Abstract
Background
Staphylococcus aureus is an opportunistic commensal bacterium that mostly colonizes the skin and soft tissues. The pathogenicity of S. aureus is due to both its ability to resist antibiotics, and the production of toxins. Here, we characterize a group of genes responsible for toxin production and antibiotic resistance of S. aureus strains isolated from skin, soft tissue, and bone related infections.
Results
A total of 136 S. aureus strains were collected from five different types of infection: furuncles, pyomyositis, abscesses, Buruli ulcers, and osteomyelitis, from hospital admissions and out-patients in Benin. All strains were resistant to benzyl penicillin, while 25% were resistant to methicillin, and all showed sensitivity to vancomycin. Panton-Valentine leukocidin (PVL) was the most commonly produced virulence factor (70%), followed by staphylococcal enterotoxin B (44%). Exfoliative toxin B was produced by 1.3% of the strains, and was only found in isolates from Buruli ulcers. The tsst-1, sec, and seh genes were rarely detected (≤1%).
Conclusions
This study provides new insight into the prevalence of toxin and antibiotic resistance genes in S. aureus strains responsible for skin, soft tissue, and bone infections. Our results showed that PVL was strongly associated with pyomyositis and osteomyelitis, and that there is a high prevalence of PVL-MRSA skin infections in Benin.
Similar content being viewed by others
Background
Staphylococcus aureus is an opportunistic pathogen that mainly colonizes the nares and skin of up to 80% of the population [1]. S. aureus is a Gram-positive cocci that is frequently isolated in hospitals, and is responsible for diverse infections and toxicoses [2]. S. aureus is the most common cause of skin and soft-tissue infections (such as impetigo, furunculosis, and abscess), as well as systemic infections (such as pneumonia and endocarditis) [3]. The threat of S. aureus is not only due to its distribution and pathogenicity [4, 5], but also because of its ability to overcome antimicrobial agents [6–8].
Virulence factors produced by S. aureus render this organism highly pathogenic. The known virulence factors include exotoxins, such as exfoliative toxins (ETs), along with toxic shock syndrome toxin-1 (TSST-1), staphylococcal enterotoxins (SEs), leukocidins (Panton-Valentine leukocidin; PVL, LukE/D), and hemolysins (α, β, γ, δ) [9]. Enterotoxins often cause food poisoning [10], while ETs (also called epidermolysins) act on the skin [11].
Among the leukocidins, PVL is an extracellular protein consisting of two subunits, F and S, which act in concert and have leucocidal and dermonecrotic functions. The PVL toxin targets the outer membrane of polymorphonuclear cells, monocytes, and macrophages [12–15]. S. aureus strains that are positive for PVL are usually associated with skin and soft-tissue infections, and were first isolated in the 1960s [16–19]. PVL-positive strains are particularly associated with furuncles, accounting for 96% of cases [11, 17, 20], and approximately 90% of PVL-positive S. aureus strains were originally isolated from furuncles. PVL has also been associated with severe infections, including necrotizing pneumonia [19, 21–24], osteomyelitis [25], and even cases of purpura fulminans [26]. PVL toxin was recently identified in Lemierre’s syndrome [27], and in a case of Fournier’s gangrene [28]. PVL has also been associated with community-acquired necrotizing and hemorrhagic pulmonary infections affecting previously healthy children and young adults [22, 29].
Several antibiotics were routinely used in the treatment of S. aureus infections, contributing to the emergence of antibiotic-resistant strains. Widespread resistance severely complicates management of S. aureus infections. S. aureus strains that are resistant to methicillin (methicillin-resistant S. aureus, MRSA) are pervasive in the hospital environment, and have recently also caused a global epidemic of community-associated S. aureus (CA-MRSA) infections [30]. The changing trend of MRSA epidemiology, showed the use of PVL locus detection as a marker of CA-MRSA isolates, alongside with non multiresistant pattern and SCCmec type IV or V [31]. Vancomycin has been used successfully for over 50 years for the treatment of S. aureus infections, particularly those caused by MRSA strains [32]. However, vancomycin-resistant S. aureus (VRSA) and vancomycin-intermediate (VISA) strains have been reported, three decades after the introduction of vancomycin [33]. The presence of resistance genes may also affect toxin production.
The production of multiple virulence factors, as well as the presence of antibiotic resistance genes, makes S. aureus a highly pathogenic microorganism. The objective of this work was to study the susceptibility profile and toxin production of S. aureus strains isolated from various skin, soft tissue, and bone infections.
Results
Prevalence of S. aureusstrains according to the sample origin
Using standard microbiological methods for identification of microorganisms; a total of 136 strains of S. aureus were collected during this study. The proportions of the strains varied depending on the five types of infection: furuncle, osteomyelitis, pyomyositis, abscess, and Buruli ulcer. Almost 37% (50/136) of the collected strains originated from abscesses, followed by strains isolated from pyomyositis patients (27%, 37/136), furuncles (14%, 19/136), Buruli ulcers (12%, 16/136), and osteomyelitis cases (10%, 14/136).
Susceptibility to antibiotics
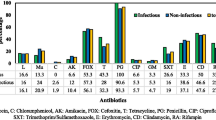
There was a wide range in the susceptibility of the isolates to the various antibiotics examined. All of the strains were resistant to benzyl penicillin, while other antibiotics (vancomycin, fusidic acid, fosfomycin, and linezolid) were active against some of the strains (Figure 1).
Global Staphylococcus aureus strains isolated from primary and secondary infections resistance profile to 22 antibiotics. Benzyl penicillin (BP), oxacillin (Ox), cefoxitin screen (Cef), gentamicin (Gen), tobramycin (Tob), kanamycin (Kan), vancomycin (Van), teicoplanin (Tei), fusidic acid (FA), fosfomycin (Fos), rifampicin (Rif), trimethopim/sulfamethoxazole (T/Sul), erythromycin (Ery), lincomycin (Lin), pristinamycin (Pri), linezolid (Line), tetracyclin (Tet).
There was no significant difference in the antibiotic resistance of the strains based on their origin (Figure 2). S. aureus strains originating from pyomyositis (Figure 2a), furuncles (Figure 2b) and osteomyelitis cases (Figure 2c) were resistant to 4/17 tested antibiotics (benzyl penicillin, rifampincin, tetracycline, and trimetroprim/sulfamethoxazol), while strains originating from abscesses and Buruli ulcer were strongly resistant to respectively 13/17 and 7/17 of the tested antibiotics (Figure 2d, e). Of the 136 isolated S. aureus strains, 34 (25%) were resistant to oxacillin (MRSA), while none of the strains showed resistance to vancomycin (VRSA). The oxacillin-resistant strains were all isolated from abscesses and Buruli ulcers.
Staphylococcus aureus strains resistance profile to 22 antibiotics according to their origin. Benzyl penicillin (BP), oxacillin (Ox), cefoxitin screen (Cef), gentamicin (Gen), tobramycin (Tob), kanamycin (Kan), vancomycin (Van), teicoplanin (Tei), fusidic acid (FA), fosfomycin (Fos), rifampicin (Rif), trimethopim/sulfamethoxazole (T/Sul), erythromycin (Ery), lincomycin (Lin), pristinamycin (Pri), linezolid (Line), tetracyclin (Tet).
Toxins production and/or presence of their encoding genes
There was a significant difference in the production and/or the presence of genes encoding the 12 toxins (p < 0.0001). Thus, a significant number of strains (70.0%) were capable of producing PVL, followed by the production of staphylococcal enterotoxin B (SEB) (44.3%). None of the strains contained the genes responsible for exfoliative toxin B (ETB) or staphylococcal enterotoxin D (SED) production, while the ability to produce staphylococcal enterotoxins C and E (SEC, SEE), as well as the toxic shock syndrome toxin (TSST), was detected in <1% of strains (Figure 3). The observed difference was related to the origin of the S. aureus strains. PVL was the most commonly produced toxin, regardless of the origin of the strains (Figure 4). PVL toxin was particularly prevalent in strains isolated from furuncles (89.5%) and pymyositis patients (89.2%). Other toxins were produced in various proportions depending on the origin of the strain (p < 0.0001). There was a significant difference in the detection of genes encoding toxins in MRSA strains (Figure 5).
Toxins production by the Staphylococcus aureus strains isolated from primary and secondary infections. PVL: Panton-Valentine Leukocidin; ETA: Exfoliative Toxin A; ETB: Exfoliative Toxin B; SEA: staphylococcal enterotoxin A; SEB: staphylococcal enterotoxin B; SEC: staphylococcal enterotoxin C; SED: staphylococcal enterotoxin D; SEE: staphylococcal enterotoxin E; SEG: staphylococcal enterotoxin G; SEH: staphylococcal enterotoxin H; SEI: staphylococcal enterotoxin I; TSST: Toxic-shock syndrome Toxin. Means ± standard deviations (SD) for three experiments are given. ***: p˂0.0001.
Specificity of the toxins production by the S. aureus strains isolated from primary and secondary infections. PVL: Panton-Valentine Leukocidin; ETA: Exfoliative Toxin A; ETB: Exfoliative Toxin B; SEA: staphylococcal enterotoxin A; SEB: staphylococcal enterotoxin B; SEC: staphylococcal enterotoxin C; SED: staphylococcal enterotoxin D; SEE: staphylococcal enterotoxin E; SEG: staphylococcal enterotoxin G; SEH: staphylococcal enterotoxin H; SEI: staphylococcal enterotoxin I; TSST: Toxic-shock syndrome Toxin. Means ± standard deviations (SD) for three experiments are given.
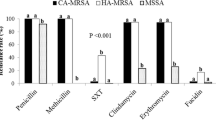
Effect of Methicillin resistance on the encoding toxins genes presence. PVL: Panton-Valentine Leukocidin; ETA: Exfoliative Toxin A; ETB: Exfoliative Toxin B; SEA: staphylococcal enterotoxin A; SEB: staphylococcal enterotoxin B; SEC: staphylococcal enterotoxin C; SED: staphylococcal enterotoxin D; SEE: staphylococcal enterotoxin E; SEG: staphylococcal enterotoxin G; SEH: staphylococcal enterotoxin H; SEI: staphylococcal enterotoxin I; TSST: Toxic-shock syndrome Toxin. Means ± standard deviations (SD) for three experiments are given. ***: P˂0.001; the other differences were not statistically significant (P˃0.05).
Discussion
The S. aureus stains analyzed in this study displayed a wide range of sensitivity to the 17 tested antibiotics. Generally, benzyl penicillin was not efficient in controlling the strains (Figure 1). This is consistent with previous reports showing high rates of S. aureus resistance (>90%) to benzyl penicillin [34, 35], suggesting that this antibiotic, one of the first to be introduced, is no longer effective against S. aureus[36]. A very high proportion of strains showed resistance to rifampicin (67%), tetracycline (60%), and trimetroprim/sulfamethoxazol (57%). This finding is consistent with previous studies performed in Africa [37–40]. The high proportion of strains showing resistance to penicillin and three other antibiotics may be explained by the practice of patient self-medication in Benin, and by the availability and low price of the antibiotics. These antibiotics can be bought without prescription, especially in developing countries. In our in vitro study, 4/17 tested antibiotics (vancomycin, fusidic acid, fosfomycin, and linezolid) were effective against all the S. aureus strains.
The collection of strains isolated from abscesses was sensitive to only four of the 17 tested antibiotics. However, strains isolated from furuncles (Figure 2b) and osteomyelitis patients (Figure 2d) were sensitive to 13 antibiotics (Figure 2). This result can likely be attributed to the origin of the strains. In our study, the samples collected from furuncles and osteomyelitis patients were from an extra-hospital community origin. Indeed, the selection pressure observed when using antibiotics in a hospital environment causes nosocomial strains to develop multi-resistance, in contrast to strains of community origin.
Of the 136 tested strains, 34 (25%) were resistant to oxacillin. This proportion of resistant strains appears to have increased steadily in Benin, compared with the recorded resistance rate of 11.6% in 1999 [40]. However, the overall proportion of oxacillin-resistant strains in Benin is still low compared to surrounding countries [41], even though methicillin is the preferred antibiotic for treatment of staphylococcal infections [42]. To efficiently control MRSA, vancomycin is recommended [43], and in our study we observed no resistance to vancomycin. Fortunately, vancomycin remains active against methicillin resistant strains of S. aureus.
In the current study of S. aureus strains isolated from skin, soft tissue, and bone related infections, PVL was the most prevalent toxin in our collection (70.0% of strains), followed by SEB (44.3%), SEG (35.5%), SEA (32.0%), SEH (28.8%), and SEI (28.9%). The genes encoding ETB and SED were not detected in any of the strains, while the SEE (0.8%), SEC (0.6%), and TSST (1%) genes were detected, but at a very low rate (Figure 3). This high detection frequency of the gene encoding PVL (p < 0.0001) was observed throughout the analysis, regardless of the origin of the sample (Figure 4). PVL appears to be a primordial toxin of S. aureus strains associated with skin, soft tissue, and bone related infections. These results are lower than the 96% of PVL-positive production strains we observed among S. aureus isolated from furuncle [20]. But the prevalence of PVL-positive S. aureus obtained in our study is higher than the 52.1% observed in Nigeria [44] and in cape Verdes Island [45]. The observe differences observed can be explain by the fact that in our study, we use various kind of strains. However, our result is close to the 72% obtained in Algeria [46]. Then, comparing with the other studies, we can say that the prevalence level of PVL ocus varies with geographical location, and clinical specimen [47] In the clinical field, PVL-positive S. aureus strains are more pathogenic than PVL-negative strains [22]. This is explained by the fact that the lytic activity of PVL directly affects monocytes, macrophages, polynuclear neutrophils, and metamyelocytes, although erythrocytes are not lysed by PVL [48]. PVL toxin is known to have a cytolytic effect, and as such polynuclear neutrophils were identified as important indicators of staphylococcal virulence [16]. Moreover, the cytolytic activity of PVL is observed at high toxin concentrations, while apoptosis is observed at low concentrations [49].
Regarding the ETs, only the gene encoding epidermolysin A (ETA) was detected, and in all cases the S. aureus strains were isolated from Buruli ulcers (Figure 4e). Such specific production of ETA by Buruli ulcers may be explained by the fact that ETs are known to be serine active proteases, with their activity highly specialized for desmoglein-1, an important epidermal protein [50, 51]. Therefore, the production of ETA by S. aureus strains from Buruli ulcers indicates a secondary role for S. aureus in the development of these ulcers, which are predominantly caused by Mycobacterium ulcerans.
Of the eight genes encoding enterotoxins, three (SEC, SED, and SEE) were not detected at all, while three others (SEA, SEB, and SEH) were detected at various proportions, regardless of the origin of the strains (Figure 4). Based on the homology of their sequences, the three detected enterotoxin genes belong to three different groups [52]. This explains the diversity of enterotoxins produced by S. aureus isolated from skin infections. Those toxins associated with food poisoning have antigenic and emetic activities [53–55]. The presence of enterotoxin genes in the strains isolated from skin, soft tissue, and bone related infections may be explained by human or environmental contamination, through the presence of open wounds. Similar observations are reported for TSST-1, which is the most prevalent toxin in cases of food poisoning [56].
Our study revealed that resistance to methicillin negatively correlates with toxin production (Figure 5). The difference in toxin production was extremely significant for PVL and some enterotoxins (B, G, and I) (p < 0.0001), and we observed that MSSA strains produced twice as many toxins as MRSA strains. These results suggest that the isolated strains were in majority Hospital acquired methicillin resistance S. aureus (HA-MRSA) because the community-acquired methicillin resistance S. aureus (CA-MRSA). Indeed, these CA-MRSA have an SCCmec type IV cassette conferring resistance to methicillin [57], and 77% of them harbor genes for Panton- Valentine leukocidin (PVL) [58, 59]. In addition, the prevalence of the genes for some toxins is higher in CA-MRSA than in HA-MRSA, suggesting that strains circulating in the community are more toxinogenic than hospital-associated strains [60]. Focusing on the duality of the observed activity between the resistance to methicillin and detection of the PVL-encoding gene, we may deduce that the resistance gene has a repressive activity against PVL. This observation was also made by Baldwin and Lowe [61], and mostly relates to HA-MRSA strains. In addition, we found that the presence of the methicillin resistance gene negatively impacts the expression of the gene encoding PVL. The emergence of MRSA in the hospital acquired strains may be viewed as disadvantageous in the selection of strains producing toxins, notably PVL. Indeed, mecA-encoded methicillin resistance involves β-lactamase production [62], which is not favorable for bacterial development [63]. Although community-acquired MRSA infections are increasingly frequent, the use of alternative antibiotics, such as vancomycin or ofloxacin/ciprofloxacin, is not appropriate because of the risk of the development of resistance to these antibiotics. Vancomycin is usually not available because of high costs and the necessity for assessing drug levels in the blood. Studies on the use of vancomycin for prophylaxis in medical centers with high rates of MRSA show that the use of this antibiotic is controversial in preventing some infections.
Conclusions
Our study showed that for S. aureus strains isolated from skin, soft tissue, and bone related infections, resistance to antibiotics depends on the origin of the strain. We attempted to perform a correlation analysis between toxin production, resistance to antibiotics, and the origin of samples. The S. aureus strains examined in this study produced a variety of toxins, with PVL, one of the most severe S. aureus toxins, being the most common amongst all of the strains. Overall, it is desirable to integrate to the current morphological and biochemical diagnostic analysis with virulence factor screening to accurately diagnose infectious disease mediated by S. aureus. This integrated diagnostic strategy will help to efficiently treat patients affected by pathogenic S. aureus strains. This study concerning skin, soft tissue, and bone related infections should be extended to include other types of infections in Benin.
Methods
Ethics statement
Ethical clearance was obtained from the Ministry of Public health of Benin Republic under protocol number: 2959/MSP/DC/SG/DRH/SPREA-05-2002. But it was important to notice that, the strains were de-identified and analyzed anonymously and the strains, not a human, were studied.
Samples collection
Clinical Staphylococcus aureus samples were collected from patients with skin, soft tissue at the National University Hospital of Cotonou (Benin) for various bacteriological screenings in routine, from November 2009 to March 2011. The incidence of secondary infections in Burili ulcer is unknown; antibiotics may be frequently prescribed for this indication. It is equally unknown which bacteria these antibiotics should target and what the sensitivity of these bacteria is. So the samples from Burili ulcer were screened for S. aureus. Theses samples were carried out during a prospective study made in a village of Lalo in Benin. Osteomyelitis and pyomyositis samples were collected during a prospective study made in a Hlagba Ouassa village in Benin. So these strains are considered as community strains and the others sample were isolated in hospital as stated previously.
S. aureus’ identification
Standard microbiological methods for identification of microorganisms were applied. All swabs were inoculated onto mannitol salt agar, incubated at 37°C and inspected visually for three days. Any suspected colony was subcultured on tryptic soy agar (bioMérieux) and identified by subsequent Gram staining, catalase test and Slidex Staph Plus (bioMérieux) and the coagulase test with the rabbit plasma [64]. Bacterial identification was performed by colony isolation on sheep blood agar plates and the automated Vitek 2 system.
Antibiotic susceptibility
Antimicrobial susceptibility was determined by the disc diffusion method of Kirby-Bauer on agar Mueller-Hinton (bioMérieux, Marcy l'Etoile, France) as recommended by the Antibiogram Committee of the French Microbiology Society (CASFM) [65]. After 24 h at 37°C, the zone of inhibition was measured. Susceptibility to vancomycin was confirmed by broth microdilution methodology as described by the Clinical and Laboratory Standards Institute (CLSI) [66]. For susceptibility to oxacillin, an inoculum of 107 CFU/ml was prepared and the plate was incubated at 37°C for 24 hours on Mueller-Hinton agar + 2% NaCl. Antibiotic disks were obtained from Biorad, Marne la Coquette, France.
The 17 tested antibiotics were: benzyl penicillin (10 UI), oxacillin (5 μg), cefoxitin screen (30 μg), gentamicin (10 UI), tobramycin (10 μg), kanamycin (30 μg), vancomycin (30 μg), teicoplanin (30 μg), fusidic acid (10 μg), fosfomycin (50 μg), rifampicin (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), erythromycin (15 μg), lincomycin (30 μg), pristinamycin (15 μg), linezolid (30 μg) and tetracyclin (30 UI).
Toxin detection
Phenotypic detection of toxins
For the phenotypic detection of toxins radial gel immunodiffusion was performed. The production of Panton-Valentine Leukocidin (PVL) and epidermolysins A (ETA) and B (ETB) were evidenced from culture supernatants after 18 h of growth in Yeast Casamino-acid Pyruvate (YCP) medium [67] by radial gel immunodiffusion in 0.6% (wt/vol) agarose with component-specific rabbit polyclonal and affinity-purified antibodies [68, 69].
Genotype detection of toxins
Presence of genes encoding for the 12 toxins, for which we don’t have antibody, was detected by Multiplex PCR using specific primers (Table 1) previously used for [70]. Then, the genes encoding for enterotoxins A (sea), B (seb), C (sec), D (sed), E (see), G (seg), H (seh), I (sei) and tsst were analyzed. Additionally, genes encoding PVL, ETA and ETB were also detected. Briefly, total DNA was purified using QIAamp® DNA Mini Kit (Qiagen, GmbH, Germany) with a Gene Amp® PCR System 9700 (Perkin-Elmer, Norwalk, USA) and amplified in a total volume of 50 μl containing 25 pmoles of each primer, 50 ng of total DNA, 1.5 mM MgCl2, 200 μM of dNTP mixture, 1× PCR reaction Buffer and 5 units of Taq™ DNA polymerase (Invitogen™). The thermal cycling conditions included an initial denaturation step (2 min at 92°C) followed by 35 cycles of amplification comprising three steps: 2 min denaturation for 92°C, 1 min annealing at 50°C, 2 min extension at 72°C. The reaction was terminated with 3 min extension at 72°C. PCR products were analysed by electrophoresis through 1.4% (wt/vol) agarose gel (Euromedex, Mundolsheim, France).
Staphylococcal enterotoxins production (SEA, SEB, SEC, SED, SEE, SEG, SEH, SEI) by Bio-Plex Assay (xMAP Multiplex assay)
The centrifuged supernatant (3ml) of S. aureus grown on BHI at 37°C (night) was recovered and diluted ½ in TBS-Tween20-IgG rabbit nonspecific at 100μg/mL, and incubated for 30 min at room temperature. The Bio-Plex assays consisted of three incubation steps that were performed into flat-bottom Multiscreen microplates (pores diameter = 1.2 μm, Millipore) according to the previous describe method [71]. Any steps were separated by three washes into TBS-Tween 20.
Statistical analysis
Microsoft Excel Spreadsheet has been used for data processing. For comparison tests of positive isolates of each patient group, we used the Student T test, and the Fischer’s test for lower number series (GraphPad Prism 5). P < 0.05 was considered statistically significant.
References
Kluytmans A, van Belkum A, Verbrugh H: Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbial Rev. 1997, 10 (3): 505-520.
Nubel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M: Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008, 105: 14130-14135.
Holmes A, Ganner M, McGuane S, Pitt TL, Cookson BD, Kearns AM: Staphylococcus aureus Isolates Carrying Panton-Valentine Genes in England and Wales: Frequency, Characterization, and Association with Clinical Desease. J Clin Microbiol. 2005, 43 (5): 2384-2390.
Baba-Moussa L, Anani L, Scheftel JM, Couturier M, Riegel P, Haikou N, Hounsou F, Monteil H, Sanni A, Prevost G: Virulence factors produced by strains of Staphylococcus aureus isolated from urinary tract infections. J Hosp Infect. 2008, 68: 32-38.
Baba-Moussa L, Ahissou H, Azokpota P, Assogba B, Atindéhou M, Anagonou S, Keller D, Sanni A, Prévost G: Toxins and adhesion factors associated with Staphylococcus aureus strains isolated from diarrheal patients in Benin. Afr J Biotechnol. 2010, 9: 604-611.
Bohach GA, Fast DJ, Nelson RD, Schlievert PM: Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990, 17: 251-272.
Breneman DL: Bacterial infection of the skin and soft tissues and their treatment. Curr Opin Infect Dis. 1993, 6: 678-682.
Murray DL, Ohlendorf DH, Schlievert PM: Staphylococcal and streptococcal superantigens: their role in human diseases. ASM News. 1995, 61: 229-235.
Dinges MM, Orwin PM, Schlievert PM: Exotoxins of Staphylococcus aureus. Clin Microbiol Rev. 2000, 13: 16-34.
Barg NL, Harris T: Toxin-mediated syndromes. The staphylococci in human disease. Edited by: Crossley KB, Archer GL. 1997, New York: Churchill Livingstone, 527-544.
Durupt F, Mayor L, Bes M, Reverdy ME, Vandenesch F, Thomas L, Etienne J: Prevalence of Staphylococcus aureus toxins and nasal carriage in furuncles and impetigo. Br J Dermatol. 2007, 157: 1161-1167.
Gladstone GP, Van Heyningen WE: Staphylococcal leucocidins. Br J Exp Pathol. 1957, 38: 123-137.
Woodin AM: Fractionation of a leucocidin from Staphylococcus aureus. Bioch J. 1959, 73: 225-237.
Szmigielski S, Sobiczewska E, Prévost G, Monteil H, Colin DA, Jeljaszewicz J: Effect of purified staphylococcal leukocidal toxins on isolated blood polymorphonuclear leukocytes and peritoneal macrophages in vitro. Zentralbl Bakteriol. 1998, 288: 383-394.
Hongo I, Baba T, Oishi K, Morimoto Y, Ito T, Hiramatsu K: Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by Panton- Valentine leukocidin. J Infec Dis. 2009, 200: 715-723.
Cribier B, Prevost G, Couppie P, Finck-Barbancon V, Grosshans E, Piemont Y: Staphylococcus aureus leukocidin: a new virulence factor in cutaneous infections? An epidemiological and experimental study. Dermatology. 1992, 185: 175-180.
Couppié P, Cribier B, Prévost G, Grosshans E, Piémont Y: Leucocidin from Staphylococcus aureus and cutaneous infections: an epidemiological study. Arch Dermatol. 1994, 130: 1208-1209.
Prevost G, Cribier B, Couppie P, Petiau P, Supersac G, Finck-Barbancon V, Monteil H, Piemont Y: Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995, 63: 4121-4129.
Lina G, Piémont Y, Godail-Gamot F, Bès M, Peter MO, Gauduchon V, Vandenesh F, Etienne J: Involvement of Panton Valentine leukocidineproducing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999, 29: 1128-1132.
Baba-Moussa L, Sina H, Scheftel JM, Moreau B, Sainte-Marie D, Kotchoni SO, Prevost G, Couppie P: Staphylococcal Panton-Valentine leucocidin as a major virulence factor associated to furuncles. PLoS One. 2011, 6: e25716-
Couppié P, Hommel D, Prévost G, Godart MC, Moreau B, Sainte- Marie D, Peneau C, Hulin A, Monteil H, Pradinaud R: Septicémie à Staphylococcus aureus, furoncle et leucocidine de Panton et Valentine: 3 observations. Ann Dermatol Venereol. 1997, 124: 684-686.
Gillet Y, Issartel B, Vanhems P, Fournet JC, Lina G, Bes M, Vandenesch F, Piémont Y, Brousse N, Floret D, Etienne J: Association between Staphylococcus aureus strains carrying gene for Panton Valentin leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002, 359: 753-759.
Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Höök M, Etienne J, Vandenesch F, Bowden MG: Staphylococcus aureus Panton Valentine leukocidin causes necrotizing pneumonia. Science. 2007, 315 (5815): 1130-1133.
Diep BA, Palazzolo-Ballance AM, Tattevin P, Basuino L, Braughton KR, Whitney AR, Chen L, Kreiswirth BN, Otto M, DeLeo FR, Chambers HF: Contribution of Panton Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PLos One. 2008, 3: e3198-
Gillet Y, Dohin B, Dumitrescu O, Lina G, Vandenesch F, Etienne J, Floret D: Osteoarticular infections with Staphylococcus aureus secreting Panton Valentine leucocidin. Arch Pediatr. 2007, 14 (Suppl 2): 102-107.
Hussain A, Robinson GO, Malkin J, Duthie M, Kearns A, Perera N: Purpura fulminans in a child secondary to Panton Valentine leukocidinproducing Staphylococcus aureus. J Med Microbiol. 2007, 56: 1407-1409.
Shivashankar GH, Murukesh N, Varma MP, Sharif IM, Glynn G: Infection by Panton Valentine leukocidin-producing Staphylococcus aureus clinically mimicking Lemierre’s syndrome. J Med Microbiol. 2008, 57: 118-120.
Burton MJ, Shah P, Swiatlo E: Community-acquired methicillin resistant Staphylococcus aureus as a cause of Fournier’s gangrene. J Med Sci. 2008, 335: 327-328.
Dumitrescu O, Boisset S, Badiou C, Bes M, Benito Y, Reverdy ME, Vandenesch F, Etienne J, Lina G: Effect of antibiotics on Staphylococcus aureus producing Panton-Valentine leukocidin. Antimicrob Agents Chemother. 2007, 51: 1515-1519.
Deleo FR, Otto M, Kreiswirth BN, Chambers HF: Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010, 375: 1557-1568.
Deurenberg RH, Stobberingh EE: The evolution of Staphylococcus aureus. Inf Genet Evol. 2008, 8: 747-763.
Holmes NE, Johnson PD, Howden BP: Relationship between Vancomycin-Resistant Staphylococcus aureus, Vancomycin-Intermediate S. aureus, High Vancomycin MIC, and Outcome in Serious S. aureus Infections. J Clin Microbiol. 2012, 50: 2548-2552.
Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Kobayashi I: Dissemination in Japanese hospitals of strains of S. aureus heterogeneously resistant to vancomycin. Lancet. 1997, 350: 1670-1673.
Denton M, O'Connell B, Bernard P, Jarlier V, Wiliams Z, Santerre Henriksen A: The EPISA Study: antimicrobial susceptibility of Staphylococcus aureus causing primary or secondary skin and soft tissue infections in the community in France, the UK and Ireland. J Antimicrobial Chemother. 2008, 61 (3): 586-588.
Elazhari M, Saile R, Dersi N, Timinouni M, Elmalki A, Zriouil SB, Hassar M, Zerouali K: Activité de 16 Antibiotiques vis-à-vis des Staphylococcus aureus communautaires à Casablanca (Maroc) et Prévalence des Souches Résistantes à la Méthicilline. Eur J Sci Res. 2009, 30: 128-137.
Cohen ML: Epidemiology of drug resistance: implications for a post-antimicrobial era. Science. 1992, 257: 1050-1055.
Sina H, Baba-Moussa F, Ahoyo TA, Mousse W, Anagonou S, Gbenou JD, Prévost G, Kotchoni SO, Baba-Moussa L: Antibiotic susceptibility and Toxins production of Staphylococcus aureus isolated from clinical samples from Benin. Afr J Microbiol Res. 2011, 5: 2797-2808.
Randrianirina F, Soares JL, Ratsima E, Carod JF, Combe P, Grosjean P, Richard V, Talarmin A: In vitro activities of 18 antimicrobial agents against Staphylococcus aureus isolates from the Institut Pasteur of Madagascar. Ann Clin Microbiol Antimicrob. 2007, 6: 5-
Kesah C, Ben Redjeb S, Odugbemi TO, Boye CS, Dosso M, Ndinya Achola JO, Koulla-Shiro S, Benbachir M, Rahal K, Borg M: Prevalence of methicillin-resistant Staphylococcus aureus in eight African hospitals and Malta. Clin Microbiol Infect. 2003, 9: 153-156.
Baba-Moussa L, Sanni A, Dagnra AY, Anagonou S, Prince-David M, Edoh V, Befort JJ, Prévost G, Monteil H: Approche épidémiologique de l'antibiorésistance et de la production de leucotoxines par les souches de Staphylococcus aureus isolées en Afrique de l'Ouest. Med Mal Infect. 1999, 29 (11): 689-696.
Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Beach M: Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin Infect Dis. 2001, 32 (suppl 2): 114-132.
Belabbès H, Elmdaghri N, Hachimi K, Marih L, Zerouali K, Benbachir M: Résistance de Staphylococcus aureus isolé des infections communautaires et hospitalieres à Casablanca. Communication brève. Med Mal Infect. 2001, 31: 25-28.
Maor Y, Hagin M, Belausov N, Keller N, Ben-David D, Rahav G: Clinical features of heteroresistant vancomycin-intermediate Staphylococcus aureus bacteremia versus those of methicillin-resistant S. aureus bacteremia. J Infect Dis. 2009, 199: 619-624.
Okon KO, Uba A, Oyawoye OM, Yusuf IZ, Adesina OO: Prevalence and Antibiotic susceptibility pattern of Panton-Valentine Leucocidin (PVL) positive Staphylococcus aureus Strains from clinical specimens in Northeastern Nigeria. Sierra Leone J Biomed Res. 2012, 4 (1): 43-52.
Aires de Sousa M, Conceicao T, de Lancastre H: Unusually high prevalence of nosocomial Panton-Valentine leukocidin –positive Staphylococcus aureus isolates in Cape Verdes Island. J Clin Microbiol. 2006, 44: 37-3793.
Campbell SJ, Deshmukl HS, Nelson CL, Bae I: Genotypic characterization of Staphylococcus aureus isolates from a multinational trial of topical drugs for skin and skin stricture infections. J Clin Microbiol. 2008, 46: 678-684.
Goering RV, Shawar RM, Scangarella NE, OHara FP, Amrine-Madsen H, West JM, Dalessandro M, Becker JA, Walsh SL, Miller LA, van Horn SF, Thomas ES, Twynholm T: Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trial. J Clin Microbiol. 2008, 9: 2842-2847.
Prévost G, Mourey L, Colin DA, Menestrina G: Staphylococcal pore-forming toxins. Curr Top Microbiol Immunol. 2001, 257: 53-83.
Genestier AL, Michallet MC, Prevost G, Bellot G, Chalabreysse L, Peyrol S, Thivolet F, Etienne J, Lina G, Vallette FM, Vandenesch F, Genestier L: Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J Clin Invest. 2005, 115: 3117-3127.
Ladhani S: Understanding the mechanism of action of the exfoliative toxins of Staphylococcus aureus. FEMS Immunol Med Microbiol. 2003, 39: 181-189.
Prevost G, Couppie P, Monteil H: Staphylococcal epidermolysins. Curr Opin Infect Dis. 2003, 16: 71-76.
Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J: Community acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003, 9: 978-984.
Morris CA, Conway HD, Everall PH: Food-poisoning due to staphylococcal enterotoxin E. Lancet. 1972, 2: 1375-1376.
Chen TR, Chiou CS, Tsen HY: Use of Novel PCR Primers Specific to the Genes of Staphylococcal Enterotoxin G, H, I for the Survey of Staphylococcus aureus Strains Isolated From Food-Poisoning Cases and Food Samples in Taiwan. Int J Food Microbiol. 2004, 92: 189-197.
Ikeda T, Tamate N, Yamaguchi K, Makino S: Mass Outbreak of Food Poisoning Disease Caused by Small Amounts of Staphylococcal Enterotoxins A and H. Appl Environ Microbiol. 2005, 71: 2793-2795.
Daum RS, Ito T, Hiramatsu K, Hussain F, Mongkolrattanothai K, Jamklang M, Boyle-Vavra S: A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J Infect Dis. 2002, 186: 1344-1347.
Naimi TS, LeDell KH, Como-Sabetti K, et al: Comparison of com-munity- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA. 2003, 290: 2976-2984.
Buckingham SC, McDougal LK, Cathey LD, et al: Emergence of com-munity-associated methicillin-resistant Staphylococcus aureus at a Memphis, Tennessee Children’s Hospital. Pediatr Infect Dis J. 2004, 23: 619-624.
Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y: Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003, 36: 53-59.
Bergdoll MS, Crass BA, Reiser RF, Robbins RN, Davis JP: A New Staphylococcal Enterotoxin, Enterotoxin F, Associated with Toxic-Shock-Syndrome Staphylococcus aureus Isolates. Lancet. 1981, 1: 1017-1021.
Baldwin LN, Lowe AD: Panton-Valentine Leukocidin associated with community acquired methicillin resistant Staphylococcus aureus: a case report and review of interim guidelines. Anaesthesia. 2008, 63: 764-766.
Chambers HF: Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997, 10: 781-791.
Labischinski H: Consequences of the interaction of beta-lactam antibiotics with penicillin binding proteins from sensitive and resistant Staphylococcus aureus strains. Med Microbiol Immunol. 1992, 181: 241-265.
Cheesbrough M: District Laboratory Practice in Tropical Countries: Part 2. 2004, Cambridge, UK: Cambridge University Press, 299-329.
Société Française de Microbiologie: Recommandations du Comité de l’Antibiogramme de la Société Française de Microbiologie. 2012, http://www.sfm-microbiologie.org/UserFiles/file/CASFM/CASFM_2012.pdf,
Clinical and Laboratory Standards Institute: Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved standard. 2009, Document M7-A8. Clinical and Laboratory Standards Institute Wayne: PA, 8, ISBN 1-56238-689-1
Gauduchon V, Werner S, Prévost G, Monteil H, Colin DA: Flow cytometric determination of Panton-Valentine leucocidin S component binding. Infect Immun. 2001, 69: 2390-2395.
Prévost G, Couppie P, Prévost P, Gayet S, Petiau P, Cribier B, Monteil H, Piemont Y: Epidemiological data on Staphylococcus aureus strains producing synergohymenotropic toxins. J Med Microbiol. 1995, 42: 237-245.
Gravet A, Colin DA, Keller D, Girardot R, Monteil H, Prevost G: Characterization of a novel structural member, LukE-LukD, of the bi-component staphylococcal leucotoxins family. FEBS Lett. 1998, 436: 202-208.
Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F: Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002, 70: 631-641.
Joubert O, Keller D, Pinck A, Monteil H, Prevost G: Sensitive and specific detection of staphylococcal epidermolysins A and B in broth cultures by flow cytometry-assisted multiplex immunoassay. J Clin Microbiol. 2005, 43: 1076-1080.
Acknowledgements
This work was supported by Benin Government which granted Haziz Sina and by grant EA-4438 from “Direction de la Recherche et des Etudes Doctorales” of Unversity of Strasbourg in France.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
Authors declare no conflict of interest.
Authors’ contributions
Conception and design of the study: LB-M and GP. Acquisition of data: HS, AT-A, WM, YB, HB. Analysis and interpretation of data: LB, GP, YS. Drafting the article: LB-M, SOK, and HS. Revising it critically for important intellectual content: LB-M, GP, SOK, YS. Final approval of the version to be submitted: All the co-authors. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Sina, H., Ahoyo, T.A., Moussaoui, W. et al. Variability of antibiotic susceptibility and toxin production of Staphylococcus aureus strains isolated from skin, soft tissue, and bone related infections. BMC Microbiol 13, 188 (2013). https://doi.org/10.1186/1471-2180-13-188
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-13-188