Abstract
Background
Composting is microbial decomposition of biodegradable materials and it is governed by physicochemical, physiological and microbiological factors. The importance of microbial communities (bacteria, actinomycetes and fungi) during composting is well established. However, the microbial diversity during composting may vary with the variety of composting materials and nutrient supplements. Therefore, it is necessary to study the diversity of microorganisms during composting of different agricultural byproducts like wheat bran, rice bran, rice husk, along with grass clippings and bulking agents. Here it has been attempted to assess the diversity of culturable bacteria during composting of agricultural byproducts.
Results
The culturable bacterial diversity was assessed during the process by isolating the most prominent bacteria. Bacterial population was found to be maximum during the mesophilic phase, but decreased during the thermophilic phase and declined further in the cooling and maturation phase of composting. The bacterial population ranged from 105 to 109 cfu g-1 compost. The predominant bacteria were characterized biochemically, followed by 16S rRNA gene sequencing. The isolated strains, both Gram-positive and Gram-negative groups belonged to the order Burkholderiales, Enterobacteriales, Actinobacteriales and Bacillales, which includes genera e.g. Staphylococcus, Serratia, Klebsiella, Enterobacter, Terribacillus, Lysinibacillus Kocuria, Microbacterium, Acidovorax and Comamonas. Genera like Kocuria, Microbacterium, Acidovorax, Comamonas and some new species of Bacillus were also identified for the first time from the compost made from agricultural byproducts.
Conclusion
The use of appropriate nitrogen amendments and bulking agents in composting resulted in good quality compost. The culture based strategy enabled us to isolate some novel bacterial isolates like Kocuria, Microbacterium, Acidovorax and Comamonas first time from agro-byproducts compost. These bacteria can be used as potential compost inoculants for accelerating composting process.
Similar content being viewed by others
Background
Lignocellulosic agricultural byproducts are well known for their use as soil conditioners in the form of compost. According to conservative estimates, around 600–700 million tones (mt) of agricultural waste including 272 mt of crop residues [1]; 40–50 mt of municipal solid waste (MSW) and 500–550 mt of animal dung [2] are available in India every year for bioconversion to compost.
Composting is an intense microbial process leading to decomposition of the most biodegradable materials for further humification [3, 4]. Successful composting depends on a number of factors that have both direct and indirect influence on the activities of the microorganisms. Tiquia et al. [5] included the type of raw material being composted, its nutrient composition and physical characteristics such as bulk density, pH, and moisture content etc. as the important factors. Moreover, Fracchia et al. [6] also observed that various other factors influenced the microbial colonization of finished products, i.e., (i) origin and composition of the initial substrates, (ii) previous process conditions and (iii) substrate quality of the finished product.
For the composting processes, the importance of microbial communities is well established [7]. Studies on bacterial population, actinobacteria and fungi during composting have been reported extensively [8]. Liu et al.[9] reported that there were several molecular approaches, which provide powerful adjuncts to the culture-dependent techniques. A known powerful tool, namely PCR has been used for bacterial identification and its classification at species level [10]. PCR targeting the 16S rRNA gene sequencing is used extensively to study the prokaryote diversity and allows identification of prokaryotes as well as the prediction of phylogenetic relationships [11]. The analyses of rRNA genes encoding for the small subunit ribosomal RNA (for bacteria, 16S rRNA) [12–14] have recently dramatically increased our knowledge about the contribution of different bacteria to various compost production phases.
Molecular approach to characterize and classify microbial communities by cultivation methods has switched to the genetic level, and the analysis of community structure has become possible only with further need to address the cultivation approach for a systematic analysis. Cultivation based techniques have allowed merely a glimpse of microbial diversity because only an estimated 1% of the naturally occurring bacteria has been isolated and characterized so far [15]. Even though, recent advances in culture independent molecular approaches based on rRNA or genomic approaches have improved the knowledge of microbial ecosystems, the isolation of bacterial species in pure culture remains to be the only way to fully characterize them, both for their physiological and catabolic properties. Moreover, the unculturable bacteria identified using recent molecular techniques cannot be used as compost inoculant for improving composting process. Therefore, culture-dependent methods are still a powerful tool. These viable fractions (grown to a detectable level on agar based medium) form only a small part of the total microorganisms, but they can still be used for comparison of data representing different times of the year or different areas [16]. So, it is imperative to study in-depth the culturable bacterial diversity so as to identify some new bacteria which can be applied for better and quick compost preparation. Besides composting, bacteria isolated from compost have been used by many researchers for others applications as well [17, 18].
In the traditional methods of composting some pathogenic bacteria survived, this was probably because of an inadequate aeration and lack of building-up of relatively high temperature. Moreover, the prevailing conditions might have prevented some of the indigenous microorganisms to colonize and degrade plant wastes. As a result, the final composts obtained from such an unimproved method are generally poor in quality. It has therefore become highly exigent to develop an alternative technique for producing good quality compost using locally available lignocellulosic biomass and bulking agents. This paper describes an attempt to identify specific microorganisms involved in the degradation of plant materials with the aim of studying the succession of bacterial population during composting in order to exploit the isolated bacteria in future for diverse uses such as compost inoculants, enzyme production, biocontrol agents.
Results
Physicochemical characteristics of compost
The pile and environmental temperatures were monitored during the entire period of composting (Figure 1). Initial temperature of the heap after mixing was 30°C. Within a week, the pile temperature reached to 37°C. However, the temperature increased to 40°C after 15 days and remained the same for four days, thereafter, which it rose to 50°C on 20th day and remained static for next few days. However, as composting proceeded, the temperature of the pile dropped to 45°C by the 30th day and fell further, but stabilized at 27°C (near to ambient) by the sixth week. After that, the pile was left uncovered for cooling for the next ten days.
During the present study, the substrates mixtures showed an initial electrical conductivity (EC) of 3.8 dS m-1. It reached upto 4.9 dS m-1 with progressive degradation upto 40 days. The pH of the compost heap remained 7.5 during first 30 days of the process, and thereafter it declined to 7.0 and continued till 50th day.
Chemical characteristics
The changes in organic carbon (C), total nitrogen (N), the C: N ratio, phosphorus and potassium varied considerably during composting (Table 1). The organic C decreased, whereas total nitrogen, phosphorus and potassium increased with time. Finally C: N ratio was observed to be stabilized at 11:1 at the end of composting during 40–50 days.
Total micronutrients
There was a significant increase in nutrients e.g. Na, Cu, Zn, Mg, S, Mn, Fe and Ca during composting. The respective average values of various metal contents varied considerably from the beginning to end of composting (Table 1).
Changes in viable bacterial population during composting
The number of mesophilic bacteria increased rapidly in first ten days, the count of mesophilic bacterial count was 1.7- 2.84 × 109cfu g-1. However, the thermophilic bacteria were dominant from 11–32 days of composting, with count in between 108 to107cfu g-1. Finally, mesophilic population stabilized between 106 to 105 cfu g-1 during the cooling and maturation phase (33–40 days).
Morphological, biochemical and molecular characterization of isolates
The most predominant bacterial isolates were picked up and morphologically different colonies were selected for further studies (Table 2). A total of thirty-three bacteria were subsequently purified and subjected to morphological, biochemical and molecular characterization. Interestingly, 84.8% isolates were Gram-positive, out of which 85.7% were rods and 14.3% cocci, whereas, the remaining 15.2% of the isolates were Gram-negative and all them were rods (Figure 2). The bacterial cultures were tentatively identified on the basis of Bergey’s Manual of Systematic Bacteriology (Table 3).
Identification of culturable bacteria isolated from compost
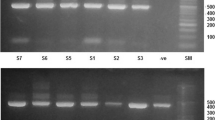
Marked changes in the profiling patterns of bacteria between the initial, mid and final stages of the composting process were observed. The changes in the structure of bacterial community were analyzed on the basis of 16S rRNA gene sequence chronometer from day one to end of composting. The amplified PCR products of bacterial 16S rRNA genes were sequenced partially.
All sequences were compared with 16S rRNA gene sequences present in the Genebank using BLAST and their percentage similarity was also compared and recorded in Table 4. The majority of the bacterial isolates (78.8%) were affiliated with Firmicutes (especially the genera Bacillus sp., Terribacillus sp. and Lysinibacillus sp. etc.), whereas only 9.1, 6.1 and 6.1% of bacterial isolates were affiliated to the members of γ-proteobacteria, β-proteobacteria and actinobacteria, respectively (Figure 3). Apart from spore forming Bacilli other genera in the compost were Staphylococcus, Serratia, Klebsiella, Enterobacter, Microbactrium, Kocuria, Acidovorax and Comamonas.
Interestingly, genera like Kocuria, Microbacterium, Acidovorax and Teribacillus have been reported for the first time from the compost population from agricultural by-products. The heat generated during composting destroyed all pathogenic bacteria in the final mature compost and was found to be free from Staphylococcus, Klebsiella, Enterobacter and Serratia. The phylogenetic affiliation of compost isolates with their accession numbers and their nearest neighbors of the GenBank database are shown in (Figure 4 and Table 4).
Neighbour-joining unrooted tree depicting the phylogenetic relationship of the dominant bacteria among the related species of the genus. Staphylococcus, Bacillus, Terribacillus, Lysinibacillus, Serratia, Klebsiella, Enterobacter, Microbacterium, Kocuria, Acidovorax and Comamonas using MEGA 5 software.
Discussion
Composting is a dynamic process affected by a large number of environmental and biological factors. Change in any of these factors greatly affects the quality of compost as well as the time required for composting. The present investigation demonstrated changes in temperature, physiochemical characteristics and bacterial population during composting process. This study also deals with the characterization of predominant bacterial genera isolated from different phases of composting.
Biddlestone and Gray [19] reported that the complexity of degraded plant materials and quality of the final product may depend upon the type of biomass. Therefore, various agricultural byproducts were used as raw material in order to provide an excellent substratum for the growth of microorganisms. All these supplements had high mineral and N content, which balance the relatively high C: N ratio of rice husk. Rice husk may supply K, Ca, Mg and other minerals along with C and silica [20]. In composting, C: N ratio was considered to be the most important parameter, as it reflects the extent of the bio-transformations that took place in the compost in chemical terms [21]. In the beginning of composting the C: N ratio of agricultural byproducts was 31.1 and it was decreased to 11.4 at the end of composting (Table 1). This decline might be because of reduction of C, which is obviously due to evolution of CO2 during degradation of organic matter and increase in N due to mineralization of organic-N compound. Brito et al. [22] also observed a decline in C: N ratio from 36 to 14 at the end of composting. The C: N ratio less than 12 during the solid phase was believed to be an indicator for the maturity of the compost [23, 24].
The temperature regime in the compost indicated that the organic materials passed through different phases like mesophilic, thermophilic, cooling and maturation (Figure 1) as already reported by Ishii et al. [25]. The temperature started dropping in the compost pile once the material was stabilized, which also indicated that the pile was becoming anaerobic and should be aerated by turning [26]. Therefore, turning was performed first on 15th day of composting, and then on every tenth day. The results indicated that processes like thorough mixing of the materials and turning enhanced the decomposition process. Moreover, if turning process failed to reheat the composting pile, it showed that the composting material was biologically stable [27].
Nutrient status of mature compost
The results showed a significant increase in minerals (w w-1) in agricultural byproducts composting (Table 1) and no gradual fluctuations were observed after 40th day. Janakiram and Sridevi [28] attempted the composting of Kattamanakku (Jatropha curcas) waste with slurries of cow dung by an aerobic composting method; the percentages of N, P, K, Na, Ca and Mg increased after 30 and 60 days of composting. The findings correlated with the present study. Similarly Felton et al. [29] reported that total P increased during the compost process. The metal concentrations like Cu, Mn and Zn increased rapidly during first 49 days of composting from swine (Susdo mesticus) feces [30]. The stability and solubility of various compounds in compost is influenced by the pH of the compost [31, 32].
Microbial population
Kell et al. [33] studied that at the simplest level, bacteria may be classified into two physiological groups: those that can, and those that cannot readily be grown to detectable levels in vitro. The viable count usually refers to the number of individual organisms in compost that can be grown to a detectable level, in vitro by forming colonies on an agar-based medium. However, the number of viable cells approximates to the number of colony forming units [34]. Changes in bacterial population were analyzed by cultivation-based method (cfu g-1) to reveal changes in the number of mesophilic and thermophilic bacteria during the composting process.
Hargerty et al. [35] reported that there was maximum increase in microbial population in the early stages of composting which was dependent on initial substrate used and environmental conditions of the composting. High content of degradable organic compound in the initial mixture might have stimulated microbial growth involved in self-heating during initial stage of composting [36]. An equivalent tendency does not occur with regard to mesophilic and thermophilic bacteria in the present study when the population density decreased from 109 to 107 cfu g-1. However from thermophilic to cooling and maturation phase, the gradual decrease in 107 to 105 cfu g-1 could be due to the unavailability of nutrients during maturation phase. During peak heating the bacterial populations declined by approximately 10-fold at 40°C and 100-fold at 50°C, followed by population growth at cooling phase, which decreased by 1000 fold as compared to the mesophilic (starting) phase of composting [7]. The Gram-positive bacteria dominated the composting process as they accounted for 84.8% of total population and the remaining 15.2% were Gram-negative as illustrated in Figure 2.
For bacteria, 16S rRNA gene sequence analysis is a widely accepted tool for molecular identification [37, 38]. Franke-Whittle et al. [39] also investigated the microbial communities in compost by using a microarray consisting of oligonucleotide probes targeting variable regions of the 16S rRNA gene. During the present investigation, thirty three bacterial isolates were cultured, out of which twenty six isolates (78.8%) belonged to class firmicutes; two isolates (6.1 %) belonged to actinobacteria; three isolates (9.0 %) belonged to class γ-proteobacteria and the remaining two isolates (6.1%) showed sequence similarity to class β-proteobacteria (Figure 3). Table 4 and Figure 4 summarizes all the bacterial taxa reported in agricultural byproduct compost based on sequence similarity, which were categorized in four main classes: Firmicutes, β-proteobacteria, γ-proteobacteria and actinobacteria in concurrence with the findings of Ntougias et al. [40] and Chandna et al. [41].
The present study determined the microbial succession of the dominating taxa and functional groups of microorganisms, as well as the total bacterial activity during composting of agricultural byproducts, using incubation, isolation, and enumeration techniques. The bacterial population showed differences between mesophilic, thermophilic and maturing stages of compost. Ryckeboer et al. [7] analyzed the bacterial diversity and found that both Gram-positive and Gram-negative bacteria increased during the cooling and maturation phases of biowaste composting in compost bin. In the present study, the level of firmicutes increased markably during mesophilic phase, and then decreased during the next phase upto cooling and maturation. The number of actinobacteria count remained stable during mesophilic and thermophilic phase of composting. Population of β-proteobacteria remained insignificant in thermophilic phase whereas, the level of γ-proteobacteria increased slightly during mesophilic phase and then decreased markably during thermophilic phase. Similarly, Fracchia et al. [6] observed the prevalence of Gram-positive organisms belonging to the firmicutes and actinobacteria.
In the present study a few Serratia, Enterobacter, Klebsiella and Staphylococcus sp. were also isolated during early phase of composting. Silva et al. [42] also found Serratia sp. in bagasse and coast-cross straw during the first stage of composting. Enterobacter sp. was predominantly present at an early stage of composting process and died off at increased temperature [43] in accordance with the present study. Moreover, Enterobacter sp. is common in soil, water and even in compost too and mainly survives as saprophytes [44]. Strauch [45] found that the Klebsiella sp. was present at the beginning of thermophilic phase till the temperature was below 60°C. Similarly, Ahlawat and Vijay [46] also isolated Staphylococcus sp. from mushroom research farm compost at a wider temperature range (43–55°C). Importantly no pathogen could be detected during the curing phase of compost produced from agricultural byproducts. Thus our composting process also resulted in the eradication of pathogens, as has been reported by Danon et al. [47].
Heating is essential to enable the development of a thermophilic population of microorganisms, which is capable of degrading the more recalcitrant compounds, to kill pathogens and weed seeds [48]. Bacillus sp. was able to survive in the compost pile due to their property to form endospores during thermophillic stage. Various researchers investigated that Bacillus sp. was a predominant genera present throughout the composting process [25, 49], and the most dominant bacterial taxon recovered from compost feedstock [50]. Additonally, Kocuria sp. was one of the isolates, cultured from present studied compost. Similarly, Vaz-Moreira et al. [51] also isolated Kocuria palustris from vermicompost from food wastes.
BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of 16S rRNA gene sequence revealed similarity to sequences of the species Comamonas kerstersii, a β-Proteobacterium of the Comamonadaceae family, as published in GenBank. Young et al. [52] isolated Comamonas sp. from food waste compost. It had the ability to metabolize complex organic compounds as energy sources for growth [53]. Moreover, Comamonadaceae, a new family encompassing the Acidovorans[54], was also recovered from agricultural byproduct compost. Pinel et al. [55] isolated β-proteobacterial Acidovorax sp. symbionts from the nephridia of four different species of earthworms. Pizl and Novokova [56] also showed the establishment of different kinds of relationship between earthworms and microbes. The nephridial symbionts form their own monophyletic group closely related to the genus Acidovorax[57]. The bacteria reduced the biodegradable organic content and help in mineralization of solid waste [58].
Conclusion
The production of high quality compost can be enhanced by biological, physiochemical properties of raw material and compost inoculants. Present study indicated the usefulness of different nitrogen amendments and bulking agents for improved composting process to prepare high quality compost. These culture-based approaches taken in this study enabled us to isolate, for the first time, Kocuria, Microbacterium, Acidovorax and Comamonas from agricultural byproducts compost. However, in order to understand better the nature of bacterial communities associated with compost, the use of sequencing of 16S rRNA genes was used to describe the complete bacterial community composition. The new genera Kocuria, Microbacterium, Acidovorax and Comamonas identified from the compost can be used as compost inoculants for accelerating the composting process. Besides being prospected for degradation, they can be evaluated for their ability to produce hydrolytic enzymes and antimicrobial compounds etc.
Methods
Site selection, raw material for composting
The experiment was carried out at University of Delhi South Campus, New Delhi, India during the month of December 2006 and January 2007. The composting pile (1.50 × 0.90 × 0.80 m3) was prepared on a clean ground surface, covered with black polyethylene. The raw materials used for composting were rice bran (15 kg), wheat bran (10 kg), rice husk (10 kg) and other additives like grass and leaves (5 kg) each; ash (2.5 kg) was used as a bulking agent. Nitrogen (N) was enriched by amending with cow dung (25 kg), mustard oil cake (10 kg), cow urine (40 l) and molasses (4 l). To eliminate the pH variation, approximately 0.6% (w w-1) of calcium oxide was added to the compost raw materials during mixing. Table 5 depicts raw materials and their properties. The pile was turned manually on the 15th day of composting and then after every 10th day.
During the composting process, the temperature in the pile (5 to 30 cm from the top) was measured daily using a dry bulb thermometer. Similarly, the environment temperature was also recorded during composting near the pile. The samples were collected at every 10th day for microbial and physicochemical analysis. The composting was terminated after 50 days. The duplicate samples were used to assess the consistency or reproducibility in the method.
Physiochemical analysis of compost
Compost pH and electrical conductivity (EC) were measured by preparing a (1:5 w v-1 compost: water) mixture as described by Rhoades [59] and Blakemore et al. [60] respectively. The percent organic carbon (C) in the compost was determined by the wet digestion method outlined by Walkley and Black [61]. Total nitrogen (N) was estimated by Kjeldahl method [62] and total sulfur according to the method of Steinbergs [63]. The potassium was estimated by ammonium-acetate method [64]. The samples were analyzed for micronutrient by atomic absorption spectrophotometer (Model 3030, Perkin-Elmer, USA). Macronutrients like calcium (Ca), magnesium (Mg) were determined following the methodology of Moral et al. [65] and sodium (Na) by using the method of Thompson and Wood [66]. The trace metals; copper (Cu), zinc (Zn), iron (Fe) and manganese (Mn) were estimated by ICP-MS (Induced coupled plasma Mass Spectrometer) as per methodology of Koplık et al. [67]; Fingerová and Koplık [68]; Jenn-Hung and Shang-Lien [30], respectively.
Isolation and enumeration of bacteria during composting
Bacteria were isolated from compost by serial dilution method by plating 100 μl of diluted suspension from each phase {the mesophile (30 and 35°C), thermophile (40 and 50°C), maturation and cooling phase (35 and 30°C) samples} were spread plated on nutrient agar (NA) plates. The plates were incubated at 30°C, 35°C, 40°C and 50°C for 24 h. Colonies were counted and populations were expressed in term of cfu g-1. Morphologically different colonies were purified on NA plates. All isolates and were preserved on slants at 4°C and glycerol stock at -20°C in 20% (v v-1). All chemicals and media were of molecular grade and procured from either Merck Pvt. Ltd or Himedia, India.
Morphological, biochemical and molecular characterization
Presumptive identification was carried out by colony morphology and use of the first stage diagnostic biochemical tests for Gram-positive and Gram-negative bacteria. Further identification was carried out by standard biochemical tests by using Himedia tests kits (Hi motility™ and Assorted™ Biochemical kit, Hi Carbohydrate™ kit, Hi IMViC™ Biochemical test kit).
Genomic DNA extraction, purification and quantification
Loopful of selected bacterial isolates were streaked and grown on NA plates at their relevant temperature and freshly grown isolates were used to inoculate in 50 ml of Luria-Bertani (LB) broth (Himedia, India). The broth cultures were grown at their respective temperature of the isolates with shaking at 200 rpm till the cultures reached OD600 of 0.4-0.5. Thereafter, cells were pelleted by centrifugation at 9167 × g for 10 min at 4°C and washed with TE buffer [10 mM Tris–HCl pH 8.0, 1 mM ethylenediaminetetraacetic acid (EDTA)] and pellets were either frozen (-20°C) for storage or used immediately for genomic DNA extraction by using the method of Sambrook & Russell [69].
DNA samples were quantified by running on agarose gel electrophoresis using 0.8% agarose gel in 1 × tris-boric acid EDTA (TBE) (89 mM tris pH 7.6, 89 mM boric acid, 2 mM EDTA) and visualized by ethidium bromide (0.5 μg ml-1) staining, to determine DNA size and to assess RNA contamination.
PCR Amplification and sequencing
Amplifications were performed in 50 μl reaction mixture containing 75 ng of template DNA, 1-unit of i-Taq™ polymerase (NEB, UK), 2 mM MgCl2 (NEB, UK) , 2 μl of 10X PCR buffer, 0.1 mM dNTP (NEB, UK), 100 ng of each forward (8f’:5’-AGAGTTTGATCCTGGCTCAG-3’ [70]), and reverse (1542r’:5′-AAGGAGGTGATCCAGCCGCA-3’ [71]) primers. The amplification was carried out using G-strom thermal cycler (Labtech, UK). Amplification programme consisted of initial cycle of denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, initial extension at 72°C for 1 min 30 sec and final extension at 72°C for 7 min. Amplified products were electrophoresed at 5 Vcm-1 through 1.5% agarose gel containing 0.5 μg ml-1 ethidium bromide in 1xTBE electrophoresis buffer with 50 bp DNA Ladder (NEB, UK). The gels were visualized under UV illumination in Gel Documentation system 2000 (Biorad, Hercules CA, USA) and stored as TIFF file format. Sizes of the amplicons were estimated in comparison with 50bp DNA ladder (NEB, UK).
Sequencing of 16S rRNA gene and phylogenetic analysis
The expected DNA band of 1.5 kb was excised from gel and purified using the gel elution kit (Sigma-Aldrich, USA) as per the manufacturer’s protocol. Sequencing reactions were carried out with a BigDye Terminator cycle sequencing kit (Applied Biosystems, USA), standard universal primer forward (8f’) and reverse (1542r’) primer and sequenced by using ABI Prism 3100 genetic analyzer (Applied Biosystems, USA). The sequences thus obtained were assembled and edited using Clone Manager Version 5 (http://www.scied.com/pr_cmbas.htm).
Database search was carried out for similar nucleotide sequences with the BLAST search of Non-reductant (NR) database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [72]). Partial length 16S rRNA gene sequences of strains closely related to the isolate were retrieved from NCBI for further analysis. For describing their phylogentic relationship, the partial 16S rRNA gene sequences were aligned by using Clustal_X [73]. A phylogenetic tree was constructed by means of neighbor-joining method using MEGA version 5 programme [74].
Nucleotide sequences accession number
The nucleotide sequences of 16S rRNA were obtained and deposited in the GenBank database (EMBL, U.K.) and the accession numbers; AM778178-AM778192, AM884572-AM884579 and FR865468-FR865475 were assigned to their respective sequences.
Abbreviations
- (cfu g-1):
-
Colony forming unit per gram
- MSW:
-
(Municipal Solid Wastes)
- PCR:
-
(Polymerase Chain Reaction)
- EC:
-
(Electrical Conductivity)
- NA:
-
(Nutrient Agar)
- LB:
-
(Luria-Bertani)
- OD:
-
(optical density)
- (NEB):
-
New England Biolab
References
Suthar S: Bioremediation of Agricultural Wastes through Vermicomposting. Biorem. J. 2009, 13 (1): 21-28. 10.1080/10889860802690513.
Rao PV, Baral SS, Dey R, Mutnuri S: Biogas generation potential by anaerobic digestion for sustainable energy development in India. Renew Sustain Energy Rev. 2010, 14 (7): 2086-2094. 10.1016/j.rser.2010.03.031.
Adani F, Genevini PL, Gasperi F, Zorzi G: Organic matter evolution index (OMEI) as a measure of composting efficiency. Comp Sci & Utiliz. 1997, 5 (2): 53-62.
Weltzien HC: Biocontrol of foliar fungal diseases with compost extracts. Microbial Ecology of Leaves. Edited by: Andrews JH, Hirano S. 1991, New York, NY, USA: Springer-Verlag, 430-450.
Tiquia SM, Richard TL, Honeyman MS: Effects of windrow turning and seasonal temperatures on composting hog manure from hoop structures. Environ Technol. 2000, 21 (9): 1037-1046. 10.1080/09593332108618048.
Fracchia L, Dohrmann AB, Martinotti MG, Tebbe CC: Bacterial diversity in finished compost and vermicompost: differences revealed by cultivation-independent analyses of PCR-amplified 16S rRNA genes. Appl Microbiol Biotechnol. 2006, 71 (6): 942-952. 10.1007/s00253-005-0228-y.
Ryckeboer J, Mergaert J, Coosemans J, Deprins K, Swings J: Microbiological Aspects of biowaste during composting in a monitored compost bin. J Appl Microbiol. 2003, 94 (1): 127-137. 10.1046/j.1365-2672.2003.01800.x.
Sundberg C, Franke-Whittle IH, Kauppi S, Yu D, Romantschuk M, Insam H, Håkan J: Characterisation of source-separated household waste intended for composting. Bioresour Technol. 2011, 102 (3): 2859-2867. 10.1016/j.biortech.2010.10.075.
Liu WT, Marsh TL, Cheng H, Forney LJ: Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997, 63 (11): 4516-4522.
Massol-Deya AA, Odelson DA, Hickey RF, Tiedje JM: Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA). Molecular microbial ecology manual. Edited by: Akkermans ADL, van Elsas JD, deBruijn FJ. 1995, Dordrecht: Kluwer, 1-8.
Pace NR: A molecular view of microbial diversity and the biosphere. Sci. 1997, 276 (5313): 734-740. 10.1126/science.276.5313.734.
Peters S, Koschinsky S, Schwieger F, Tebbe CC: Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes. Appl Environ Microbiol. 2000, 66 (3): 930-936. 10.1128/AEM.66.3.930-936.2000.
Dees PM, Ghiorse WC: Microbial diversity in hot synthetic compost as revealed by PCR-amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiol Ecol. 2001, 35 (2): 207-216. 10.1111/j.1574-6941.2001.tb00805.x.
Stackebrandt E: Phylogeny Based on 16S rRNA/DNA. eLS. 2009, Chichester: John Wiley & Sons Ltd, http://onlinelibrary.wiley.com/doi/10.1002/9780470015902.a0000462.pub2/abstract. (http://onlinelibrary.wiley.com/book/10.1002/047001590X)
Muyzer G: Genetic fingerprinting of microbial communities: present status and future perspective. Proceedings of the 8th International Symposium on Microbial Ecology. Edited by: Bell CR, Brylinsky M, Johnson-Green P. 1999, Halifax, Nova Scotia: Atlantic Canada Society for Microbial Ecology, 1-10.
Van Es FB, Meyer Reil LA: Biornass and metabolic activity of heterotrophic marine bacteria. Advances in microbial ecology. Edited by: Marshall KC. 1982, New York, USA: Plenum Publishing Corp, lll-170. 6
Denizci AA, Kazan D, Erarslan A: Bacillus marmarensis sp. nov., an alkaliphilic, protease-producing bacterium isolated from mushroom compost. Int J Sys Evol Microbiol. 2010, 60 (7): 1590-1594. 10.1099/ijs.0.012369-0.
Bandounas L, Wierckx NJP, de Winde JH, Ruijssenaars HJ: Isolation and characterization of novel bacterial strains exhibiting ligninolytic potential. BMC Biotechnology. 2011, 11: 94-10.1186/1472-6750-11-94.
Biddlestone AJ, Gray KR: Composting. Comprehensive Biotechnology: The Principles, Applications, and Regulations of Biotechnology in Industry, Agriculture, and Medicine. Edited by: Moo-Young M. 1985, Oxford: Pergamon Press, 1059-1070.
Hashim AB, Aminuddin H, Siva KB: Nutrient content in rice husk ash of some Malaysian rice varieties. Pert J Trop Agric Sci. 1996, 19 (1): 77-80.
Saber M, Mohammed Z, Badr-el-Din S, Awad N: Composting certain agricultural residues to potting soils. J Ecol Nat Environ. 2011, 3 (3): 78-84.
Brito LM, Coutinho J, Smith SR: Methods to improve the composting process of the solid fraction of dairy cattle slurry. Bioresour Technol. 2008, 99 (18): 8955-8960. 10.1016/j.biortech.2008.05.005.
Bernal MP, Paredes C, Sanchez-Monedero MA, Cegarra J: Maturity and stability parameters of composts prepared with a, wide range of organic wastes. Biores Technol. 1998, 63 (1): 91-99. 10.1016/S0960-8524(97)00084-9.
Iglesias-Jimenez E, Garcia PV, Espino M, Hernadez JM: City refuse compost as a phosphorus source to overcome the P-fixation capacity of sesquioxide-rich soils. Plant and Soil. 1993, 148: 115-127. 10.1007/BF02185391.
Ishii K, Fukui M, Takii S: Microbial succession during a composting process as evaluated by denaturing gradient gel electrophoresis analysis. J Appl Microbiol. 2000, 89 (5): 768-777. 10.1046/j.1365-2672.2000.01177.x.
Adegunloye DV, Adetuyi FC, Akinosoye FA, Doyeni MO: Microbial analysis of compost using cowdung as booster. Pak J Nut. 2007, 6 (5): 506-510. 10.3923/pjn.2007.506.510.
Adegunloye DV, Adetuyi FC: Composting of food wastes using cow and pig dung as booster. Afr J Bas & Appl Sci. 2009, 1 (3–4): 70-75.
Janakiram T, Sridevi K: Conversion of Waste into Wealth: A Study in Solid Waste Management. E-Journal of Chemistry. 2010, 7 (4): 1340-1345. 10.1155/2010/549185. (http://www.e-journals.net/)
Felton GK, Carr LE, Prigge CE, Bouwkamp JC: Nitrogen and phosphorous dynamics in cocomposted yard trimmings and broiler litter. Comp Sci Utiliz. 2004, 12 (4): 349-355.
Jenn-Hung H, Shang-Lien L: Effect of composting on characterization and leaching of copper, manganese and zinc from swine manure. Environ Poll. 2011, 114 (1): 119-127.
Willson GB: Organic Waste Processing loa Q: Combining raw materials for composting. Biocycl. 1989, 30 (5): 82-85.
Paulin B, O’Malley P: Compost production and use in horticulture. 2008, Department of Agriculture and Food, Government of Western Australia, Bulletin 4746 ISSN 1833 7236 (http://www.agric.wa.gov.au/objtwr/imported_assets/content/hort/compost_bulletin08.pdf)
Kell DB, Kaprelyants AS, Weichart DH, Harwood CR, Barer MR: Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Ant von Leeuwen. 1998, 73 (2): 169-187. 10.1023/A:1000664013047.
Postgate JR: Viable counts and viability. Meth Microbiol. 1969, 1: 611-628.
Hargerty DJ, Pavoni JL, Heer JE: Solid Waste Management. 1999, New York: Van Nostrand Reinhold, 12-13.
Golueke CG: Bacteriology of composting. Biocycl. 1992, 33: 55-57.
Kolbert CP, Persing DH: Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr Opin Microbiol. 1999, 2 (3): 299-305. 10.1016/S1369-5274(99)80052-6.
Olson JC, Cuff CF, Lukomski S, Lukomska E, Canizales Y, Wu B, Crout RJ, Thomas JG, McNeil DW, Weyant RJ, Marazita ML, Paster BJ, Elliott T: Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health. 2011, 11: 1-7. 10.1186/1472-6831-11-1.
Franke-Whittle IH, Knapp BA, Fuchs J, Kaufmann R, Insam H: Application of COMPOCHIP microarray to investigate the bacterial communities of different composts. Microb Ecol. 2009, 57 (3): 510-521. 10.1007/s00248-008-9435-2.
Ntougias S, Zervakis GI, Kavroulakis N, Ehaliotis C, Papadopoulou KK: Bacterial diversity in spent mushroom compost assessed by amplified rDNA restriction analysis and sequencing of cultivated isolates. Syst Appl Microbiol. 2004, 27 (6): 746-754. 10.1078/0723202042369857.
Chandna P, Mallik S, Kuhad RC: Assessment of bacterial diversity in agricultural by-product compost by sequencing of cultivated isolates and amplified rDNA restriction analysis. Appl Microbiol Biotechnol. 2012, 10.1007/s00253-012-4434-0.
Silva CF, Azevedo RS, Braga C, Silva R, Dias ES, Schwan RF: Microbial diversity in a baggase-based compost prepared for the production of Agaricus brasiliensis. Braz J Microbiol. 2009, 40 (3): 590-600. 10.1590/S1517-83822009000300023.
Gbolagade JS: Bacteria associated with compost used for cultivation of Nigerian edible mushrooms Pleurotus tuber-regium (Fr.) Singer, and Lentinus squarrosulus (Berk.). Afr J of Biotech. 2006, 5 (4): 338-342.
Murray PR, Drew WL, Kobayashi GS, Thompson JH: Medical Microbiology. 1990, PA: Mosby Publ Philadelphia
Strauch D: Occurrence of microorganisms pathogenic for man and animals in source separated biowaste and compost – importance, controls, limits, epidemiology. “The Science of Composting”. Edited by: de Bertoldi M, Sequi P, Lemmes B, Tizano P. 1996, London: CEC, Blackie Academic and Professional, 224-232.
Ahlawat OP, Vijay B: Potential of thermophilic bacteria as microbial inoculant for commercial scale white button mushroom (Agaricus bisporus) compost production. J Sci & Ind Res. 2010, 69 (12): 948-955.
Danon M, Franke-Whittle IH, Insam H, Chen Y, Hadar Y: Molecular analysis of bacterial community succession during prolonged compost curing. FEMS Microbiol. Ecol. 2008, 65 (1): 133-144. 10.1111/j.1574-6941.2008.00506.x.
Boulter JI, Boland GJ, Trevors JT: Compost: a study of the development process and end-product potential for suppression of turfgrass disease. W J Microbio Biotech. 2000, 16 (2): 115-134. 10.1023/A:1008901420646.
Kumar A, Prakash A, Johri BN: Bacillus as PGPR in Crop Ecosystem. Bacteria in Agrobiology: Crop Ecosystems. Edited by: Maheshwari DK. 2011, Berlin Heidelberg: Springer-Verlag, 10.1007/978-3-642-18357-7_2, #.
Ryckeboer J, Mergaert J, Vaes K, Klammer S, De Clercq D, Coosemans J, Insam H, Swings J: A survey of bacteria and fungi occurring during composting and self-heating processes. Ann Microbiol. 2003, 53 (4): 349-410.
Vaz-Moreira I, Silva ME, Manaia CM, Nunes OC: Diversity of Bacterial Isolates from Commercial and Homemade Composts. Microbial Ecol. 2008, 55 (4): 714-722. 10.1007/s00248-007-9314-2.
Young CC, Chou JH, Arun AB, Yen WS, Sheu SY, Shen FT, Lai WA, Rekha PD, Chen WM: Comamonas composti sp. nov., isolated from food waste compost. Int J Syst Bacteriol. 2008, 58 (1): 251-256.
Quinteros R, Goodwin S, Lenz RW, Park WH: Extracellular degradation of medium chain length poly (β-hydroxyalkanoates) by Comamonas sp. Int J Biolog Macromol. 1999, 25 (1–3): 135-143.
Willems A, De Ley J, Gillis M, Kersters K: Comamonadaceae, a new family encompassing the acidovorans rRNA complex, including Variovorax paradoxus gen. nov., comb. nov., for Alcaligenes paradoxus (Davis 1969). Int J Syst Evol Microbiol. 1991, 41 (3): 445-450.
Pinel N, Davidson SK, Stahl DA: Verminephrobacter eiseniae gen. nov., sp. nov., a nephridial symbiont of the earthworm Eisenia foetida (Savigny). Int J Syst Evol Microbiol. 2008, 58 (9): 2147-2157. 10.1099/ijs.0.65174-0.
Pizl V, Novokova A: Interactions between microfungi and Eisenia andrei (Oligochaeta) during cattle manure vermicomposting. Pedobiologia. 2003, 47 (5–6): 895-899.
Schramm A, Davidson S, Dodsworth J, Drake H, Stahl D, Dubilier N: Acidovorax-like symbionts in the nephridia of earthworms. Environ Microbiol. 2003, 5 (9): 804-809. 10.1046/j.1462-2920.2003.00474.x.
Alidadi H, Parvaresh AR, Shahmansouri MR, Pourmoghadas H: Combined compost and vermicomposting process in the treatment and bioconversion of sludge. Iran J Environ Heal Sci Eng. 2005, 2 (4): 251-254.
Rhoades JD: Salinity: electrical conductivity and total dissolved solids. Methods of Soil Analysis. Part 3. Chemical Methods. Edited by: Sparks DL. 1996, Madison: SSSA, 417-435.
Blakemore LC, Searle PL, Daily BK: Methods for chemical analysis of soils. 1981, New Zealand Soil Bureau Report IDA. D5C
Walkley AJ, Black CA: Estimation of soil organic carbon by chromic acid titration method. Soil Sci. 1934, 37: 29-38. 10.1097/00010694-193401000-00003.
Kjeldahl J: A new method for the estimation of nitrogen in organic compounds. Z. Anal Chem. 1883, 22: 366-
Steinbergs A: A method for the determination of total sulphur in soils. Analyst (London). 1955, 80: 457-461. 10.1039/an9558000457.
Anonymous: Guide to the interpretation of analytical data for loam less compost. Ministry of Agriculture, Fisheries and Food, No. 25. ADAS. 1988, United Kingdome: Agricultural Development and Advisory Service
Moral R, Navarro-Pedreno J, Gomez I, Mataix J: Distribution and accumulation of heavy metals (Cd, Ni and Cr) in tomato plant. Environ Bulletin. 1994, 3: 395-399.
Thompson M, Wood SJ: Atomic absorption methods in applied geochemistry. Atomic Absorption Spectrometry. Edited by: Cantle JE. 1982, Amsterdam: Elsevier, 261-284.
Koplı´k R, Curdova E, Suchanek M: Trace element analysis in CRM of plant origin by inductively coupled plasma mass spectrometry. Fresenius’ J Anal Chem. 1998, 300: 449-451.
Fingerová H, Koplı ´k R: Study of minerals and trace elements. Fresenius J Anal Chem. 1993, 63 (5–6): 545-549.
Sambrook J, Russell DW: Molecular Cloning. 2001, New York: Cold Spring Harbor Laboratory Press, 3
DeLong EF: Archaea in coastal marine sediments. Proc Natl Acad Sci. 1992, 89: 5685-5689. 10.1073/pnas.89.12.5685.
Wilmotte A, van-der Auwera G, de Wachter R: Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett. 1993, 317 (1–2): 96-100.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Acid Res. 1997, 25 (17): 3389-3402. 10.1093/nar/25.17.3389.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acid Res. 1997, 25 (24): 4876-4882. 10.1093/nar/25.24.4876.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA 5: molecular evolutionary genetic analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011, 10.1093/molbev.msr121.
Acknowledgement
Authors thank Ms. Urvashi Kuhad, Department of Modern Indian Languages and Literary Studies, University of Delhi, Delhi, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RCK, PC, LN and SS planned the study. PC performed the experiments. PC and RCK analyzed the results. RCK, PC, LN and SS drafted the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chandna, P., Nain, L., Singh, S. et al. Assessment of bacterial diversity during composting of agricultural byproducts. BMC Microbiol 13, 99 (2013). https://doi.org/10.1186/1471-2180-13-99
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-13-99