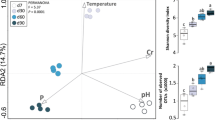
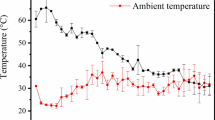
Abstract
Bioplastic waste may play a key role in shaping the microbial communities in composting environment. The present study aims to determine the possible influence of bioplastics on microbial structure of aerobic organic waste treatments and to assess the evolution of microbial community during thermophilic and maturation phases of composting, comparing microorganisms in compost and attached on bioplastics surface. Composting process was simulated in a 2-month lab-scale test. Mater-Bi®, PBAT, PLA and LDPE were considered as benchmark. A qualitative screening was done with DGGE, and then the bacterial community profile was disclosed with 16S rRNA amplicon analysis. The 16S rRNA profile in bioplastics-associated communities was found to largely differed from that of the initial compost (positive control). Moreover, as expected, during the thermophilic phase there was a prevalence of better heat-resistant phyla compared to the mesophilic phase. Some specialists were found to be plastic nature dependent, in particular Streptomyces, Pseudomonas, Aeribacillus, Schlegellela and Cohnella were among the most abundant genera. Finally, to select bacterial species capable of growing on the tested bioplastics, an enrichment method was applied. The method disclosed the possibility to culture some specific bioplastics degrading species, opening the frontiers for bioaugmentation practice in industrial composting.






Similar content being viewed by others
References
Calabrò PS, Grosso M (2018) Bioplastics and waste management. Waste Manag 78:800–801. https://doi.org/10.1016/j.wasman.2018.06.054
Consorzio Italiano Compostatori (2017) Dati di settore, rapporto CIC 2017. 69–71
Lucas N, Bienaime C, Belloy C et al (2008) Polymer biodegradation: mechanisms and estimation techniques—a review. Chemosphere 73:429–442. https://doi.org/10.1016/j.chemosphere.2008.06.064
Shah AA, Hasan F, Hameed A, Ahmed S (2008) Biological degradation of plastics: a comprehensive review. Biotechnol Adv 26:246–265. https://doi.org/10.1016/j.biotechadv.2007.12.005
Kumaravel S, Hema R, Lakshmi R (2010) Production of polyhydroxybutyrate (Bioplastic) and its biodegradation by pseudomonas lemoignei and Aspergillus niger. E J Chem 7:1–4. https://doi.org/10.1155/2010/148547
Tokiwa Y, Calabia BP (2004) Degradation of microbial polyesters. Biotechnol Lett 26:1181–1189. https://doi.org/10.1023/B:BILE.0000036599.15302.e5
Folino A, Karageorgiou A, Calabrò PS, Komilis D (2020) Biodegradation of wasted bioplastics in natural and industrial environments: a review. Sustainability. https://doi.org/10.3390/su12156030
Emadian SM, Onay TT, Demirel B (2017) Biodegradation of bioplastics in natural environments. Waste Manag 59:526–536. https://doi.org/10.1016/j.wasman.2016.10.006
Dey S, Rout AK, Behera BK, Ghosh K (2022) Plastisphere community assemblage of aquatic environment: plastic-microbe interaction, role in degradation and characterization technologies. Environ Microbiomes 17:1–21. https://doi.org/10.1186/s40793-022-00430-4
Zettler ER, Mincer TJ, Amaral-Zettler LA (2013) Life in the “plastisphere”: microbial communities on plastic marine debris. Environ Sci Technol 47:7137–7146. https://doi.org/10.1021/es401288x
Bandini F, Misci C, Taskin E et al (2020) Biopolymers modulate microbial communities in municipal organic waste digestion. FEMS Microbiol Ecol 96:1–13. https://doi.org/10.1093/femsec/fiaa183
Gu JD, Mitchell R, Ford TE (2003) Microbiological deterioration and degradation of synthetic polymeric materials: recent research advances. Int Biodeterior Biodegrad 52:69–91. https://doi.org/10.1016/S0964-8305(02)00177-4
Rocha-Santos TAP, Duarte AC (2017) Characterization and analysis of microplastics, vol 75, (ISBN 9780444638984)
Ruggero F, Gori R, Lubello C (2019) Methodologies to assess biodegradation of bioplastics during aerobic composting and anaerobic digestion: a review. Waste Manag Res. https://doi.org/10.1177/0734242X19854127
Massardier-Nageotte V, Pestre C, Cruard-Pradet T, Bayard R (2006) Aerobic and anaerobic biodegradability of polymer films and physico-chemical characterization. Polym Degrad Stab 91:620–627. https://doi.org/10.1016/j.polymdegradstab.2005.02.029
EN 13432 (2000) Packaging—Requirements for packaging recoverable through composting and biodegradation—Test scheme and evaluation criteria for the final acceptance of packaging
ISO 20200 (2015) Test, Standards Publication Plastics—Determination of the degree of disintegration of plastic materials under simulated composting conditions in a laboratory-scale
ASTM D5338 (2011) Standard test method for determining aerobic biodegradation of plastic materials under controlled composting conditions, incorporating thermophilic temperatures. ASTM International
European Commission (2017) Report from the commission to the european parliament, the council, the european economic and social committee and the committee of the regions. Off J Eur Union COM (2017):1–14
Bastioli C (1998) Properties and applications of Mater-Bi starch-based materials. Polym Degrad Stab 59:263–272
ISO 14855-2 (2018) Determination of the ultimate aerobic biodegradability of plastic materials under controlled composting conditions - Method by analysis of evolved carbon dioxide
ISO 11465 (1993) Soil quality—Determination of dry matter and water content on a mass basis—Gravimetric method
Ruggero F, Onderwater RCA, Carretti E, et al (2021) Degradation of film and rigid bioplastics during the thermophilic phase and the maturation phase of simulated composting. J Polym Environ 29:3015–3028
Barbier FF, Chabikwa TG, Ahsan MU et al (2019) A phenol/chloroform-free method to extract nucleic acids from recalcitrant, woody tropical species for gene expression and sequencing. Plant Methods 15:9–12. https://doi.org/10.1186/s13007-019-0447-3
Wang Y, Naumann U, Wright ST, Warton DI (2012) Mvabund- an R package for model-based analysis of multivariate abundance data. Methods Ecol Evol 3:471–474. https://doi.org/10.1111/j.2041-210X.2012.00190.x
Dixon P (2003) Computer program review VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930
Yoshida S, Hiraga K, Takehana T et al (2016) A bacterium that degrades and assimilates poly(ethylene terephthalate). Science (80- ). https://doi.org/10.1126/science.aaf8305
Delacuvellerie A, Cyriaque V, Gobert S et al (2019) The plastisphere in marine ecosystem hosts potential specific microbial degraders including Alcanivorax borkumensis as a key player for the low-density polyethylene degradation. J Hazard Mater 380:120899. https://doi.org/10.1016/j.jhazmat.2019.120899
Bonhomme S, Cuer A, Delort AM et al (2003) Environmental biodegradation of polyethylene. Polym Degrad Stab 81:441–452. https://doi.org/10.1016/S0141-3910(03)00129-0
Arrieta MP, López J, Rayón E, Jiménez A (2014) Disintegrability under composting conditions of plasticized PLA–PHB blends. Polym Degrad Stab 108:307–318. https://doi.org/10.1016/j.polymdegradstab.2014.01.034
Poli A, Laezza G, Gul-Guven R et al (2011) Geobacillus galactosidasius sp. nov., a new thermophilic galactosidase-producing bacterium isolated from compost. Syst Appl Microbiol 34:419–423. https://doi.org/10.1016/j.syapm.2011.03.009
Hatayama K, Shoun H, Ueda Y, Nakamura A (2006) Tuberibacillus calidus gen. nov., sp. nov., isolated from a compost pile and reclassification of Bacillus naganoensis Tomimura et al. 1990 as Pullulanibacillus naganoensis gen. nov., comb. Nov. and Bacillus laevolacticus Andersch et al. 1994 as Sporolacto. Int J Syst Evol Microbiol 56:2545–2551. https://doi.org/10.1099/ijs.0.64303-0
Han SI, Lee JC, Lee HJ, Whang KS (2013) Planifilum composti sp. nov., a thermophile isolated from compost. Int J Syst Evol Microbiol 63:4557–4561. https://doi.org/10.1099/ijs.0.053199-0
Liu H, Huang Y, Duan W et al (2020) Microbial community composition turnover and function in the mesophilic phase predetermine chicken manure composting efficiency. Bioresour Technol 313:123658. https://doi.org/10.1016/j.biortech.2020.123658
Wang X, Pan S, Zhang Z et al (2017) Effects of the feeding ratio of food waste on fed-batch aerobic composting and its microbial community. Bioresour Technol 224:397–404. https://doi.org/10.1016/j.biortech.2016.11.076
Chen Z, Zhang S, Wen Q, Zheng J (2015) Effect of aeration rate on composting of penicillin mycelial dreg. J Environ Sci 37:172–178
Sangeetha T, Guo Z, Liu W et al (2017) Energy recovery evaluation in an up flow microbial electrolysis coupled anaerobic digestion (ME-AD) reactor: Role of electrode positions and hydraulic retention times. Appl Energy 206:1214–1224. https://doi.org/10.1016/j.apenergy.2017.10.026
Goodfellow M, Maldonado LA, Quintana ET (2005) Reclassification of Nonomuraea flexuosa (Meyer 1989) Zhang et al. 1998 as Thermopolyspora flexuosa gen. nov., comb. nov., nom. rev. Int J Syst Evol Microbiol 55:1979–1983. https://doi.org/10.1099/ijs.0.63559-0
Zhang M, Jia H, Weng Y, Li C (2019) Biodegradable PLA/PBAT mulch on microbial community structure in different soils. Int Biodeterior Biodegrad 145:104817. https://doi.org/10.1016/j.ibiod.2019.104817
Yoon JH, Kang SJ, Kim W, Oh TK (2007) Pigmentiphaga daeguensis sp. nov., isolated from wastewater of a dye works, and emended description of the genus Pigmentiphaga. Int J Syst Evol Microbiol 57:1188–1191. https://doi.org/10.1099/ijs.0.64901-0
Cerda A, Artola A, Font X et al (2018) Composting of food wastes: status and challenges. Bioresour Technol 248:57–67
Zhong XZ, Ma SC, Wang SP et al (2018) A comparative study of composting the solid fraction of dairy manure with or without bulking material: performance and microbial community dynamics. Bioresour Technol 247:443–452. https://doi.org/10.1016/j.biortech.2017.09.116
Kong W, Sun B, Zhang J et al (2020) Metagenomic analysis revealed the succession of microbiota and metabolic function in corncob composting for preparation of cultivation medium for Pleurotus ostreatus. Bioresour Technol 306:123156. https://doi.org/10.1016/j.biortech.2020.123156
Aliabadi N, Aminzadeh S, Karkhane AA, Haghbeen K (2016) Thermostable chitinase from Cohnella sp. A01: isolation and product optimization. Brazilian J Microbiol 47:931–940. https://doi.org/10.1016/j.bjm.2016.07.009
Narancic T, Cerrone F, Beagan N, O’Connor KE (2020) Recent advances in bioplastics: application and biodegradation. Polymers (Basel). https://doi.org/10.3390/POLYM12040920
Quitadamo A, Massardier V, Iovine V et al (2019) Effect of cellulosicwaste derived filler on the biodegradation and thermal properties of HDPE and PLA composites. Processes. https://doi.org/10.3390/pr7100647
Anbarasan S, Wahlström R, Hummel M et al (2017) High stability and low competitive inhibition of thermophilic Thermopolyspora flexuosa GH10 xylanase in biomass-dissolving ionic liquids. Appl Microbiol Biotechnol 101:1487–1498. https://doi.org/10.1007/s00253-016-7922-9
Peelman N, Ragaert P, De Meulenaer B et al (2013) Application of bioplastics for food packaging. Trends Food Sci Technol 32:128–141. https://doi.org/10.1016/j.tifs.2013.06.003
Lee B, Pometto AL, Fratzke A, Bailey TB (1991) Biodegradation of degradable plastic polyethylene by Phanerochaete and Streptomyces species. Appl Environ Microbiol 57:678–685. https://doi.org/10.1128/aem.57.3.678-685.1991
Storey S, Chualain DN, Doyle O et al (2015) Comparison of bacterial succession in green waste composts amended with inorganic fertiliser and wastewater treatment plant sludge. Bioresour Technol 179:71–77. https://doi.org/10.1016/j.biortech.2014.11.107
Xu L, Yi M, Yi H et al (2018) Manure and mineral fertilization change enzyme activity and bacterial community in millet rhizosphere soils. World J Microbiol Biotechnol. https://doi.org/10.1007/s11274-017-2394-3
Maeda K, Hanajima D, Morioka R, Osada T (2010) Characterization and spatial distribution of bacterial communities within passively aerated cattle manure composting piles. Bioresour Technol 101:9631–9637. https://doi.org/10.1016/j.biortech.2010.07.057
Boyandin AN, Prudnikova SV, Karpov VA et al (2013) Microbial degradation of polyhydroxyalkanoates in tropical soils. Int Biodeterior Biodegrad 83:77–84. https://doi.org/10.1016/j.ibiod.2013.04.014
Ghosh SK, Pal S, Ray S (2013) Study of microbes having potentiality for biodegradation of plastics. Environ Sci Pollut Res Int 20:4339–4355. https://doi.org/10.1007/s11356-013-1706-x
Nakei MD (2015) Isolation and identification of plastics-degrading microorganisms from soils of Morogoro, Tanzania
Zhang H, McGill E, Gomez CO et al (2017) Disintegration of compostable foodware and packaging and its effect on microbial activity and community composition in municipal composting. Int Biodeterior Biodegrad 125:157–165. https://doi.org/10.1016/j.ibiod.2017.09.011
Ramarao N, Tran SL, Marin M, Vidic J (2020) Advanced methods for detection of Bacillus cereus and its pathogenic factors. Sensors (Switzerland). https://doi.org/10.3390/s20092667
Khalil AB, Sivakumar N, Arslan M et al (2018) Insights into Brevibacillus borstelensis AK1 through whole genome sequencing: a thermophilic bacterium isolated from a hot spring in Saudi Arabia. Biomed Res Int. https://doi.org/10.1155/2018/5862437
Hermabessiere L, Himber C, Boricaud B et al (2018) Optimization, performance, and application of a pyrolysis-GC/MS method for the identification of microplastics. Anal Bioanal Chem 410(25):6663–6676. https://doi.org/10.1007/s00216-018-1279-0
Manachini PL, Mora D, Nicastro G et al (2000) Bacillus thermodenitrificans sp. nov., nom. rev. Int J Syst Evol Microbiol 50:1331–1337. https://doi.org/10.1099/00207713-50-3-1331
Nazina TN, Tourova TP, Poltaraus AB et al (2001) Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus th. Int J Syst Evol Microbiol 51:433–446
Inoue M, Tanimura A, Ogami Y, et al (2019) Draft Genome Sequence of Parageobacillus thermoglucosidasius. Microbiol Resour Announc 8(5). https://doi.org/10.1128/mra.01666-18
Wang YQ, Yuan Y, Yu Z et al (2013) Bacillus borbori sp. Nov., Isolated from an electrochemically active biofilm. Curr Microbiol 67:718–724. https://doi.org/10.1007/s00284-013-0426-2
Yildirim V, Baltaci MO, Ozgencli I et al (2017) Purification and biochemical characterization of a novel thermostable serine alkaline protease from Aeribacillus pallidus C10: a potential additive for detergents. J Enzyme Inhib Med Chem 32:468–477. https://doi.org/10.1080/14756366.2016.1261131
Hadad D, Geresh S, Sivan A (2005) Biodegradation of polyethylene by the thermophilic bacterium Brevibacillus borstelensis. J Appl Microbiol 98:1093–1100. https://doi.org/10.1111/j.1365-2672.2005.02553.x
Owusu-Darko R, Allam M, Mtshali S et al (2017) Draft genome sequence of Bacillus oleronius DSM 9356 isolated from the termite Reticulitermes santonensis. Genom Data 12:76–78. https://doi.org/10.1016/j.gdata.2017.03.005
Szkaradkiewicz A, Chudzicka-Strugala I, Karpiński TM et al (2012) Bacillus oleronius and Demodex mite infestation in patients with chronic blepharitis. Clin Microbiol Infect 18:1020–1025. https://doi.org/10.1111/j.1469-0691.2011.03704.x
Sen R, Tripathy S, Padhi SK et al (2015) Assessment of genetic diversity of Bacillus spp. isolated from eutrophic fish culture pond. 3 Biotech 5:393–400. https://doi.org/10.1007/s13205-014-0234-9
Ivanova N, Sorokin A, Anderson I et al (2003) Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87–91. https://doi.org/10.1038/nature01582
Mols M, De Been M, Zwietering MH et al (2007) Metabolic capacity of Bacillus cereus strains ATCC 14579 and ATCC 10987 interlinked with comparative genomics. Environ Microbiol 9:2933–2944. https://doi.org/10.1111/j.1462-2920.2007.01404.x
Alenezi FN, Rekik I, Bouket AC et al (2017) Increased biological activity of Aneurinibacillus migulanus strains correlates with the production of new gramicidin secondary metabolites. Front Microbiol 8:1–11. https://doi.org/10.3389/fmicb.2017.00517
Takagi H, Shida O, Kadowaki K et al (1993) Characterization of Bacillus brevis with descriptions of Bacillus migulanus sp. nov., Bacillus choshinensis sp. nov., Bacillus parabrevis sp. nov., and Bacillus galactophilus sp. nov. Int J Syst Bacteriol 43:221–231. https://doi.org/10.1099/00207713-43-2-221
Hsu KJ, Tseng M, Don TM, Yang MK (2012) Biodegradation of poly(β-hydroxybutyrate) by a novel isolate of streptomyces bangladeshensis 77T-4. Bot Stud 53:307–313
Trinh Tan F, Cooper DG, Marić M, Nicell JA (2008) Biodegradation of a synthetic co-polyester by aerobic mesophilic microorganisms. Polym Degrad Stab 93:1479–1485. https://doi.org/10.1016/j.polymdegradstab.2008.05.005
Jeszeová L, Puškárová A, Bučková M, Kraková L, Grivalský T, Danko M, Mosnáčková K, Chmela Š, Pangallo D (2018) Microbial communities responsible for the degradation of poly(lactic acid)/poly(3-hydroxybutyrate) blend mulches in soil burial respirometric tests. World J Microbiol Biotechnol 34:101. https://doi.org/10.1007/s11274-018-2483-y
Teeraphatpornchai T, Nakajima-Kambe T, Shigeno-Akutsu Y, Nakayama M, Nomura NTN, Uchiyama H (2003) Isolation and characterization of a bacterium that degrades PBSA. Biotechnol Lett 25:23–28
Pattanasuttichonlakul W, Sombatsompop N, Prapagdee B (2018) Accelerating biodegradation of PLA using microbial consortium from dairy wastewater sludge combined with PLA-degrading bacterium. Int Biodeterior Biodegrad 132:74–83. https://doi.org/10.1016/j.ibiod.2018.05.014
Acknowledgements
The experimental composting tests were carried out at Materia Nova (MaNo) in Parc Initialis, Avenue Nicolas Copernic 3, 7000 Mons, Belgium. SEM from the Department of Biology of Marine Organisms and Biomimetics (UMONS) was utilized. Microbiological analyses were partially carried out in the joint laboratory of Proteomics and Microbiology of UMONS and Materia Nova and partially performed by Novogene.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ruggero, F., Roosa, S., Onderwater, R. et al. Characterization of bacterial communities responsible for bioplastics degradation during the thermophilic and the maturation phases of composting. J Mater Cycles Waste Manag 25, 3270–3285 (2023). https://doi.org/10.1007/s10163-023-01751-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10163-023-01751-3