Abstract
Background
Insertion Sequence (IS) elements are mobile genetic elements widely distributed among bacteria. Their activities cause mutations, promoting genetic diversity and sometimes adaptation. Previous studies have examined their copy number and distribution in Escherichia coli K-12 and natural isolates. Here, we map most of the IS elements in E. coli B and compare their locations with the published genomes of K-12 and O157:H7.
Results
The genomic locations of IS elements reveal numerous differences between B, K-12, and O157:H7. IS elements occur in hok-sok loci (homologous to plasmid stabilization systems) in both B and K-12, whereas these same loci lack IS elements in O157:H7. IS elements in B and K-12 are often found in locations corresponding to O157:H7-specific sequences, which suggests IS involvement in chromosomal rearrangements including the incorporation of foreign DNA. Some sequences specific to B are identified, as reported previously for O157:H7. The extent of nucleotide sequence divergence between B and K-12 is <2% for most sequences adjacent to IS elements. By contrast, B and K-12 share only a few IS locations besides those in hok-sok loci. Several phenotypic features of B are explained by IS elements, including differential porin expression from K-12.
Conclusions
These data reveal a high level of IS activity since E. coli B, K-12, and O157:H7 diverged from a common ancestor, including IS association with deletions and incorporation of horizontally acquired genes as well as transpositions. These findings indicate the important role of IS elements in genome plasticity and divergence.
Similar content being viewed by others
Background
Insertion sequence (IS) elements are generally small (700 to 2,500 bp) DNA sequences that carry genetic information related to their transposition and its regulation. A recent database includes about 500 IS elements from almost 80 bacterial genera and about 160 species [1]. IS elements were first discovered because of their mutagenic activity. Insertion of an IS element can lead to gene inactivation and strong polar mutation [2]. Activation of cryptic genes [3], over-expression [4] or silencing of adjacent genes [5] may also be consequences of IS insertions. More global chromosome rearrangements can occur by recombination between two copies of homologous IS elements, leading to inversions [6] or deletions [7].
The copy number and distribution of IS elements vary among natural isolates of E. coli [8, 9]. Investigation of the distribution of seven IS elements in the ECOR collection of E. coli strains suggests host regulation of IS transposition that varies in strength depending on the element, as well as more harmful effects on fitness with increasing copy number [10, 11]. The mapping of IS elements in E. coli K-12 suggests a non-random chromosomal distribution, with regions of low and high density [12–14].
Although some IS-mediated mutations have been demonstrated to be beneficial [15–17], it remains controversial whether these elements should, on balance, be viewed as genomic parasites or as beneficial agents that promote adaptive evolution [18, 19]. IS elements are also widely seen as important for horizontal gene transfer, including especially the movement of acquired genes from extrachromosomal elements into the chromosome. As whole-genome sequences are made available, it becomes more feasible to compare the precise distribution of IS elements in different strains of the same species. On the basis of the known types of IS elements present in E. coli K-12 [14], we have mapped almost all of the IS insertion sites for an E. coli B derivative strain, and we compare this map with the ones derived from the genome sequences of E. coli K-12 [14] and O157:H7 EDL933 [20] and Sakai [21]. The sequences we obtained for regions adjacent to the IS elements in B augment existing data on the sequence divergence between E. coli B and K-12. These new data and comparative analyses are of particular interest for three reasons. First, E. coli B has been, after K-12, the second most widely studied E. coli strain. Second, the longest-running evolution experiment uses E. coli B, and the IS map developed here will facilitate the analysis of genomic changes that have occurred during the thousands of generations of this experiment [22–26]. Third, genome sequencing projects initially focused on a single genotype of the species of interest, but this constraint is disappearing. For new intra-specific genomic comparisons to be most efficient and useful, more data must be obtained on the co-linearity of genomes among individuals within the same species. IS elements and their associated mutagenic activities may substantially influence deviations from co-linearity.
Results and Discussion
Numbers and clustering of IS elements in B versus K-12
The E. coli B strain used as the ancestor for the long-term evolution experiment, and hence in this work, was originally described as Bc251 T6R StrR rm111 Ara-[27]. Twenty copies of IS1, 1 copy of IS2, 5 copies of IS3, 1 copy of IS4, 1 copy of IS30, 5 copies of IS150, and 5 copies of IS186 were detected in this strain, while no IS5 element was found [24]. Sequences adjacent to these IS elements were cloned, sequenced, and compared to the E. coli K-12 genomic sequence [14].
Copy number of IS elements provides a first criterion for comparing the genomes of E. coli B and K-12, and pronounced differences are observed. One difference is the absence of IS5 in E. coli B. It is unknown whether this difference reflects selection against this element in B, a lack of historical opportunity to invade B, or some other factor. The copy number of the other elements is also quite variable: in K-12 and B, respectively, IS1 has 6 and 20 copies, IS2 has 11 and 1 copies, IS3 has 5 copies in each strain, IS4 has 1 copy in each strain, IS30 has 4 and 1 copies, IS150 has 1 and 5 copies, and IS186 has 3 and 5 copies. High variability in copy number was also reported among natural isolates of E. coli [10].
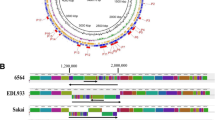
In B, there are apparent clusters of IS elements in the chromosomal regions between 12 and 15 min, 58 and 65 min, and 97 and 0.5 min (Figure 1). The 12–15 min region corresponds to a region of high IS density in K-12 [13]. On the other hand, no IS elements were found in B between 65 and 75 min, nor between 85 and 97 min. Chromosomal regions devoid of IS elements were reported in K-12 between 53 and 67 min, and between 83 and 92 min [12–14, 28]; the 83–92 min region corresponds to a low IS-density region in B, but IS elements are not underrepresented in the 53–67 min region of B.
Location of IS elements on the E. coli B chromosome. IS elements are represented by black dots. The position of the different IS elements is given in minutes. For clarity, in the two regions (between 8 and 15 min, and near 98 min) where several IS elements are closely located, their number is also given. The precise data are given in Table 1 (see Additional file: Table 1).
Genomic locations of IS elements in B versus K-12
Thirty-five of the 38 IS insertion sites were mapped for this B strain by sequencing their adjacent regions. (We were unable to obtain reliable sequences adjacent to the other three elements.) In terms of the precise locations of IS elements, there are only a few commonalities between B and K-12 (see Additional file: Table 1). In the nmpC gene (12.4 min), the rfb gene cluster (45.2 min), and the gat operon (46.9 min), IS elements are found in both strains, but the specific element or the exact site of insertion are different. The single IS4 element in each strain is in the same position, between two genes of unknown function (97 min). Also, IS3-1 (23.6 min) shares one endpoint (see below) with IS3-D in K-12, near a gene of unknown function.
Besides the single IS4 element, there are only three cases in which IS locations are identical for B and K-12. Interestingly, in all three of these other cases, the insertions are in loci (0.3, 13.1, and 80.2 min) homologous to the hok-sok plasmid stabilization systems [29]. Five hok-sok loci were reported in K-12, whereas six are present in B [29]. E. coli B has two additional insertions in one of these loci (IS1-5 and IS150-1 at 13.1 min). Moreover, IS150-3 and IS186-5 are both inserted in hokX-sokX (62.2 min), a locus that is absent from E. coli K-12. For the IS1 elements mapped in some natural isolates of E. coli [28], the only site in common with our study was the hokC-sokC locus (at 0.3 min), where IS186-1 is present in E. coli B.
From these data, it is clear that IS elements are found in most hok-sok loci in E. coli B and K-12. By contrast, no such elements are found in the corresponding loci of O157:H7 [20, 21], nor in some strains from the ECOR collection [29]. We suggest this discrepancy between IS-inactivated hok-sok loci in the laboratory strains B and K-12, and apparently active hok-sok loci in recent isolates from nature, may indicate that these genes have an important function in nature but not under typical laboratory conditions. The abundance of insertions in hok-sok loci in laboratory strains may even reflect inadvertent selection to disrupt their function (for example, if expression of these genes reduces growth rate).
Association of IS elements in B and K-12 with O157:H7 islands
IS insertion sites in E. coli B share no locations with the two E. coli O157:H7 strains that have been sequenced, with one partial exception. The IS3-5 element in B shows, on one side only, homology with an O-island of O157:H7 EDL933. This homologous region is aligned with a bacteriophage structural gene. For 13 other IS elements in B, O157:H7 EDL933-specific sequences (phage DNA or an O-island [20]) are found at the corresponding locations (see Additional file: Table 1). Three such B elements (IS1-5, IS150-1, and IS186-3) are in the same region (13.1 min) and correspond to the same O-island. IS4 and IS1-17 (97 and 97.5 min, respectively) also correspond to a single O-island. Thus, 10 independent regions in all are found to carry IS elements in E. coli B, but phage DNA or O-islands in O157:H7 EDL933. When the same comparison is performed with O157:H7 Sakai [21], only 12 of the 13 elements (9 independent regions) in B correspond to O-specific sequences, with the IS3-3 in B located at the site that corresponds to this difference between the two O157:H7 strains.
The same comparison between E. coli K-12 and E. coli O157:H7 EDL933 reveals that 31 of 43 IS locations in K-12 correspond to O157:H7-specific regions. Again, some K-12 elements lie in the same chromosomal regions, but there are still 17 separate regions that exhibit this cross-genomic association between IS elements in one strain and strain-specific sequences in the other. Some of these IS-bearing regions are the same in B and K-12, including around 6–8, 12, 23, 30–32, 45, 65, and 96–98 min. These chromosomal regions include genomic fragments that have been reported to carry horizontally acquired genes in K-12 [30]. The presence of IS elements in these regions suggests their possible involvement in moving foreign DNA into the chromosome. The same 31 IS locations were also different between K-12 and O157:H7 Sakai [21]. However, for two of these locations, no specific sequences are found in the Sakai strain, but instead several genes present in K-12 are deleted in the Sakai strain.
B-islands contain DNA sequences specific to E. coli B
Just as genomic regions specific to E. coli O157:H7 have been called O-islands, B-islands can be defined as regions present in B but absent from K-12, O157:H7, or both. Two IS elements (IS1-11 and IS3-4) are located in regions of the B chromosome that are absent from both O157:H7 and K-12. A third B element (IS3-5) is located in a region that is homologous with an O-island of O157:H7 EDL933 on one side of the IS element, but not on the other side. A fourth element (IS1-17) is inserted in a region homologous to K-12 on one side and to a complex region of a pathogenicity island in Shigella flexneri [31] on the other side. A fifth B element (IS186-5) is inserted in a gene absent from K-12 but present in O157:H7. We now examine these five cases in greater detail.
IS1-11
This element is in a gene designated wbbD from yet another E. coli strain, O7:K1. That gene encodes a putative galactosyltransferase and is located in the rfb cluster (45.2 min on the K-12 map), which is involved in synthesis of the O-specific chain of the outer membrane LPS. The sequences adjacent to IS1-11 have 100% identity to 615 bp covering parts of wbbD and manC (formerly called orf275 and rfbM, respectively [32]); the latter gene encodes GDP-mannose pyrophosphorylase. E. coli B does not express O-specific side-chain LPS, and IS1-11 may be responsible for this lack of function. Interestingly, E. coli K-12 harbors an IS5 element in the last rfb gene, wbbL, which encodes a rhamnosyltransferase, thus accounting for its own absence of O-antigen production [33]. O-antigens vary widely among gram-negative bacteria, with horizontal transfer and recombination contributing to allelic diversity for the rfb genes [34]. Given the presence of the wbbD gene in E. coli B, and the similarity of the corresponding rfb region in B and E. coli O7 [35], it is possible that B acquired the rfb gene cluster by horizontal transfer from O7, with its subsequent inactivation in B by IS1 transposition. It is also worth noting the role of IS elements in inactivating these gene functions in both laboratory strains.
IS3-4 and IS3-5
The sequences adjacent to the B element IS3-4 show no compelling similarity to anything in the genomic databases, except for a short alignment with a sequence from an unknown open reading frame of the Salmonella typhimurium pathogenicity island 2. Extending this adjacent sequence would help elucidate whether this might represent an IS-inactivated pathogenicity island. The sequences adjacent to the B element IS3-5 show, on one side only, very high similarity (91/92 nucleotides) with a bacteriophage structural gene present in an O-island of O157:H7 EDL933, while no homology is detected on the other side of the insertion site. Along with the inactivation of the O-antigen rfb gene cluster (described above), these insertions suggest that some ancestor of E. coli B may once have been a pathogen that later reverted to commensal status, and they also suggest a role for IS elements in that change in ecological niche. The GC content of the sequences around IS3-4 and IS3-5 is lower than is typical for E. coli genes, which might reflect acquisition through horizontal transfer.
The target-site duplications at the extremities of both IS3-4 and IS3-5 suggest there were no further rearrangements after these insertions. By contrast, all three other IS3 elements in E. coli B indicate rearrangements at their boundaries. IS3-1 (23.6 min), in particular, reveals a deletion of the region between ycdT and ycdV, two genes of unknown function. In K-12, IS3-D is at the same position near ycdT, whereas no element is near ycdV. Thus, the IS3-1 element in B may have resulted from an IS3 transposition near ycdV, followed by a recombination event between the new copy and the one near ycdT, which led to the deletion of the intervening sequence.
IS1-17
The sequence adjacent to one side of the IS1-17 element in B corresponds to sgcR (97.5 min). However, the other adjacent sequence is similar to a region of a Shigella flexneri pathogenicity island that contains the remnants of various IS elements [31]. No target-site duplication was found at the extremities of IS1-17, which suggests that a further rearrangement occurred at this locus. Our data also indicate that this chromosomal region contains several other IS elements in B, suggesting that this region may have been acquired by horizontal gene transfer. This region was previously reported to harbor horizontally transferred DNA in other E. coli genomes [30].
IS186-5
This B element is located in hokX-sokX, a locus that is absent from K-12 but present in O157:H7. As noted above, hok-sok loci are derived from plasmid stabilization systems, but most have been inactivated by IS elements in laboratory strains. Five such loci were reported in K-12 [29]. A sixth one, hokX-sokX, was reported in E. coli B [29], and that one we have found is also inactivated by this IS element.
Overall sequence similarity between E. coli B and K-12
Previous research has shown that E. coli B and K-12 are fairly closely related to one another, relative to the tremendous diversity that exists within this species as a whole [36, 37]. Multi-locus enzyme electrophoresis puts both of these strains in the A group [37], whereas that same approach places O157:H7 in group E [38].
The nucleotide sequences for the regions adjacent to the IS elements in our E. coli B strain permit two levels of comparison with E. coli K-12. The first is that of sequence similarity. An earlier summary of data from 33 genes found >95% similarity for all except one of them [36]; and M. Travisano (pers. comm.) has sequenced four additional regions around crr, cya, fruR, and ptsHI in E. coli B, all of which also show high similarity to K-12. For the 32 IS sites in the B chromosome that share one or both adjacent sequences with K-12 (only one in common with the earlier summary), 29 exhibit >98% similarity and all but two are >95% similar (see Additional file: Table 1). The hokC-sokC locus in B has only 85% similarity to K-12, although they share an IS186 inserted at the exact same site. The low similarity here may reflect rapid divergence owing to the absence of any selective constraint on this inactivated gene. The fucA gene from B is about 94% similar to K-12, with the start of the gene being only 86% similar whereas the end is >99% similar. This abrupt transition in sequence similarity suggests that one of these two strains, since they diverged from their common ancestor, underwent a recombination event in this region with some more distantly related strain [36]. Overall, there is striking contrast between the high level of gene-sequence similarity of E. coli B and K-12, on the one hand, and the few cases of overlap in the location of their IS elements, on the other hand (see Additional file: Table 1).
A second level of comparison is the gene order along the chromosome. While the genetic map for K-12 is well established [39], we have only local sequences in the vicinity of IS elements for E. coli B. Nonetheless, in 14 cases, an IS element in B was located between two genes. Except for one case (IS1-17, discussed above) in which we found a Shigella flexneri-related sequence on one side, the same pair of genes are immediately adjacent, and have the same relative orientation, in K-12 as in B.
Phenotypic properties of the B strain associated with its IS elements
Several IS insertions found in this study appear to be responsible for three phenotypes that either distinguish E. coli B from K-12 or are properties of this particular B strain. First, the IS1-13 element in B is associated with a deletion of the upstream promoter and first 114 bp of ompC (49.8 min). This deletion also eliminates micF, which negatively controls expression of ompF [40, 41]. The corresponding outer-membrane proteins, OmpC and OmpF, are the two major porins of K-12, with their relative expression regulated by osmolarity [42, 43]. By contrast, E. coli B expresses only OmpF [44–46], which evidently is a consequence of the deletion associated with IS1-13.
Second, the particular B strain with which we work was isolated by Seymour Lederberg [27] as a restriction-modification deficient mutant following treatment with the mutagen 1-methyl-3-nitro-1-nitrosoguanidine. This mutation was called rm111 and genetic mapping indicated that it was a few minutes counterclockwise to the thr-ara-leu region (0.1, 1.5, and 1.7 min) on the K-12 map. We found the IS1-19 element at 98.6 min, with an accompanying deletion that removes 93 bp of the 5' end of the mcrD gene. That gene is involved in the 5-methylcytosine restriction system, and this position fits with Lederberg's mapping of the rm111 mutation, which together suggest that this element is responsible for the restriction-modification deficiency.
Third, as described in detail elsewhere [17], this strain of E. coli B is able to grow on ribose as a sole carbon source, but the Rbs+ phenotype is genetically unstable. This genetic instability is a consequence of the IS150-5 element located immediately upstream of rbsD (84.7 min), which evidently promotes deletions of this adjacent region [17].
Conclusions
In this study, we mapped most of the IS elements present in the E. coli B genome. The E. coli B strain with which we have worked has 38 known IS elements [24], including 20 copies of IS1; 5 copies each of IS3, IS150, and IS186; and 1 copy each of IS2, IS4, and IS30. No IS5 element is present. We compared the resulting data with two other E. coli strains, K-12 and O157:H7, whose genomes are completely sequenced, with the following main results.
1) The genomic locations of IS elements show few commonalities between E. coli B and K-12. The most striking common feature is the presence of IS elements, often identical, in multiple hok-sok loci in both B and K-12. By contrast with this situation in these two laboratory strains, IS elements are absent from these loci in strains more recently isolated from nature, including O157:H7.
2) Many of the insertion sites of IS elements in E. coli B and K-12 correspond to phage DNA or O-islands in O157:H7. This association indicates that these regions of the chromosome underwent chromosomal rearrangements, in particular either the insertion or deletion of the DNA regions that distinguish the different strains; in the case of insertions, the large-scale differences also imply horizontal transfer. IS elements are well known, of course, to play important roles in gene deletion events and in the incorporation of foreign DNA into the chromosome during and following horizontal gene transfer.
3) A few sequences adjacent to IS elements in E. coli B reveal chromosomal regions that are absent from K-12, O157:H7, or both. One such B-island contains a gene, wbbD, that is identical to a gene involved in O-antigen synthesis from O7:K1, but the IS element precludes this function in B.
4) Despite the pronounced differences in IS number and locations between E. coli B and K-12, most adjacent DNA sequences show >98% similarity. The available evidence also indicates that local gene order along the two chromosomes is largely the same, except for islands of strain-specific DNA.
5) Some phenotypic features of E. coli B are explained by inactivation of the relevant genes by IS elements, most notably the absence of expression of the OmpC porin and the correspondingly elevated expression of the OmpF porin.
6) These observations, taken together, indicate a high level of IS activity since E. coli strains B, K-12, and O157:H7 diverged from their common ancestor, including transpositions, IS-associated deletions, and horizontal transfer events.
Materials and methods
Strains and culture conditions
The strain used in this study is an E. coli B derivative, originally designated Bc251 T6R StrR rm111 Ara-[27]. For cloning experiments, we used E. coli strain TOP10 (Invitrogen). Cultures grew in Luria-Bertani medium [47] supplemented with 12 g/l agar for plating cells. When appropriate, the medium was supplemented with kanamycin (50 μg/ml) or Xgal (40 μg/ml).
DNA handling
Genomic DNA was prepared from 1.5-ml cultures as described elsewhere [48]. Plasmid DNA was prepared from 1.5-ml cultures using the Qiaprep Spin Miniprep kit (Qiagen), according to the manufacturer's recommendations. All restriction enzymes were purchased from Life Technologies.
Characterization of sequences adjacent to IS elements by inverse PCR
Sequences adjacent to the various IS elements were cloned as described previously [25]. Briefly, genomic DNA of the E. coli B strain was digested with Eco RV for all IS elements, except IS150; we used Hin cII for IS150, because Eco RV cuts within that element. Fragments were separated on a 0.8% agarose gel, with Pst I- and Hin dIII-digested Lambda DNA as size markers. Gel fractions carrying the different IS-containing fragments were cut and the DNA was purified. These fragments were self-ligated with T4 DNA ligase (Roche) at 5 to 10 μg/ml, and the ligated mixtures were used as templates in PCR experiments. The primer pairs used for inverse-PCR to amplify sequences adjacent to the different IS elements are listed in Table 2. All primer pairs are near the extremities of the corresponding IS and are directed outward. PCR reactions were performed using Expand Taq DNA polymerase (Roche), according to the manufacturer's recommendations. The PCR products containing sequences adjacent to IS elements were cloned using the Topo TA Cloning kit (Invitrogen). The fragments containing adjacent sequences were used as probes in subsequent hybridization experiments to confirm that the right fragments were cloned. These cloned adjacent sequences were then sequenced using the same primers as for the PCR experiments. Sequences were compared with databases using the BLAST program [49]. All map positions are reported based on the genome sequence of E. coli K-12 [14].
Hybridization experiments
DNA fragments used as probes were cold-labeled and hybridizations were performed with the DIG-labeling and detection kit sold by Roche. All hybridizations and washes were done at 68°C under high stringency conditions. Sequences adjacent to IS elements were used to probe reference membranes that had been previously probed with the corresponding IS elements themselves. These membranes carry Eco RV- and Hin cII-digested genomic DNA from the E. coli B strain. The membranes were probed with the IS elements, stripped and re-probed with adjacent sequences to demonstrate that the correct sequences were cloned.
Abbreviations
- IS:
-
insertion sequence
- LPS:
-
lipopolysaccharide.
References
Mahillon J, Chandler M: Insertion sequences. Microbiol Mol Biol Rev. 1998, 62: 725-774.
Jordan E, Saedler H, Starlinger P: O0 and strong polar mutations in the gal operon are insertions. Mol Gen Genet. 1968, 102: 353-363.
Reynolds AE, Felton J, Wright A: Insertion of DNA activates the cryptic bgl operon in Escherichia coli K12. Nature. 1981, 293: 625-629.
Ciampi MS, Schmid MB, Roth JR: Transposon Tn10 provides a promoter for transcription of djacent sequences. Proc Natl Acad Sci USA. 1982, 79: 5016-5020.
Blasband AJ, Marcotte JR, Schnaitman CA: Structure of the lc and nmpC outer membrane porin protein genes of lambdoid bacteriophage. J Biol Chem. 1986, 261: 12723-12732.
Xia X-M, Enomoto M: A naturally occurring large chromosomal inversion in Escherichia coli K12. Mol Gen Genet. 1986, 205: 376-379.
Reif HJ, Saedler H: IS1 is involved in deletion formation in the gal region of Escherichia coli K12. Mol Gen Genet. 1975, 137: 17-28.
Green L, Miller RD, Dykhuizen DE, Hartl DL: Distribution of DNA insertion element IS5 in natural isolates of Escherichia coli. Proc Natl Acad Sci USA. 1984, 81: 4500-4504.
Sawyer SA, Hartl DL: Distribution of transposable elements in prokaryotes. Theor Pop Biol. 1986, 30: 1-16.
Sawyer SA, Dykhuizen DE, DuBose RF, Green L, Mutangadura-Mhlanga T, Wolczyk DF, Hartl DL: Distribution and abundance of insertion sequences among natural isolates of Escherichia coli. Genetics. 1987, 115: 51-63.
Hall BG, Parker LL, Betts PW, DuBose RF, Sawyer SA, Hartl DL: IS103, a new insertion element in Escherichia coli: characterization and distribution in natural populations. Genetics. 1989, 121: 423-431.
Birkenbihl RP, Vielmetter W: Complete maps of IS1, IS2, IS3, IS4, IS5, IS30, and IS150 locations in Escherichia coli K12. Mol Gen Genet. 1989, 220: 147-153.
Deonier RC: Native insertion sequence elements: locations, distributions, and sequence relationships. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Edited by: Neidhardt FC. 1996, Washington, DC, ASM Press, 2000-2012.
Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF: The complete genome sequence of Escherichia coli K-12. Science. 1997, 277: 1453-1462. 10.1126/science.277.5331.1453.
Chao L, McBroom SM: Evolution of transposable elements-an IS10 insertion increases fitness in Escherichia coli. Mol Biol Evol. 1985, 2: 359-369.
Treves DS, Manning S, Adams J: Repeated evolution of an acetate-crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998, 15: 789-797.
Cooper VS, Schneider D, Blot M, Lenski RE: Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. J Bacteriol. 2000, 183: 2834-2841. 10.1128/JB.183.9.2834-2841.2001.
Blot M: Transposable elements and adaptation of host bacteria. Genetica. 1994, 93: 5-12.
Charlesworth B, Sniegowski P, Stephan W: The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994, 371: 215-220. 10.1038/371215a0.
Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA: Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001, 409: 529-533. 10.1002/(SICI)1096-9861(19990712)409:4<529::AID-CNE2>3.0.CO;2-N.
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T: Genome sequence and comparative analysis of enterohemorrhagic E. coli O157:H7. DNA Res. 2001, 8: 11-22.
Lenski RE, Rose MR, Simpson SC, Tadler SC: Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991, 138: 1315-1341. 10.1086/285289.
Lenski RE, Travisano M: Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994, 91: 6808-6814.
Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M: Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci USA. 1999, 96: 3807-3812. 10.1073/pnas.96.7.3807.
Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M: Long-term experimental evolution in Escherichia coli. IX. Characterization of IS-mediated mutations and rearrangements. Genetics. 2000, 156: 477-488.
Cooper VS, Lenski RE: The population genetics of ecological specialization in evolving E. coli populations. Nature. 2000, 407: 736-739. 10.1038/35037572.
Lederberg S: Genetics of host-controlled restriction and modification of deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1966, 91: 1029-1036.
Boyd EF, Hartl DL: Nonrandom location of IS1 elements in the genomes of natural isolates of Escherichia coli. Mol Biol Evol. 1997, 14: 725-732.
Pedersen K, Gerdes K: Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999, 32: 1090-1102. 10.1046/j.1365-2958.1999.01431.x.
Lawrence JG, Ochman H: Molecular archaeology of the E. coli genome. Proc Natl Acad Sci USA. 1998, 95: 9413-9417. 10.1073/pnas.95.16.9413.
Luck SN, Turner SA, Rajakumar K, Sakellaris H, Adler B: Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect Immun. 2001, 69: 6012-6021. 10.1128/IAI.69.10.6012-6021.2001.
Marolda CL, Valvano MA: Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995, 177: 5539-5546.
Raetz CRH: Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Edited by: Neidhardt FC. 1996, Washington, DC, 1035-1063.
Selander RK, Li J, Nelson K: Evolutionary genetics of Salmonella enterica. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Edited by: Neidhardt FC. 1996, Washington, DC, ASM Press, 2691-2707.
Marolda CL, Valvano MA: Identification, expression, and DNA sequence of the GDP-mannose biosynthesis genes encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1). J Bacteriol. 1993, 175: 148-158.
Milkman R: Recombinational exchange among clonal populations. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Edited by: Neidhardt FC. 1996, Washington, DC, ASM Press, 2663-2684.
Herzer PJ, Inouye S, Inouye M, Whittam TS: Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990, 172: 6175-6181.
Tarr PI, Schoening LM, Yea Y-L, Ward TR, Jelacic S, Whittam TS: Acquisition of the rfb-gnd cluster in evolution of Escherichia coli O55 and O157. J Bacteriol. 2000, 182: 6183-6191. 10.1128/JB.182.21.6183-6191.2000.
Rudd KE: Linkage map of Escherichia coli K-12, edition 10: the physical map. Microbiol Mol Biol Rev. 1998, 62: 985-1019.
Chou JH, Greenberg JT, Demple B: Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J Bacteriol. 1993, 175: 1026-1031.
Ramani N, Hedeshian H, Freundlich M: micF antisense RNA has a major role in osmoregulation of OmpF in Escherichia coli. J Bacteriol. 1994, 176: 5005-5010.
Kawaji H, Mizuno T, Mizushima S: Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J. Bacteriol. 1979, 140: 843-847.
Nikaido H: Outer membrane. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Edited by: Neidhardt FC. 1996, Washington, DC, ASM Press, 29-47.
Pugsley AP, Rosenbusch JP: OmpF porin synthesis in Escherichia coli strains B and K-12 carrying heterologous ompB and/or ompF loci. FEMS Microbiol Lett. 1983, 16: 143-147. 10.1016/0378-1097(83)90001-0.
Travisano M, Lenski RE: Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996, 143: 15-26.
Prilipov A, Phale PS, Koebnik R, Widmer C, Rosenbusch JP: Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli B-E. J Bacteriol. 1998, 180: 3388-3392.
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning: a laboratory manual, edition 2. Cold Spring Harbor, Cold Spring Harbor Press. 1989
Nichols BP, Shafiq O, Meiners V: Sequence analysis of Tn10 insertion sites in a collection of E. coli strains used for genetic mapping and strain construction. J Bacteriol. 1998, 180: 6408-6411.
Altschul S, Stephen F, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25: 3389-3402. 10.1093/nar/25.17.3389.
Acknowledgements
We thank Michael Travisano for sharing unpublished sequence data. This research was supported by grants from the French CNRS and CEA to M.B., and from the U.S. National Science Foundation to R.E.L.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author's contributions
DS was primarily responsible for designing and performing the molecular experiments, preparing the manuscript, and generally coordinating this project. ED and JD performed some experiments, and EC provided technical assistance. This project grew out of a collaboration between REL and MB to investigate the role of IS elements in the evolution of E. coli B, and they provided both scientific input and help with the manuscript.
All authors read and approved the final manuscript.
Electronic supplementary material
12866_2002_52_MOESM1_ESM.doc
Additional file 1: Chromosomal locations of IS elements in E. coli B strain. Data are provided on the positional correspondence between IS elements in B and those in K-12, and on the similarity of adjacent sequences in B with those from K-12. Comparisons with the O157:H7 EDL933 and Sakai map positions are also included. (DOC 32 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Schneider, D., Duperchy, E., Depeyrot, J. et al. Genomic comparisons among Escherichia coli strains B, K-12, and O157:H7 using IS elements as molecular markers. BMC Microbiol 2, 18 (2002). https://doi.org/10.1186/1471-2180-2-18
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-2-18