex and CIRAD-FLHOR, Station de Neufchateâu, Sainte Marie, Capesterre-Belle-Eau, Guadeloupe, France. Rhizoctonia solani RSR1 (sheath blight of rice), Magnaporthe grisea MGS (blast of rice), Sarocladium oryzae SONS (sheath rot of rice), Botrytis cinerea BCTNAU (Blight of tobacco), Macrophomina phaseolina MPS (charcoal rot of ground nut), Pestalotia theae PTS (leaf spot of tea), Colletotrichum falcatum (red rot of sugarcane), C. capsici CPS (fruit rot of chili), C. gleosporoides CGS (Anthracnose of mango) were obtained from the Microbial Culture Collection (MCC), Department of Biotechnology, Pondicherry University, Puducherry. These phytopathogenic fungi are major production constraints of rice, banana and other crops and therefore, selected for screening antagonism. Briefly, bacterial plugs (6 mm diameter) were removed from a 48 h culture and were transferred to the center of PDA plates, which had been inoculated with fungal spore suspension (106 conidia ml-1). Assay plates were incubated at 28°C for 3 days and growth inhibition appeared around the bacterial plugs was measured. Assays were done with three replicates.
Screening of putative antibiotic producing strains by polymerase chain reaction
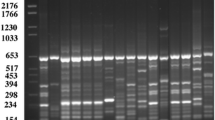
Detection of the genes that encode for the production of antibiotics such as, 2,4-diacetylphloroglucinol (DAPG), phenazine-1-carboxylic acid (PCA), phenazine-1-carboxamide (PCN), pyrrolnitrin (PRN) and pyoluteorin (PLT) was done by PCR using gene-specific primers. Reference strains, Pseudomonas fluorescens Pf5, P. fluorescens 2–79, P. aureofaciens 30–84 (now considered as P. chlororaphis) and P. aeruginosa PAO1 were kindly supplied by Linda S. Thomashow, USDA, Washington State University, Pullman and P. fluorescens CHAO was kindly supplied by Geneviève Défago, Swiss Federal Institute of Technology, Zurich. Oligonucleotide primers were synthesized by Integrated DNA Technologies Inc. (Coralville, IA, USA). The primer sets and the amplification conditions for the screening of genes encoding antibiotics are listed in Table 1. PCR reaction (50 μl) contained 50 pM of each primer, 50 ng of genomic DNA, 1× Taq DNA polymerase buffer, 0.5 U of Taq DNA polymerase (Promega, Madison, WI, USA), 0.2 mM of each deoxynucleotide triphosphate, and 1.5 mM MgCl2. Amplification was performed in a DNA thermal cycler. A 5 μl aliquot of each amplified product was electrophoresed on a 0.7% agarose gel in 1× TAE buffer at 50 V for 45 min, stained with ethidium bromide and the PCR products were visualized with a UV transilluminator.
Production and quantification of antifungal metabolites and analytical methods
Antifungal metabolites were extracted as described [6, 36, 47]. Reference strains and antibiotic standards were kindly provided by Linda S. Thomashow, USDA, Washington State University, Pullman; and Geneviève Défago, Swiss Federal Institute of Technology, Zurich. Thin layer chromatography (TLC) was carried out on silica gel G60 plate (20 × 20 cm; 0.25 mm thick, Selecto Scientific, GA, USA). The plates were activated at 110°C for 30 min, cooled and spotted with ethanol solution containing standard antibiotic (0.5 μg) and 20 μl of the extract. Separation was performed using the solvent system: chloroform-methanol (9:1 vol/vol) for PCA and DAPG or chloroform-acetone (9:1 vol/vol) for PLT and PRN. The corresponding spots by PCA, and DAPG were detected by UV irradiation at 254 nm [48]. PLT spots were detected by spraying with an aqueous 0.5% (wt/vol) Fast Blue RR salt solution 0.5% (wt/vol) and the PRN spots were detected by spraying the TLC plates with 2% p-dimethylaminobenzaldehyde dissolved in the ethanol-sulfuric acid (1:1 vol/vol). Detection and quantitative determination of antibiotics was done by analytical HPLC (Shimadzu, Kyoto, Japan) as described [36]. Production of hydrogen cyanide (HCN) was determined by growing bacteria on KB agar medium supplemented with 4.4 g l-1 glycine. A piece of filter paper impregnated with picric acid and sodium carbonate (0.5% and 2%, respectively) was placed in the lid of each Petri dish. Petri dish was sealed with parafilm and incubated at 28°C for 96 h. The production of cyanide was identified by the change in the color of filter paper from yellow to orange-brown [47].
Results
Isolation and screening of phosphate solubilizing bacteria and estimation of phosphate solubilization
Of the 443 strains, 80 strains (18%) produced phosphate solubilization on Pikovskaya's agar medium by inducing clear zones (Fig. 1). The solubilization of tri-calcium phosphate was estimated for all strains. Soluble phosphate was estimated to be 29 to 105 μg ml-1 on 10 days of inoculation. A significant reduction in pH of Pikovskaya's liquid medium from pH 7.4 to pH 4.8 was observed on 10 days of incubation [See Additional file 2].
Phenotypic characterization of phosphate solubilizing bacteria
Out of 443 fluorescent pseudomonad strains tested, 80 strains showed phosphate solubilization potential. These phosphate solubilizing strains were Gram-negative, motile, rod shaped and tested positive for cytochrome oxidase, arginine dihydrolase, strains showed variability for traits such as gelatin hydrolysis, levan production and growth at 4°C and 42°C. All phosphate solubilizing strains utilized dextrose, galactose, mannose and citrate but exhibited varying degree of utilization towards other carbon sources such as lactose, xylose, fructose, melibiose, L-arabinose, glycerol, ribose, α-methyl-D-mannoside, xylitol, esculin, D-arabinose, malonate, sorbose, trehalose, sorbitol, mannitol, adonitol and glucosamine. These strains did not utilize maltose, sucrose, inulin, salicin, dulcitol, inositol, α-methyl-D-gluconate, rhamnose, cellobiose, melazitose, xylitol and ONPG. Numerical analysis of phenotypic characteristics revealed a high degree of polymorphism. All phosphate solubilizing strains were grouped into 3 major phenons at 0.76 similarity coefficient level (Fig. 2). The similarity coefficient range among phosphate solubilizing strains was 0.57 to 1.00. Phenons I, II and III consist a total of 61, 3 and 16 strains, respectively.
Phenogram of 80 phosphate solubilizing fluorescent pseudomonads based on their carbon source utilization profiles. The clustering was done using sequential, agglomerative, hierarchical and nested (SAHN) method. The pairwise coefficient of similarity (Dice) was used for clustering with the UPGMA algorithm using NTSYSpc2.02a software. The phenogram resulted into 3 major phenon at 0.76 smilarity coefficient. The experiment was done with three replicates.
BOX-PCR based genotypic analysis
The cluster analysis based on the pair-wise coefficient similarity with UPGMA of BOX-PCR resulted into 3 distinct genomic clusters and at a 80% similarity coefficient generated 26 distinct BOX profiles (Fig. 3). A total of 20 strains were grouped into cluster I which shared 40% similarity with other strains. Cluster III consisted of 2 strains and a large number of strains grouped in cluster II comprising 58 strains. All the strains showed wide variations in fingerprinting pattern due to their high degree of genetic variability and distributed into different clusters. On the basis of these results, present study identified a high degree of genetic variability among different species of phosphate solubilizing fluorescent pseudomonads.
Cluster analyses of BOX-PCR fingerprints showing the genotypic diversity of phosphate solubilizing fluorescent pseudomonads. Dendrogram was obtained from the similarity coefficient (Dice) calculations and clustering was done using unweighted pair-grouping method based on arithmetic averages (UPGMA) algorithm using BIOGENE software v11.02. The dendrogram resulted into 3 major clusters and 26 distinct BOX profiles.
16S rRNA gene amplification, sequencing and phylogenetic tree analysis
On the basis of phylogenetic analysis of 16S rRNA gene (600 bp), species of pseudomonads such as Pseudomonas monteilii, P. putida, P. plecoglossicida, P. fluorescens, P. fulva, P. mosselii and P. aeruginosa were identified. Phylogenetic analyses of the 80 fluorescent pseudomonad strains based on the NJ method with 1000 bootstrap sampling were resulted into 3 major clusters (Fig. 4). Of the 80 strains, cluster I formed with 58 strains, cluster II formed with 8 strains and cluster III formed with 14 strains. A total of 36 strains belong to P. monteilii, 10 strains belong to P. plecoglossicida, 12 strains belong to P. putida, 7 strains belong to P. fluorescens, 1 strain belongs to P. fulva, 12 strains belong to P. aeruginosa and 2 strains belong to P. mosselli (See Additional file 1; Fig. 4).
Phylogenetic analyses of phosphate solubilizing strains of fluorescent pseudomonads based on the nucleotide sequence of 16S rRNA. The multiple sequence alignment was done in CLUSTAL program. The pair-wise evolutionary distances were calculated using Kimura2-parameter model. The phylogenetic tree was constructed by neighbor-joining (NJ) method with 1000 replicates using bootstrap. A total of 7 reference fluorescent pseudomonad strains were used for the tree construction.
Production of plant growth promoting enzymes, hormones and fungal cell wall degrading enzymes
All the strains tested positive for the production of siderophore on the basis of change in colour of the CAS medium from blue to orange. Of the 80 phosphate solubilizing strains, 39 strains (49%) showed positive for the production of plant growth-promoting hormone, IAA. The ACC deaminase was observed in 13 strains (16%) by their growth on Dworking and Foster (DF) minimal medium containing ACC. Of the 80 strains, 22 strains (28%) showed proteolytic activity by inducing clear zones around the cells on skim milk agar medium, 7 strains (9%) showed chitinase activity by inducing clear zones around the cells on chitin agar medium, 33 strains (41%) produced pectinase and 25 strains (31%) produced cellulase [See Additional file 1].
Test for antagonism
Of the 80 phosphate solubilizing strains, 33 strains (41%) showed antifungal activity towards phytopathogenic fungi used in the study [See Additional file 3]. Strains induced growth-free inhibition zones (diameter) ranging from 10 to 36 mm towards phytopathogenic fungi.
Screening of putative antibiotic producing strains by PCR
Genomic DNA of strains when tested as templates using gene-specific primers, 12 strains (FPB16, FPB17, FPB51, FP2, FP3, FP5, FP7 FP10, FP14, Pw70, Pw71 and Pw72) amplified the DNA fragment of 745-bp of DAPG, 10 strains (FPB15, FPB16, FPB74, FPB75, FP2, FP3, FP5, FP7, FP14 and FP15) amplified the DNA fragment of 719-bp of PRN, 6 strains (FP2, FP3, FP5, FP7, FP9 and FP14) amplified DNA fragment of 779-bp of PLT and 6 strains (FPB18, FPB52, FPB75, Pw60, Pw61 and FP15) amplified DNA fragment of 1,150-bp of PCA (C, D).
Production quantitative determination of antifungal metabolites
The putative antibiotic strains identified by PCR were subjected to fermentation and antifungal metabolites such as DAPG (yellowish white), PCA (greenish yellow), PRN (light yellow) and PLT (yellowish white) were detected in the production cultures. TLC and HPLC analyses confirmed the production of PCA, DAPG, PLT and PRN by the strains. The retardation factor (Rf) values were found to be 0.77 for DAPG, 0.53 for PCA, 0.80 for PRN, 0.50 for PLT as determined by co-migration with pure standards. Strains grown in fermentation cultures yielded PCA (20 to 1124 μg ml-1), DAPG (13 to 93 μg ml-1), PLT (3 to 9 μg ml-1) and PRN (3 to 41 μg ml-1). HPLC analyses of active fractions of antibiotics yielded similar retention time to that of the standard antibiotics. PCA was detected at 257 nm with a retention time 4.94 min, DAPG was detected at 270 nm with a retention time 10.77 min, PLT was detected at 255 nm with a retention time of 17.6 min and PRN was detected at 220 nm with a retention time of 27.5 min. Of the 80 strains, 20 strains (25%) produced HCN [See Additional file 1].
Discussion
Phosphorus is considered as an essential macronutrient and a great portion of phosphorus from chemical fertilizers becomes insoluble by its conversion into calcium or magnesium salts in soils and become unavailable to plants. Soil microorganisms involve to transform the insoluble forms of phosphorus into soluble forms and thus influence the subsequent availability of phosphate to plant roots are considered essential [49, 50]. Phosphate solubilizing microorganisms have been employed in agriculture and horticulture and have been considered very important due to their potential of ecological amelioration. It is believed that microbial mediated solubilization of insoluble phosphates in soil is through the release of organic acids microbial metabolites [51–53]. However, in addition to acid production, other mechanisms can cause phosphate solubilization [54]. Phosphate solubilization has been reported to depend on the structural complexicity and particle size of phosphates and the quantity of organic acid secreted by microbes [55].
Fluorescent pseudomonads often predominant among plant rhizosphere associated bacteria [24, 56]. Plant growth promoting rhizobacteria are classified into two different groups such as strains that have the capability of synthesizing phytohormones and strains that have the ability to suppress the growth of phytopathogens [57]. Fluorescent pseudomonads enhance plant growth by improving soil nutrient status, producing plant growth hormones, enzymes and suppressing the growth of phytopathogenic fungi [56, 58]. Plant growth promoting rhizobacterial types of fluorescent pseudomonads use one or more mechanisms of direct or indirect in improving plant growth. These mechanisms can probably be active simultaneously or sequentially at different stages of plant growth. This group of bacteria exhibits multiple functional traits such as solubilizing of inorganic phosphate and iron, production of vitamins, phytohormones and antimicrobial metabolites. They are capable of improving plant nutrients uptake, tolerance to stress, salinity, metal toxicity and pesticide. Fluorescent pseudomonad strains such as P. fluorescens NJ101 [14], P. fluorescens EM85 [59], P. fluorescens [60], Pseudomonas spp. [61, 62], P. chlororaphis, P. savastanoi, P. pickettii [23] and P. corrugata [63] have been reported as phosphate solubilizers.
In the present investigation, out of 443 fluorescent pseudomonad strains screened, 80 strains have been identified as phosphate solubilizers. These strains were taxonomically described as different fluorescent pseudomonad species such as P. monteilli, P. putida, P. plecoglossicida, P. fluoresens, P. fulva, P. monteilli and P. aeruginosa on the basis of 16S rRNA gene sequencing and subsequent molecular phylogeny analysis. Phenotypic analyses as well as 16S rRNA and BOX-PCR based genotypic analyses revealed a high degree of diversity among phosphate solubilizing bacteria reported in this study. Significant decline in the pH of the culture medium by strains was observed during mineral phosphate solubilization, which suggested the microbial production of organic acids [64]. Although phosphate solubilization is not necessarily correlated with acidity, from the data present in this study relationship could be ascertained between the acidity of medium and the release of soluble phosphates. Estimation of phosphate solubilization of strains by other methods has been reported to be between 200 to 805 μg ml-1 [65]. In an earlier study, P. fluorescens strain NJ-101 isolated from agricultural soil was reported to release 74.6 μg ml-1 soluble phosphate from inorganic phosphate [14] and in the present study, up to 105.3 μg ml-1 soluble phosphate was estimated. We have found that 49% of the strains produced IAA and 16% of the strains produced ACC deaminase. It is reported that the ACC deaminase producing bacteria increase root elongation and seed germination by lowering plant ethylene levels [24, 66]. Specific strains of fluorescent pseudomonad bacteria indirectly influence the plant health by preventing the deleterious effects of phytopathogenic microorganisms through the production of antibiotics, cell wall degrading enzymes, HCN metabolite and siderophores [56, 57]. Production of HCN by P. fluorescens CHAO was recognized as a biocontrol factor, against plant pathogenic fungi [67].
Production of antibiotics by fluorescent pseudomonads considered important in suppression of phytopathogens. Recently, P. aeruginosa PUPa3, a new strain from rice rhizosphere with potential for fungal antibiosis and biofertilizing traits has been identified from our laboratory [25]. All the strains reported in this study produced hydroxamate siderophores as evidenced on FeCl3 amended CAS agar medium, production of an array of phytohormones and antifungal metabolites. Microbial production of antibiotics, PCA (2 to 3 mg ml-1), DAPG (0.5 to 3 mg ml-1), PLT (1.5 to 2 μg ml-1) and PRN (0.54 mg ml-1) by biofertilizing and biocontrol strains have been reported in earlier studies [68–70]. In the present study the production of PCA (20 to 1124 μg ml-1), DAPG (13 to 93 μg ml-1), PLT (3 to 9 μg ml-1) and PRN (3 to 41 μg ml-1) by phosphate solubilizing fluorescent pseudomonads has been reported. Strain efficiency and variations in the fermentation conditions often result in an alteration in antibiotic production. Considering the quantity of antibiotics by plant growth promoting and biocontrol strains of fluorescent pseduomonads reported by other inverstigators [68–70], strains reported in this study may be considered as non-pathogenic to plants and antagonistic bacteria against phytopathogenic fungi. Strains reported in this study utilized several carbon sources as identified by Hi-carbohydrate™ kit test. Utilization of variety of carbon sources by the strains may play an important role in adapting to a variety of crop plants and soil types.
The IAA hormone is known to have dual role in influencing plant growth, by involving in the biocontrol together with glutathione-s-transferases in defense-related plant reactions and inhibits the germination of spore and growth of mycelium of different pathogenic fungi [71, 72]. Martinez Noel et al. (2001) showed that the IAA supply to excised potato leaves reduced the severity of the disease provoked by Phytophthora infestans [73]. In present investigation, we have identified the spectrum of bacterial antagonism by measuring the inhibition zones of mycelial radial growth in plate assays. These antagonistic strains also showed production of IAA, fungal cell wall degrading enzymes, such as cellulases, proteases and chitinases which are known to be involved in antagonistic activity against phytopathogenic fungi and insects [74, 75]. Selective microbial producers of chitinase are also reported to be the efficient phosphate solubilizers [76]. Phosphate solubilizing fluorescent pseudomonad strains reported in this study may solubilize insoluble compounds due to the excretion of organic acids. Production of antimicrobial metabolites and organic acids is essential to decrease soil pH, which plays a major role in solubilization of phosphates and other nutrients.
Characterization of phosphate solubilizing fluorescent pseudomonad bacteria is required to study their ecological role in soil. Fluorescent pseudomonad strains reported in this study with phosphate solubilization potential and ability to excrete phytohormones and antimicrobial metabolites may be used as plant growth promoting bacteria and biocontrol agents in sustainable agriculture.
Conclusion
Phosphate solubilizing fluorescent pseudomonad bacterial strains with their multifunctional properties will attract more attention in the field of biofertilization and biological control. Present investigation revealed the microbial diversity of fluorescent pseudomonads with innate potential of mineralizing phosphate, plant growth promoting traits and biocontrol properties. Knowledge generated on biodiversity of phosphate solubilizing bacteria will be useful to design strategies to use these strains as inoculants in sustainable and organic agriculture.
References
Rodriguez H, Fraga R: Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999, 17: 319-339. 10.1016/S0734-9750(99)00014-2.
Richardson AE: Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Aust J Plant Physiol. 2001, 28: 8797-906.
Kim KY, Jordan D, McDonald GA: Effect of phosphate-solubilizing bacteria and vesicular-arbuscular mycorrhizeae on tomato growth and soil microbial activity. Biol Fertil Soils. 1998, 26: 79-87. 10.1007/s003740050347.
Penrose DM, Glick B: Methods for isolating and characterizing ACC deaminase containing plant growth promoting rhizobacteria. Physiol Plant. 2002, 118: 10-15. 10.1034/j.1399-3054.2003.00086.x.
Ahn T, Ka J, Lee G, Song H: Microcosm study for revegetation of barren land with wild plants by some plant growth-promoting rhizobacteria. J Micobiol Biotech. 2007, 17: 52-57. 10.1159/000098158.
O'Sullivan DJ, O'Gara F: Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992, 56: 662-676.
Pal SS: Interaction of an acid tolerant strain of phosphate solubilizing bacteria with a few acid tolerant crops. Plant Soil. 1998, 198: 169-177. 10.1023/A:1004318814385.
Richardson AE, Hadobas PA, Hayes JE: Extracellular secretion of Aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J. 2001, 25: 641-649. 10.1046/j.1365-313x.2001.00998.x.
Halvorson HO, Keynan A, Kornberg HL: Utilization of calcium phosphates for microbial growth at alkaline pH. Soil Biol Biochem. 1990, 22: 887-890. 10.1016/0038-0717(90)90125-J.
Goldstein AH, Liu ST: Molecular cloning and regulation of a mineral phosphate solubilizing gene from Erwinia herbicola. Biotechnology. 1987, 5: 72-74. 10.1038/nbt0187-72.
Liu TS, Lee LY, Tai CY, Hung CH, Chang YS, Wolfram JH, Rogers R, Goldstein AH: Cloning of an Erwinia carotovora gene necessary for gluconic acid production and enhanced mineral phosphate solubilization in Escherichia coli HB101: Nucleotide sequence and probable involvement in biosynthesis of the coenzyme pyrroloquinoline quinine. J Bacteriol. 1992, 174: 5814-5819.
Rodriguez H, Gonzalez T, Selman G: Expression of a mineral phosphate solubilizing gene from Erwinia herbicola in two rhizobacterial strains. J Biotechnol. 2000, 84: 155-161. 10.1016/S0168-1656(00)00347-3.
Goldstein AH, Rogers RD, Mead G: Separating phosphate from ores via bioprocessing. Biotechnology. 1993, 11: 1250-1254.
Bano N, Musarrat J: Characterization of a novel carbofuran degrading Pseudomonas sp. with collateral biocontrol and plant growth promoting potential. FEMS Microbiol Lett. 2004, 231: 13-17. 10.1016/S0378-1097(03)00894-2.
Ravindra Naik P, Sakthivel N: Functional characterization of a novel hydrocarbonoclastic Pseudomonas sp. strain PUP6 with plant-growth-promoting traits and antifungal potential. Res Microbiol. 2006, 157: 538-546. 10.1016/j.resmic.2005.11.009.
Ramamoorthy V, Viswanathan R, Raguchander T, Prakasam V, Samiyappan R: Induction of systemic resistance by plant growth promoting rhizobacteria in crop plants against pests and diseases. Crop Protection. 2001, 20: 1-11. 10.1016/S0261-2194(00)00056-9.
Renwick A, Campbell R, Coe S: Assessment of in vivo screening systems for potential biocontrol agents of Gaeumannomyces graminis. Plant Pathol. 1991, 40: 524-532. 10.1111/j.1365-3059.1991.tb02415.x.
Zahnder GW, Murphy JF, Sikora EJ, Kloepper JW: Application of rhizobacteria for induced resistance. Eur J Plant Pathol. 2001, 107: 39-50. 10.1023/A:1008732400383.
Bano N, Musarrat J: Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential biocontrol agent. Curr Microbiol. 2003, 46: 324-328. 10.1007/s00284-002-3857-8.
Charudattan R: The mycoherbicide approach with plant pathogens. Microbial control of weeds. Edited by: TeBeast DO. 1991, New York: Chapman and Hall, 24-57.
Gurusiddaiah S, Gealy DG: Isolation and characterization of metabolites from Pseduomonas fluorescens-D7 for control of downy brome (Bromus techtorum). Weed Sci. 1994, 42: 492-501.
Kremer RJ, Begonia MFT, Lynn S, Lanham ET: Characterization of rhizobacteria associated with weed seedlings. Appl Environ Microbiol. 1990, 56 (6): 1646-1655.
Cattelan AJ, Hartel PG, Furhmann FF: Screening for plant growth promoting rhizobacteria to promote early soybean growth. Soil Sci Soc Am J. 1999, 63: 1670-1680.
Glick BR, Karaturovic DM, Newell PC: A novel procedure for rapid isolation of plant growth promoting pseudomonas. Can J Microbiol. 1995, 41: 533-536.
Sunish Kumar R, Ayyadurai N, Pandiaraja P, Reddy AV, Venkateswarlu Y, Prakash O, Sakthivel N: Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity and biofertilizing traits. J Appl Microbiol. 2005, 98 (1): 145-154. 10.1111/j.1365-2672.2004.02435.x.
Villegas J, Fortin JA: Phosphorus solubilization and pH changes as a result of the interactions between soil bacteria and arbuscular mycorrhizal fungi on a medium containing NO3- as nitrogen source. Can J Bot. 2001, 80: 571-576. 10.1139/b02-038.
Katznelson H, Bose B: Metabolic activity and phosphate-dissolving capability of bacterial isolates from wheat roots, rhizosphere, and non-rhizosphere soil. Can J Microbiol. 1959, 5: 79-85.
King JE: The colorimetric determination of phosphorus. Biochem J. 1936, 26 (2): 292-297.
Gerhardt P, Murray RGE, Costilow RN, Nester EW, Wood WA, Krieg NR, Phillips GB, (ed): Manual of methods for general bacteriology. 1981, ASM Press: Washington DC
Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST: Bergey's Manual of Determinative Bacteriology. 1994, Williams & Wilkins: MD, Baltimore
Ryu E: A simple method of differentiating between Gram positive and Gram-negative organisms without staining. Kitasato Arch Exp. 1940, 17: 58-63.
Sneath PHA, Sokal RR: Numerical taxonomy. The principles of numerical classification. Edited by: Kennedy D, Park RB. 1973, Freemann: San Fransisco, 573-
Louws FJ, Fulbright DW, Stephens CT, de Bruijn FJ: Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994, 60: 2286-2295.
Weisburg WG, Barns SM, Lane DJ: 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991, 173: 697-703.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG: ClustalW and ClustalX version 2. Bioinform. 2007, 23: 2947-2948. 10.1093/bioinformatics/btm404.
Ayyadurai N, Ravindra Naik P, Sakthivel N: Functional characterization of antagonistic fluorescent pseudomonads associated with rhizospheric soil of rice (Oryza sativa L.). J Microbiol Biotechnol. 2007, 17: 919-927.
Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980, 16: 111-120. 10.1007/BF01731581.
Kumar S, Tamura K, Nei M: MEGA3: Integrated software for molecular evolutionary genetic analysis and sequence alignment. Brief Bioinform. 2004, 5 (2): 150-163. 10.1093/bib/5.2.150.
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4: 406-425.
Bric JM, Bostock RM, Silverstone SE: Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991, 57: 535-538.
Gordon SA, Weber RP: Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951, 26: 192-195. 10.1104/pp.26.1.192.
Arnow LE: Colorimetric determination of the component of 3,4-hydroxyphenyl-alanine-tyrosine mixture. J Biol Chem. 1937, 118: 531-537.
Neilands JB: Siderophore: structure and function of microbial iron transport compounds. J Biol Chem. 1995, 270: 26723-26726.
Schwyn B, Neiland JB: Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987, 160: 47-56. 10.1016/0003-2697(87)90612-9.
Smibert RM, Krieg NR: Phenotypic characterization. Methods for General and Molecular Bacteriology. Edited by: Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994, American Society of Microbiology: Washington DC, 607-654.
Sakthivel N, Gnanamanickam SS: Evaluation of Pseudomonas fluorescens for suppression of sheath rot disease and for enhancement of grain yields in rice(Oryza sativa L.). Appl Environ Microbiol. 1987, 53: 2056-2059.
Bakker WA, Schippers B: Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas sp. mediated plant growth stimulation. Soil Biol Biochem. 1987, 19: 451-457. 10.1016/0038-0717(87)90037-X.
Thomashow LS, Weller DM, Bonsall RF, Pierson LS: Production of the antibiotic phenazine-1-carboxylic acid of fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990, 56: 908-912.
Illumer P, Schinner F: Solubilization of inorganic calcium phosphate-solubilization mechanisms. Soil Biol Biochem. 1995, 27: 257-263. 10.1016/0038-0717(94)00190-C.
Richardson AE, Hadobas PA, Hayes JE, O'Hara JE, Simpson RJ: Utilization of phosphorus by pasture plants supplied with myo-inositol hexaphosphate is enhanced by the presence of soil microorganisms. Plant Soil. 2001, 229: 47-56. 10.1023/A:1004871704173.
Gyaneshwar P, Kumar GN, Parekh LJ: Effect of buffering on the phosphate-solubilizing ability of microorganisms. W J Microbiol Biotechnol. 1998, 14: 669-673. 10.1023/A:1008852718733.
Carrillo AE, Li CY, Bashan Y: Increased acidification in the rhizosphere of cactus seedlings induced by Azospirillum brasilense. Naturwissenschaften. 2002, 89: 428-432. 10.1007/s00114-002-0347-6.
Rodriguez H, Gonzalez T, Goire I, Bashan Y: Gluconic acid production and phosphate solubilization by the plant growth promoting bacterium Azospirillum spp. Naturwissenschaften. 2004, 91: 552-555. 10.1007/s00114-004-0566-0.
Nautiyal CS, Bhadauria S, Kumar P, Lal H, Mondal R, Verma D: Stress induced phosphate solubilization in bacteria isolated from alkaline soils. FEMS Microbiol Lett. 2000, 182: 291-296. 10.1111/j.1574-6968.2000.tb08910.x.
Gaur AC: Phosphate solubilizing microorganisms as bio-fertilizers. 1990, Omega Scientific Publication: New Delhi, 176-
Antoun H, Kloepper JW: Plant growth promoting rhizobacteria. Encyclopedia of Genetics. Edited by: Brenner S, Miller JF. 2001, Academic Press, 1477-1480.
Bashan Y, Holguin G: Proposal for the division of plant growth-promoting rhizobacteria into two classifications: biocontrol-PGPB (plant growth-promoting bacteria) and PGPB. Soil Biol Biochem. 1998, 30: 1225-1228. 10.1016/S0038-0717(97)00187-9.
Albert F, Anderson AJ: The effect of Pseudomonas putida colonization on root surface peroxidases. Plant Physiol. 1987, 85: 535-541. 10.1104/pp.85.2.537.
Dey R, Pal KK, Bhatt DM, Chauha SM: Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiological Research. 2004, 159: 371-394. 10.1016/j.micres.2004.08.004.
Dalla GC: Esperienze di lotta biologicacontrola fusariosi vascolare del garofano (Trials on the biological control of vascular kilt of carnations). Ann Inst Sper Floric. 1986, 17: 3012-
Lehinos V: Effects of pH and glucose on auxin production of phosphate-solubilizing rhizobacteria in vitro. Microbiol Res. 1994, 149: 135-138.
Lehinos V, Vacek O: Biosynthesis of auxin by phosphate-solubilizing rhizobacteria from wheat (Triticum aestivum) and rye (Secale cereale). Microbiol Res. 1994, 149: 31-35.
Pandey A, Palani LMS: Isolation of Pseudomonas corrugata from Sikkim himalayas. World J Microbiol Biotechnol. 1998, 14: 411-413. 10.1023/A:1008825514148.
Chen YP, Rekha PD, Arun AB, Shen FD, Lai WA, Young CC: Phosphate solubilizing bacteria from subtropical soil and their tri-calcium phosphate solubilizing abilities. App Soil Ecol. 2006, 34: 33-41. 10.1016/j.apsoil.2005.12.002.
Nautiyal CS: An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1990, 170: 265-270. 10.1111/j.1574-6968.1999.tb13383.x.
Belimov AA, Safronova VI, Sergeyeva TA, Egorova TN, Matveyeva VA, Tsyganov VE, Borisov AY, Tikhonovich IA, Kluge C, Preisfeld A, Dietz KJ, Stepanovik VV: Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can J Microbiol. 2001, 47: 642-652. 10.1139/cjm-47-7-642.
Voisard C, Keel C, Haas D, Defago G: Cyanide production in Pseudomonas fluorescens helps suppress balck root rot of tobacco under gnotobiotic conditions. EMBO J. 1981, 8 (2): 351-358.
Brodhajen M, Paulsen I, Loper JE: Reciprocal regulation of pyoluteorin production with membrane transporter gene expression in Pseudomonas fluorescens Pf-5. Appl Environ Microbiol. 2005, 71: 6900-6909. 10.1128/AEM.71.11.6900-6909.2005.
El-Banna N, Winkelmann G: Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomyces. J Appl Microbiol. 1998, 85: 69-78. 10.1046/j.1365-2672.1998.00473.x.
Haas D, Keel C: Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol. 2003, 41: 117-153. 10.1146/annurev.phyto.41.052002.095656.
Brown AE, Hamilton JTG: Indole-3-ethanol produced by Zygorrhynchusmoelleri, and indole-3-acetic acid analogue with antifungal activity. Mycol Res. 1993, 96: 71-74.
Strittmatter HK: Pathogen-defence gene prp1-1 from potato encodes an auxin-responsive glutathione-s-transferase. Eur J Biochem. 1994, 226: 619-626. 10.1111/j.1432-1033.1994.tb20088.x.
Martinez Noel GMA, Madrid EA, Botín R, Lamattina L: Indole acetic acid attenuates disease severity in potato-Phytophthora infestans interaction and inhibits the pathogen growth in vitro. Plant Physiol Biochem. 2001, 39: 815-823. 10.1016/S0981-9428(01)01298-0.
Chernin I, Ismailov Z, Haran S, Chet I: Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl Environ Microbiol. 1995, 61: 1720-1726.
Dunn C, Crowley JJ, Moenne-Loccoz Y, Dowling DN, de Bruijn FJ, O'Gara F: Biological control of Pythium ultinum by Stenotrophomonas maltophilia W18 is mediated by an extracellular proteolytic activity. Microbiology. 1997, 143: 3921-3931.
Krishnaraj PU, Goldstein AH: Cloning of a Serratia marcescens DNA fragment that induces quinoprotein glucose dehydrogenase-mediated gluconic acid production in Escherichia coli in the presence of stationary phase Serratia marcescens. FEMS Microbiol Lett. 2001, 205: 215-220. 10.1111/j.1574-6968.2001.tb10950.x.
Raaijmakers J, Weller DM, Thomashow LS: Frequency of antibiotic producing Pseudomonas spp. in natural environments. Appl Environ Microbiol. 1997, 63: 881-887.
Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS: Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001, 183: 6454-6465. 10.1128/JB.183.21.6454-6465.2001.
Mavrodi OV, Gardener BBM, Mavrodi DV, Bonsall RF, Weller DM, Thomashow LS: Genetic diversity of Phl D from 2,4-diacetylphloroglucinol-producing fluorescent Pseudomonas spp. Phytopathology. 2001, 91: 35-43. 10.1094/PHYTO.2001.91.1.35.
Acknowledgements
The financial support from Department of Biotechnology (DBT), New Delhi, India, is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
RN performed most experiments and crucial in writing the manuscript, GR performed estimation of phosphate solubilization; BN performed biochemical characterization, NS was vital in developing the key concepts and interpretation of results and approved draft of this manuscript.
Electronic supplementary material
12866_2008_663_MOESM1_ESM.doc
Additional file 1: Biochemical characterization and taxonomic identification of 80 phosphate solubilizing fluorescent pseudomonads. The data provided represent the taxonomic identification of strains on the basis of 16S rRNA nucleotide sequence based phylogenetic analysis, plant growth promoting traits (production of IAA, ACC deaminase and protease), plant growth affecting traits (production of pectinase and cellulase), biocontrol traits (production of chitinase, HCN and antagonism towards phytopathogenic fungi) and genotypic grouping on the basis of BOX-PCR fingerprint pattern. (DOC 251 KB)
12866_2008_663_MOESM2_ESM.doc
Additional file 2: Tricalcium phosphate solubilization by the strains of fluorescent pseudomonads. The data provided represent the estimation of soluble phosphate liberated from tricalcuim phosphate subtrate and reduction in pH of the medium due to microbial phosphate solubilization. (DOC 240 KB)
12866_2008_663_MOESM3_ESM.doc
Additional file 3: Antifungal activity of phosphate solubilizing fluorescent pseudomonads. The data provided represent the broad-spectrum antagonism towards important phytopathogenic fungi tested and possible antifungal potential of fluorescent pseudomonads. (DOC 81 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Naik, P.R., Raman, G., Narayanan, K.B. et al. Assessment of genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads isolated from rhizospheric soil. BMC Microbiol 8, 230 (2008). https://doi.org/10.1186/1471-2180-8-230
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-8-230