Abstract
Background
Helicobacter pylori is known to be a gastric pathogen of humans. Eradication regimens for H. pylori infection have some side effects, compliance problems, relapses, and antibiotic resistance. Therefore, the need for alternative therapies for H. pylori infections is of special interest. We have previously shown that polyphenols from almond skins are active against a range of food-borne pathogens. The aim of this study was to evaluate the antibacterial effects of natural almond skins before and after simulated human digestion and the pure flavonoid compounds epicatechin, naringenin and protocatechuic acid against H. pylori.
Results
H. pylori strains were isolated from gastric biopsy samples following standard microbiology procedures. Also, cagA and vacA genes were identified using PCR. Susceptibility studies on 34 strains of H. pylori, including two reference strains (ATCC 43504, ATCC 49503), were performed by the standard agar dilution method.
Natural almond skin was the most effective compound against H. pylori (MIC range, 64 to 128 μg/ml), followed by natural skin post gastric digestion (MIC range, 128 to 512 μg/ml), and natural almond skin post gastric plus duodenal digestion (MIC range, 256 to 512 μg/ml). Amongst the pure flavonoid compounds, protocatechuic acid showed the greatest activity (MIC range, 128 to 512 μg/ml) against H. pylori strains.
Conclusions
Polyphenols from almond skins were effective in vitro against H. pylori, irrespective of genotype status and could therefore be used in combination with antibiotics as a novel strategy for antibiotic resistance.
Similar content being viewed by others
Background
Helicobacter pylori is a microaerophilic Gram-negative bacterium which colonizes the human gastric mucosa. It is known to be a gastric pathogen of humans associated with chronic gastritis, peptic ulcers, atrophic gastritis, intestinal metaplasia and lymphoma or cancer development [1, 2]. Approximately 50% of the world population is infected with H. pylori, with prevalence rates ranging from 20% to more than 80% in certain countries [3]. H. pylori has been identified as group 1 carcinogen by the International Agency for Research on Cancer [4]. The observation that only a subset of infected individuals develops severe gastroduodenal diseases may depend on the virulence of the infecting organism. Amongst the different genetic determinants involved in H. pylori virulence are the cytotoxin-associated gene (cagA) and the vacuolating cytotoxin gene (vacA). VacA, which is present in all H. pylori strains, contains at least two variable parts relevant to virulence [5]. The s region encoding the signal peptide exists as s1 or s2 allelic types, and the m region (middle) occurs as m1 and m2 allelic types [6]. CagA, which is not present in every H. pylori strain [7], is a marker for a pathogenicity island (PAI) [8] associated with more severe clinical outcomes [9]. It has also been demonstrated that CagA is required to disrupt the organization of apical junctions and perturb epithelial differentiation [10]. Type s1/m1 strains produce a higher level of cytotoxin activity than other genotypes. A strong association between cagA and vacA signal sequence type s1 has been reported [5]. Strains carrying s1 m1 mosaic combination secrete vacuolating cytotoxin in contrast to those with s2 m2 activity [11].
The standard treatment for H. pylori related disease is a combination of antimicrobial agents and anti-acid agents [12]. However, side effects for these regimes are common and a major concern is the development of antimicrobial resistance [13]. As a result, several naturally occurring substances have been investigated as potential alternatives for the treatment of H. pylori infection [14–18].
Almonds (Prunus dulcis D.A. Webb) are a rich source of nutrients and phytochemicals such as vitamin E, monounsatured fatty acids and polyunsatured fatty acids [19]. Other health promoting compounds mainly present in almond skins are polyphenols which have been shown to be bioaccessible during simulated digestion in the gut [20, 21]. Among polyphenols, flavonoids are secondary metabolites well documented for their biological effects, including anticancer, antiviral, antimutagenic, anti-inflammatory and antimicrobial activities [22–24]. We have previously demonstrated that polyphenols from almond skins are active against Gram-positive bacteria including Staphylococcus aureus and Listeria monocytogenes and the Gram-negative Salmonella enterica[25]. Natural almond skins also induced a significant decrease in Herpes simplex virus type 2 replication [26]. The antioxidant and anti-inflammatory potential of almond skin polyphenols has also been demonstrated using an experimental model of inflammatory bowel disease [27].
The aim of the present study was to investigate the antimicrobial properties of natural almond skins before and after simulated human digestion in the upper GI tract and the pure flavonoid compounds epicatechin, naringenin and protocatechuic acid against H. pylori strains isolated from gastric biopsies of subjects attending an outpatient clinic in Southern Italy. Their clinical relevance has also been elucidated.
Methods
Almond skins
Natural almond skins (NS) were prepared from Californian almonds by treatment with liquid nitrogen as previously reported [20].
In vitro digestion studies
The protocol used to simulate digestion of natural almond skins under gastric and duodenal conditions in vitro has been previously described [21].
Briefly, for the gastric digestion, 1.5 g of NS was suspended in 12.4 mL acidic saline (150 mM NaCl, pH 2.5) and readjusted to pH 2.5 with HCl. Phosphatidylcholine (Lipid Products, UK) vesicle suspension, pepsin (Sigma, UK) and gastric lipase analogue (Amano Enzyme, Japan) were added so that the final concentrations were 2.4 mmol/L, 146 U/mL and 60 U/mL, respectively. Gastric digestion was performed in a shaking incubator (170 rpm, 37°C) for 2 h.
For the simulated gastric plus duodenal digestion, the pH was raised to 6.5 by addition of NaOH and the following enzymes were added: α-chymotrypsin (Sigma, 5.9 U/mL), trypsin (Sigma, 104 U/mL), colipase (Sigma, 3.2 μg/mL), pancreatic lipase (Sigma, 54 U/mL), and α-amylase (Sigma, 25 U/mL) in the presence of sodium taurocholate (4 mmol/L) and sodium glycodeoxycholate (4 mmol/L). Gastric plus duodenal digestion was performed in a shaking incubator (170 rpm, 37°C) for 1 h.
Almond skin extracts
Polyphenol-rich extracts from NS, NS post in vitro gastric digestion (NS G) and NS post in vitro gastric plus duodenal digestion (NS G + D) were prepared as previously described and their composition has been previously reported [21].
Patients, H. pyloristrains and culture conditions
Two reference American Type Culture Collection strains of H. pylori (ATCC 43504 and ATCC 49503) and thirty two clinical isolates recovered from gastric biopsy samples of dyspeptic adults (23 women, 9 men; average age, 51 years) undergoing digestive endoscopy at the Endoscopy Unit of the Department of Internal Medicine of the University of Messina, Messina, Italy, were used in this study. None of the patients had previously undergone eradication therapy. All study subjects gave their informed consent and the study was approved by the local ethical committee (Comitato Etico Scientifico A.O.U. Policlinico “G. Martino” Messina, Italy). Diagnosis of peptic ulcer (PU) and non-ulcer dyspepsia (NUD) or gastritis was based on endoscopic examination of the stomach and duodenum. Biopsy samples were taken for each patient for culture. Isolates were derived from patients suffering from gastritis (n = 27; 84.37%), or NUD (n = 5; 15.62%).
Gastric biopsy specimens for culture were placed in the sterile screw-capped tubes containing 0.5 ml sterile saline and transported to the microbiology laboratory within 2 h. The samples were soaked and sowed in selective (Pylori agar, BioMérieux) and non-selective (Columbia agar with 7% horse blood, CB, Oxoid) culture media. Cultures were incubated for 7 days at 37°C under microaerophilic conditions. Grown bacteria were identified as H. pylori by typical morphology, Gram staining results and positive reactions to oxidase, catalase, and urease activities. The cagA and vacA status as a virulence factors have been determined in all strains by PCR method.
All strains were harvested by suspension in Brucella broth (Difco) supplemented with 10% fetal bovine serum (BB, Euroclone) and 30% glycerol and stored in liquid nitrogen until used.
DNA extraction from H. pyloriisolates
DNA was extracted from H. pylori isolates using the QIAamp DNA Mini Kit (Qiagen, Milan, Italy) according to the manufacturer’s instructions. Briefly, one colony was harvested from an agar plate and added to an appropriate volume of phosphate-buffered saline homogenized by vortexing. Twenty microliters of a proteinase K solution (20 mg/mL) and 200 μL of buffer AL provided in the kit were then added, followed by incubation at 56°C for 10 min. Next, 200 μL of ethanol (96%) were added. The mixture was then loaded onto the QIAamp spin column provided in the kit and centrifuged at 6000 g for 1 min. The QIAamp spin column was placed in a 2-mL collection microtube, and the tube containing the mixture was discarded. The column material was washed (500 μL each) with the first washing buffer (buffer AW1) and with the second washing buffer (buffer AW2) provided in the kit. Finally, the DNA was eluted with 150 μL of a third buffer (buffer AE) provided in the kit.
Oligonucleotide primers
The primers targeting the vacA gene (region m and region s) and cagA genes [28] used in the PCR assay for the analysis of H. pylori isolates, are reported in Table 1. The primers were synthesised by MWG-Biotech AG (Mannheim, Germany).
PCR conditions
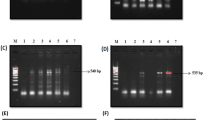
The amplification was performed using a PCR SprintThermal Cycler (Hybaid, Ashford, UK) and carried out in 50 μL reaction volume containing 200 μmol/L (each) dNTP, 0.1 μmol/L (each) primer, 1X PCR buffer, 50 mmol/L KCl, 10 mmol/L Tris–HCl pH 8.8, 0.1% Triton X-100, 50 mmol/L MgCl2, 2 U of Taq DNA polymerase and 5 μL of template DNA or water for the negative control. The temperature profile for the PCR was as follows: an initial step of 4 min at 95°C, followed by a denaturation step for 1 min at 95°C, an annealing step for 1 min at 52°C (for vacA PCR) or 59°C (for cagA PCR), and a primer extension step for 1 min at 72°C. After the 35th cycle, the extension step was prolonged for 10 min in order to complete synthesis of all strands after which the samples were kept at 4°C until analysis. A negative control lacking of the DNA template was included in each experiment. The H. pylori strains used as positive controls in the PCR tests included H. pylori ATCC 43504 and H. pylori ATCC 49503. Detection of PCR products was performed by gel electrophoresis. Samples (5 μL) of final PCR products were loaded onto 1.5% agarose gel and subjected to electrophoresis in 1X TAE (0.04 mol/L Tris–acetate, 0.001 mol/L EDTA) buffer for 60–90 min at 100 V. The gels were stained with ethidium bromide and photographed under UV light trans-illumination. A 100-bp DNA ladder (BioLab New England, Celbio, Milan, Italy) was included on each gel as a molecular size standard.
Susceptibility testing
The minimum inhibitory concentration (MIC) was assayed by the standard agar dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [29] using CB. Twofold serial dilutions of the compound tested ranging from 0.016 μg/mL to 1.024 μg/mL were used. Frozen stock cultures were thawed and subcultured on CB and grown for 3 days under microaerophilic conditions. Bacterial growth was taken from the plates, resuspended in BB and grown under shaking (125 rpm) at 37°C for 24 h. H. pylori cultures in the exponential phase of growth were diluted with BB to contain about 5 × 107 CFU/mL by adjusting the turbidity of the suspension to match the MacFarland no. 1 standard. Ten-microliter aliquots of the suspension were inoculated on CB containing twofold serial dilutions of the compound tested. Compound-free CB media were included in each experiment to confirm the viability of the inoculum and to observe the growth of any contaminants. CB incorporating twofold serial dilutions of the solvent dimethil sulfoxide was included as a growth control to ensure that the viability of the H. pylori strains was not affected by the dimethil sulfoxide used to dissolve the compound. All plates were incubated at 37°C in a microaerophilic atmosphere and examined after 3 days. For quality control, H. pylori ATCC strains 43504 and 49503 were tested in each run. Amoxicillin (Sigma Aldrich S.r.l., Italy), and clarithromycin (Abbott S.p.A., Italy), were used as control compounds for comparative analyses. According to CLSI breakpoints, the resistance breakpoints were 0.5 μg/mL for amoxicillin and 1 μg/mL for clarithromycin [29]. The MIC was considered the lowest concentration at which the compound inhibited the development of visible bacterial growth on the agar plates. All MIC determinations were performed in duplicate for each strain.
Results
To type the H. pylori strains isolated from the patients examined in this study, we amplified by PCR different alleles of the genes of the two major virulence factors of this bacteria, cagA and vacA. The amplification results are shown in Table 2. Fifteen out of 32 H. pylori isolates were cagA positive, representing 55.5% (15/27) of the isolates recovered from patients with gastritis. No strain identified from patients with NUD was cagA positive. The prevalence of the allelic variants of s1 and m1 of vacA was higher in the strains isolated from patients with gastritis compared with the strains isolated from NUD patients (77.8% versus 60%, and 63% vs 40%, respectively). When the cagA and vacA genotypes were combined and analyzed in relation to the clinical outcome (Table 3), the cagA + strains with the allelic variant s1m1 of vacA were only present in the strains isolated from gastritis patients (53.3%).
The MIC values of natural almond skin (NS), NS post in vitro gastric digestion (NS G) and NS post in vitro gastric plus duodenal digestion (NS G + D) against 34 H. pylori strains including 2 ATCC H. pylori strains are shown in Table 4. Results of negative controls containing DMSO (maximum 1% v/v) indicated the complete absence of inhibition of all the H. pylori strains tested (data not shown). All extracts inhibited the growth of both the clinical isolates and the reference strains. As expected, NS was the most effective (MIC range, 64 to 128 μg/mL), followed by NS G (MIC range, 128 to 512 μg/mL) and NS G + D (MIC range, 256 to 512 μg/mL). MIC values of 64, 128 and 256 μg/mL NS, NS G and NS G + D, respectively, inhibited the growth of 50% of the H. pylori tested strains. These results clearly confirm that all three polyphenol- rich extracts acted as good growth inhibitors against H. pylori with different virulence irrespective of the cagA and vacA status. In other words, there was no difference in the suppression of growth between the 8 H. pylori clinical isolates harboring the cagA+/vacAs1/m1 genotype, including the quality control strains (ATCC 43504 and 49503), and the other H. pylori genotypes.
The MIC results of epicathechin, naringerin and protocatechuic acid against H. pylori strains are reported in Table 5. Protocatechuic acid showed the greatest activity with MIC values of 128 μg/mL and 256 μg/mL against 50% and 90% of the tested strains, respectively. Epicatechin was the least effective compound against H. pylori (MIC of 512 μg/mL against 50% of the H. pylori strains).
All H. pylori strains tested were susceptible to amoxicillin (MIC90 0.25 μg/mL; range between 0.016 – 0.25 μg/mL). The MIC90 value of clarithromycin against H. pylori isolates was 0.5 μg/mL with MIC values ranging between 0.016 and 4 μg/mL. Two (6%) out of 32 isolates tested were clarithromycin resistant, one of which was isolated from patients suffering from gastritis harbouring the cagA+/vacAs1/m1 genotype.
The two clarithromycin-resistant strains were inhibited by almond skin extracts (NS, NS G, NS G + D) at 128 μg/mL; the MIC values of pure compounds (epicatechin, naringenin, protocatechuic acid) against these two strains were 256, 256, and 128 μg/mL, respectively.
Quality control MICs were within acceptable limits for all antimicrobial susceptibility testing.
Discussion
The results reported in the present paper demonstrated that polyphenols present in almond skins are effective against H. pylori strains, both ATCC and clinical isolates. As previously reported [21, 26], NS was the most active against the tested strains. This result could be due to the highest polyphenols concentration in NS, whereas a decrease in the total phenolic content was observed post in vitro gastric and post in vitro gastric plus duodenal digestion [21]. Catechin, epicatechin, kaempferol (aglycone and conjugated) and isorhamnetin (aglycone and conjugated) were the major compounds identified in NS [21], leading to assume the combination of these polyphenols was responsible for the higher activity against H. pylori. Quercetin and kaempferol were shown to be active against a CagA + and a CagA- strain of H. pylori and a relationship between antimicrobial potential and antioxidant activity was only reported for the CagA- G 21 strain [18]. The same authors have also recently reported an increased susceptibility to resveratrol of H. pylori strains isolated from patients suffering from gastric carcinomas [30]. The investigation of the isolated compounds in the present work demonstrated that protocatechuic acid was more active than naringenin and epicatechin and the effectiveness of protocathechic acid against H. pylori in broth and stomach homogenates from mice has also been demonstrated by Liu et al. [31], with no differences between antibiotic susceptible and resistant strains. Other investigations have reported promising effect of natural compounds, such as hydrolysable tannins and lignans, on the proliferation of H. pylori and the prevention of gastric carcinogenesis [32, 33]. Reports on the mechanism of action of a range of flavonoids have shown that isoflavones and chalcones inhibited the urease secreted by H. pylori to survive the acidic conditions found in the stomach [34, 35]. Other flavonoids may also be responsible for the neutralization of the vacA via interference of the toll-like receptor 4 signaling induced by H. pylori[36, 37]. A recent study reported that the antimicrobial potential of the oligopeptide C12K-2 against H. pylori has a dual mode of action on both membrane and cytoplasmatic components [38]. Although the rate of resistance to clarithromycin has significantly increased in several countries (13), the observed resistance to this antibiotic in the H. pylori isolates tested in the present work was surprisingly low (6%).
Conclusions
In conclusion, we have shown that polyphenols from almond skins were effective in vitro against H. pylori, irrespective of the bacterial genotype which is independent of the presence of the cagA, and could therefore be used in combination with antibiotics as a novel strategy for antibiotic resistance.
References
Ferreira AC, Isomoto H, Moriyama M, Fujioka T, Machado JC, Yamaoka Y: Helicobacter and gastric malignancies. Helicobacter. 2008, 13: 28-34.
Kandulski A, Selgrad M, Malfertheiner P: Helicobacter pylori infection: a clinical overview. Dig Liver Dis. 2008, 40: 619-626. 10.1016/j.dld.2008.02.026.
Minami M, Ando T, Hashikawa SN, Torii K, Hasegawa T, Israel DA, Ina K, Kusugami K, Goto H, Ohta M: Effect of glycine on Helicobacter pylori in vitro. Antimicrob Agents Chemother. 2004, 48: 3782-3788. 10.1128/AAC.48.10.3782-3788.2004.
Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R: Helicobacter pylori virulence and genetic geography. Science. 1999, 284: 1328-1333. 10.1126/science.284.5418.1328.
Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL: Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995, 270: 17771-17777. 10.1074/jbc.270.30.17771.
van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W: Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998, 115: 58-66. 10.1016/S0016-5085(98)70365-8.
Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R: Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993, 90: 5791-5795. 10.1073/pnas.90.12.5791.
Akopyants NS, Clifton SW, Kersulyte D, Crabtree JE, Youree BE, Reece CA, Bukanov NO, Drazek ES, Roe BA, Berg DE: Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998, 28: 37-53.
Peek RM, Blaser MJ, Mays DJ, Forsyth MH, Cover TL, Song SY, Krishna U, Pietenpol JA: Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 1999, 59: 6124-6131.
Bagnoli F, Buti L, Tompkins L, Covacci A, Amieva MR: Helicobacter pylori CagA induces a transition from polarized top invasive phenotypes in MDCK cells. Proc Natl Acad Sci USA. 2005, 102: 16339-16344. 10.1073/pnas.0502598102.
Kusters JG, van Vliet AH, Kuipers EJ: Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006, 19: 449-490. 10.1128/CMR.00054-05.
Graham DY: Therapy of Helicobacter pylori: current status and issues. Gastroenterology. 2000, 118 (Suppl 1): S2-S8.
Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG: Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006, 6: 699-709. 10.1016/S1473-3099(06)70627-2.
Bae EA, Han MJ, Kim DH: In vitro anti-Helicobacter pylori activity of some flavonoids and their metabolites. Planta Med. 1999, 65: 442-443. 10.1055/s-2006-960805.
Fukai T, Marumo A, Kaitou K, Kanda T, Tereda S, Nomura T: Anti-Helicobacter pylori flavonoids from licorice extract. Life Sci. 2002, 71: 1449-1463. 10.1016/S0024-3205(02)01864-7.
Nostro A, Cellini L, Di Bartolomeo S, Di Campli E, Grande R, Cannatelli MA, Marzio L, Alonzo V: Antibacterial effect of plants extracts against Helicobacter pylori. Phytother Res. 2005, 19: 198-202. 10.1002/ptr.1640.
Shin JE, Kim JM, Bae EA, Hyun YJ, Kim DH: In vitro inhibitory effect of flavonoids on growth, infection and vacuolation of Helicobacter pylori. Planta Med. 2005, 71: 197-201. 10.1055/s-2005-837816.
Martini S, D’Addario C, Colacevich A, Focardi S, Borghini F, Santucci A, Figura N, Rossi C: Antimicrobial activity against Helicobacter pylori strains and antioxidant properties of blackberry leaves (Rubus ulmifolius) and isolated compounds. Int J Antimicrob Agents. 2009, 34: 50-59. 10.1016/j.ijantimicag.2009.01.010.
Mandalari G, Faulks RM, Rich GT, Lo Turco V, Picout DR, Lo Curto RB, Bisignano G, Dugo P, Dugo G, Waldron KW, Ellis PR, Wickham MS: Release of protein, lipid, and vitamin E from almond seeds during digestion. J Agric Food Chem. 2008, 56: 3409-3416. 10.1021/jf073393v.
Mandalari G, Tomaino A, Arcoraci T, Martorana M, Lo Turco V, Cacciola F, Rich GT, Bisignano C, Saija A, Dugo P, Cross KL, Parker ML, Waldron KW, Wickham MS J: Characterization of polyphenols, lipids and dietary fibre from almond skins (Amygdalus communis L.). J Food Comp Anal. 2010, 23: 166-174. 10.1016/j.jfca.2009.08.015.
Mandalari G, Tomaino A, Rich GT, Lo Curto R, Arcoraci T, Martorana M, Bisignano C, Saija A, Parker ML, Waldron KW, Wickham MSJ: Polyphenol and nutrient release from skin of almonds during simulated human digestion. Food Chem. 2010, 122: 1083-1088. 10.1016/j.foodchem.2010.03.079.
Vuorela S, Kreander K, Karonen M, Nieminen R, Hämäläinen M, Galkin A, Laitinen L, Salminen JP, Moilanen E, Pihlaja K, Vuorela H, Vuorela P, Heinonen M: Preclinical evaluation of rapeseed, raspberry, and pine bark phenolics for health related effects. J Agric Food Chem. 2005, 53: 5922-5931. 10.1021/jf050554r.
Marino A, Bellinghieri V, Nostro A, Miceli N, Taviano MF, Guvenc A, Bisignano G: In vitro effect of branch extracts of Juniperus species from Turkey on Staphylococcus aureus biofilm. FEMS Immunol Med Microbiol. 2010, 59: 470-476.
Miceli N, Trovato A, Marino A, Bellinghieri V, Melchini A, Dugo P, Cacciola F, Donato P, Mondello L, Guvenc A, De Pasquale R, Taviano MF: Phenolic composition and biological activities of Juniperus drupacea Labill. Berries from Turkey. Food Chem Toxicol. 2011, 49: 2600-2608. 10.1016/j.fct.2011.07.004.
Mandalari G, Bisignano C, D’Arrigo M, Ginestra G, Arena A, Tomaino A, Wickham MS: Antimicrobial potential of polyphenols extracted from almond skins. Lett Appl Microbiol. 2010, 51: 83-89.
Arena A, Bisignano C, Stassi G, Mandalari G, Wickham MSJ, Bisignano G: Immunomodulatory and antiviral activity of almond skins. Immunol Lett. 2010, 132: 18-23. 10.1016/j.imlet.2010.04.010.
Mandalari G, Bisignano C, Genovese T, Mazzon E, Wickham MS, Paterniti I, Cuzzocrea S: Natural almond skin reduced oxidative stress and inflammation in an experimental model of inflammatory bowel disease. Int Immunopharmacol. 2011, 11: 915-924. 10.1016/j.intimp.2011.02.003.
Faundez G, Troncoso M, Figueroa G: cagA and vacA in strains of Helicobacter pylori from ulcer and non-ulcerative dyspepsia patients. BMC Gastroenterol. 2002, 2: 20-10.1186/1471-230X-2-20.
Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. M100-S22. 2012, Wayne: PA: CLSI
Martini S, Bonechi C, Rossi C, Figura N: Increased susceptibility to resveratrol of Helicobacter pylori strains isolated from patients with gastric carcinoma. J Nat Prod. 2011, 74: 2257-2260. 10.1021/np100761u.
Liu W, Hsu C, Yin M: In vitro anti-Helicobacter pylori activity of diallyl sulphides and protocathecuic acid. Phytother Res. 2008, 22: 53-57. 10.1002/ptr.2259.
Funatogawa K, Hayashi S, Shimomura H, Yoshida T, Hatano T, Ito H, Hirai Y: Antibacterial activity of hydrolysable tannins derived from medicinal plants against Helicobacter pylori. Microbiol Immunol. 2004, 48: 251-261.
Toyoda T, Tsukamoto T, Mizoshita T, Nishibe S, Deyama T, Takenata Y, Hirano N, Tanaka H, Takasu S, Ban H, Kumagai T, Inada K, Utsunomiya H, Tatematsu M: Inhibitory effect of nordihydroguaiaretic acid, a plant lignan, on Helicobacter pylori-associated gastric carcinogenesis in Mongolian gerbils. CancSci. 2007, 98: 1689-1695.
Xiao Z-P, Shi D-H, Li H-Q, Zhang L-N, Xu C, Zhu H-L: Polyphenols based on isoflavones as inhibitors of Helicobacter pylori urease. Bioorg Med Chem. 2007, 15: 3703-3710. 10.1016/j.bmc.2007.03.045.
Ansari FL, Umbreen S, Hussain L, Makhmoor T, Nawaz SA, Lodhi MA, Khan SN, Shaheen F, Choudhary MI, Atta-ur-Rahman: Syntheses and biological activities of chalcones and 1,5-benzothiazepine derivatives: promising new free-radical scavengers, and esterases, ureases and α-glucosidase inhibitors. Chem Biodivers. 2005, 2: 487-496. 10.1002/cbdv.200590029.
Tombola F, Campello S, De Luca L, Ruggiero P, Del Giudice G, Papini E, Zoratti M: Plant polyphenols inhibit VacA, a toxin secreted by the gastric pathogen Helicobacter pylori. FEBS Lett. 2003, 543: 184-189. 10.1016/S0014-5793(03)00443-5.
Lee KM, Yeo M, Choue JS, Jin JH, Park SJ, Cheong JY, Lee KJ, Kim JH, Hahm KB: Protective mechanism of epigallocatechin-3-gallate against Helicobacter pylori-induced gastric epithelial cytotoxicity via the blockage of TLR-4 signalling. Helicobacter. 2004, 9: 632-642. 10.1111/j.1083-4389.2004.00281.x.
Makobongo MO, Gancz H, Carpenter BM, McDaniel DP, Merrel DS: The oligo-acyl lysyl antimicrobial peptide C12K-2 exhibits a dual mechanism of action and demonstrated strong in vivo efficacy against Helicobacter pylori. Antimicrob Ag Chemother. 2012, 56: 378-390. 10.1128/AAC.00689-11.
Acknowledgements
We thank Dr Karen Lapsley from the Almond Board California for supplying the almonds.
This study was supported by the Almond Board of California and the University of Messina, Italy.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors have received a research grant from the Almond Board of California.
Authors’ contribution
CB, MTF, GM conceived the study and participated in its design. EL, AF, SZ carried out the experiments and performed the data analyses. EL and SZ participated in the isolation of clinical strains. EL carried out the PCR amplification. GM coordinated, supervised the study and critically revised the manuscript. CB, AF, EL, SZ, MTF, GM drafted the manuscript. All authors have read and approved the final manuscript.
Carlo Bisignano, Angela Filocamo, Erminia La Camera, Sebastiana Zummo, Maria Teresa Fera and Giuseppina Mandalari contributed equally to this work.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bisignano, C., Filocamo, A., La Camera, E. et al. Antibacterial activities of almond skins on cagA-positive and-negative clinical isolates of Helicobacter pylori. BMC Microbiol 13, 103 (2013). https://doi.org/10.1186/1471-2180-13-103
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-13-103