Abstract
Background
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative pathogen that must successfully adapt to the broad fluctuations in the concentration of dissolved dioxygen encountered in the host. In Escherichia coli, ArcA (Aerobic Respiratory Control) helps the cells to sense and respond to the presence of dioxygen. The global role of ArcA in E. coli is well characterized; however, little is known about its role in anaerobically grown S. Typhimurium.
Results
We compared the transcriptional profiles of the virulent wild-type (WT) strain (ATCC 14028s) and its isogenic arcA mutant grown under anaerobic conditions. We found that ArcA directly or indirectly regulates 392 genes (8.5% of the genome); of these, 138 genes are poorly characterized. Regulation by ArcA in S. Typhimurium is similar, but distinct from that in E. coli. Thus, genes/operons involved in core metabolic pathways (e.g., succinyl-CoA, fatty acid degradation, cytochrome oxidase complexes, flagellar biosynthesis, motility, and chemotaxis) were regulated similarly in the two organisms. However, genes/operons present in both organisms, but regulated differently by ArcA in S. Typhimurium included those coding for ethanolamine utilization, lactate transport and metabolism, and succinate dehydrogenases. Salmonella-specific genes/operons regulated by ArcA included those required for propanediol utilization, flagellar genes (mcpAC, cheV), Gifsy-1 prophage genes, and three SPI-3 genes (mgtBC, slsA, STM3784). In agreement with our microarray data, the arcA mutant was non-motile, lacked flagella, and was as virulent in mice as the WT. Additionally, we identified a set of 120 genes whose regulation was shared with the anaerobic redox regulator, Fnr.
Conclusion(s)
We have identified the ArcA regulon in anaerobically grown S. Typhimurium. Our results demonstrated that in S. Typhimurium, ArcA serves as a transcriptional regulator coordinating cellular metabolism, flagella biosynthesis, and motility. Furthermore, ArcA and Fnr share in the regulation of 120 S. Typhimurium genes.
Similar content being viewed by others
Background
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a Gram-negative intracellular pathogen that causes gastroenteritis in the human host. Although non life-threatening in healthy adults, it can be fatal for children and immunocompromised individuals. The infection proceeds via two main stages: invasion and systemic infection. During the invasion stage, the pathogen adheres and colonizes the intestines gaining access to the epithelial cells. Subsequently, Salmonella crosses the epithelial cells and gets internalized by the macrophages where it reproduces and stealthily spreads in the host and causes systemic infection [1–4]. Clearly, Salmonella must adapt quickly to the diverse environments it encounters. In fact, from the gastrointestinal tract to the intracellular milieu, it is challenged with fluctuations in oxygen concentration and with numerous host-immune defenses including a battery of reactive oxygen (ROS) and nitrogen species (RNS) and antimicrobial peptides that reduce its ability to colonize the host [1–4].
In Escherichia coli, ArcA (Aerobic Respiratory Control) is one of the main transcriptional regulators involved in the metabolic shift from anaerobic to aerobic conditions and controlling the enzymatic defenses of bacteria against ROS. ArcA is a cytosolic response regulator of a two-component global regulatory system, ArcA/ArcB, where ArcB is a transmembrane histidine kinase sensor. ArcB transfers a phosphoryl group to ArcA, activating it, and inducing or repressing a large and diverse number of operons including the tricarboxylic acid cycle (TCA), terminal oxidases, dehydrogenases of the flavoprotein class, the glyoxylate shunt, and fatty acid degradation [5–17]. Due to its importance in diverse energy metabolic processes, the ArcA regulon has been thoroughly characterized in E. coli [5, 12, 18]. Conversely, very little is known about the regulatory network controlled by ArcA in S. Typhimurium under anaerobic conditions.
Although E. coli and S. Typhimurium share a very high genomic similarity (~75-80%) [19], we previously discovered that the Fnr (Fumarate Nitrate Reductase) regulon of S. Typhimurium is markedly different from the one identified in E. coli [20]. Due to the complementary roles of ArcA and Fnr in the regulation of cellular metabolism and adaptation to changes in redox, we hypothesized that the ArcA regulon of S. Typhimurium will also differ from that of E. coli. The results indicate that in S. Typhimurium, as in E. coli, the ArcA regulon includes the core metabolic and energy functions as well as motility. However, Salmonella-specific genes/operons regulated by ArcA include newly identified flagellar genes (mcpAC, cheV), Gifsy-1 prophage genes, a few SPI-3 genes (mgtBC, slsA, STM3784), and those for propanediol utilization. Furthermore, the arcA mutant was non-motile and was as virulent as the isogenic wild-type strain. We also identified 120 genes that were regulated by the anaerobic regulator, Fnr, as well as by ArcA.
Methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Table 1. Wild-type (WT) S. Typhimurium (14028s) and its isogenic arcA mutant (NC 980) were used throughout. P22 phage was used to move the arcA::Tn10 mutation from S. Typhimurium LT2 (TT17442) [21] to strain 14028 s. Transductants were plated on Evans Blue Uranine (EBU) agar and the arcA mutant was tested for its inability to grow on toluidine blue agar [22]. The Tn10 insertion junctions of the arcA mutant were confirmed by PCR and DNA sequencing. Additionally, the absence of the ArcA protein in the mutant was confirmed by Western blotting (Additional file 1: Figure S1 - lane 3).
Unless stated otherwise, the WT and the arcA mutant were grown anaerobically at 37°C in MOPS-buffered (100 mM, pH 7.4) LB broth supplemented with 20 mM D-xylose (LB-MOPS-X). MOPS was used in the medium to avoid the indirect effects of pH, while xylose was used to avoid the effects of catabolite repression [12]. An anaerobic chamber (Coy, Ann Harbor, MI) with anaerobic gas mixture (10% H2, 5% CO2, and 85% N2) was used as previously described [20]. All solutions were anaerobically pre-equilibrated in the chamber for 48 h before use. Overnight cultures (15-18 h) were used to inoculate fresh media. Aerobic growth was carried-out using LB or LB-MOPS-X as specified (volume of the culture: flask ratio = 1:5, shaking at 200 rpm using an orbital shaker). Growth kinetic experiments were performed on the WT and the arcA mutant in triplicate under both aerobic and anaerobic conditions.
Construction of parcA
For complementation studies, a low-copy-number plasmid, expressing arcA (parcA, NC 989) was constructed. The complete arcA sequence starting from 180 bp upstream from the start codon (ATG) until the stop codon (TAA) of arcA [i.e., 897 bp fragment] was amplified from the WT strain using the following primers (Integrated DNA Technologies, Coralville, IA):
arcA-Forward 5'- TCGATCCCGGGTACCCACGACCAAGCTAATG-3' and arcA-Reverse 5'-CTACCTCCCGGGTTAATCCTGCAGGTCGCCG -3' [SmaI site underlined]. The PCR product was digested with SmaI and ligated into the pACYC177 (New England BioLabs, Ipswich, MA) vector that was also cut with SmaI. Thus, in the new plasmid (parcA) the Kanr gene in pACYC177 was disrupted by the insertion of arcA. Plasmid DNA (parcA) was first transformed into a restriction deficient strain of E. coli [ER2925 (New England BioLabs)], which was subsequently purified and transformed into and maintained in the S. Typhimurium arcA mutant, thus generating NC989 (Table 1).
Transformations were carried-out using the calcium chloride method [23]. Plasmid DNA and genomic DNA were isolated using the Qiagen Mini Spin isolation kit (Qiagen, Valencia, CA) and the DNAeasy Tissue Kit (Qiagen), respectively. Transformants containing parcA (NC989) were confirmed for Ampr (130 μg/ml) and Kans (50 μg/ml) on LB plates and the presence of parcA was confirmed via PCR and restriction analysis. The expression of ArcA was confirmed by Western blot analysis (Additional file 1: Figure S1 - lane 4).
RNA isolation
Overnight anaerobic cultures of the WT or the arcA mutant were used to inoculate three independent flasks for each strain. Every flask contained 150 ml of LB-MOPS-X equilibrated in the anaerobic gas mix for the previous 48 h. The three independent cultures of each strain were grown to an OD600 = 0.30-0.35, pooled, and treated with RNAlater (Qiagen, Valencia, CA) to fix the cells and preserve the quality of the RNA. Total RNA was extracted using RNeasy RNA extraction kit (Qiagen) and the samples were treated with RNase-free DNase (Invitrogen, Carlsbad, CA). The absence of contaminating DNA and the quality of the RNA was confirmed by the lack of PCR amplification of known genes (i.e.: fnr) and by using agarose-gel electrophoresis. Aliquots of the RNA samples were kept at -80°C for use in the microarray and the qRT-PCR studies.
Microarray studies
S. Typhimurium microarray slides were prepared and used as previously described [24]. For the hybridizations, the SuperScript™ Indirect cDNA Labeling System (Invitrogen) was used to synthesize the cDNA from the RNA prepared from the WT and arcA mutant strains. Dye swapping was performed to avoid dye-associated effects on cDNA synthesis. Slide hybridizations and scanning were carried-out using the same protocols and equipment as previously described [20].
Data analysis
Cy3 and Cy5 values for each spot were normalized over the total intensity for each dye, to account for differences in total intensity between the two scanned images. The consistency of the data obtained from the microarray analysis was evaluated using two methods: (i) a pair-wise comparison, calculated with a two-tailed Student's t test and analyzed by the MEAN and TTEST procedures of SAS-STAT statistical software (SAS Institute, Cary, NC) [the effective degrees of freedom for the t test were calculated as previously described [25]; and (ii) a regularized t test followed by a posterior probability of differential expression [PPDE (p)] method. The signal intensity at each spot from the arcA mutant and the WT were background-subtracted, normalized, and used to calculate the ratio of gene expression between the two strains. All replicas were combined and the median expression ratio and standard deviations calculated for ORFs showing ≥ 2.5-fold change.
Microarray data
The microarray data are accessible via GEO Accession Number GSE24564 at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24564.
qRT-PCR
qRT-PCR [26] was used to validate the microarray data [27]. Seventeen genes were randomly chosen (Table 2) from the differentially expressed genes. Primers (Integrated DNA Technologies, Coralville, IA) were designed and qRT-PCRs were carried-out using QuantiTectTM SYBR® Green RT-PCR Kit (Qiagen), an iCycler™ (Bio-Rad, Hercules, CA), and the data were analyzed by the Bio-Rad Optical System Software - Version 3.1, according to the manufacturer specifications. The cycling conditions comprised 30 min of a reverse transcriptase reaction at 50°C, 15 min of polymerase inactivation at 95°C, and 40 cycles each of 94°C for 15 sec for melting, 51°C for 30 sec for annealing, and 72°C for 30 sec for extension followed by 31 cycles each at 65°C for 10 sec to obtain the melt curve. To ensure accurate quantification of the mRNA levels, three amplifications of each gene were made using 1:5:25 dilutions of the total RNA. Measured mRNA levels were normalized to the mRNA levels of the housekeeping gene, rpoD (σ70) . Normalized values were used to calculate the ratios of the expression levels [20] in the arcA mutant relative to the WT.
Logo graph and promoter analysis
The information matrix for the generation of the ArcA logo was produced using the alignment of the E. coli ArcA binding sequences, available at http://arep.med.harvard.edu/ecoli_matrices/[28]. The alignment of the ArcA motifs from this website did not include the motifs present in the sodA and mutM promoters [29, 30], therefore they were included in our analysis. To account for differences in nucleotide usage or slight variations in consensus sequences, a second alignment was built for S. Typhimurium using the 5'-regions of the homologous genes originally used to build the E. coli information matrix. The Salmonella alignment was used to prepare a new information matrice using the Patser software (version 3d), available at http://rsat.ulb.ac.be/rsat/[31] and graphed using the Weblogo software (version 2.8.1, 2004-10-18), available at http://weblogo.berkeley.edu/[32].
Swarming motility assay and electron microscopy
The swarming of the WT and the arcA mutant were evaluated under anoxic conditions. Ten microliters of anaerobically grown cells (i.e., from 16 h cultures) were spotted onto LB-MOPS-X agar (0.6% agar) plates and incubated anaerobically at 37°C for 24 h. The diameter of the growth halo was used as a measure of swarming. Scanning electron microscopy (SEM) was used to examine the morphology of the extracellular surfaces, while transmission electron microscopy (TEM) and negative staining were used to visualize the flagella of the anaerobically grown WT and arcA mutant as previously described [20].
Pathogenicity studies
For single infectionassays, six to eight week old female C57BL/6 mice bred in the University of Colorado School of Medicine animal facility according to Institutional Animal Care and Use Committee guidelines were used in this study. WT and arcA mutant Salmonella were grown in LB-MOPS-X broth to stationary phase for about 20 h. For intraperitoneal (i.p.) challenge, two groups of five mice per strain (WT and arcA mutant) were inoculated with 250 CFU in 500 μl PBS/mouse. Mortality was scored over a 15- to 30-day period. Competitive infection assays were carried-out as described [33] with modifications. The strains were separately grown overnight in LB broth at 37°C with shaking at 200 rpm. Tetracycline (10 μg/ml) was used to propagate and isolate the arcA mutant. Bacterial (i. e.: WT and arcA mutant) cultures were diluted in phosphate-buffered saline (PBS) and mixed to produce a 1:1 inoculum ratio. Groups of mice were infected either i.p. or orally (p.o.). Prior to oral infection, food and water were withheld from the mice for 4 h and the bacterial cocktail was administered to the mice by allowing them to drink 20 μl from the end of a pipette tip.
On day 4 or day 6 after i.p. or p.o. infection, respectively, mice were euthanized and mesenteric lymph nodes (MLN), liver and spleen collected for bacterial enumeration. The tissues were homogenized in sterile PBS and 10-fold serial dilutions were plated on LB agar medium with or without 10 μg/mL tetracycline to distinguish the WT (Tets) from the arcA mutant (Tetr). The number of CFUs of S. Typhimurium 14028 s per organ was calculated by subtracting the number of CFU/ml on the LB-Tet plates from the number of CFU/ml on the corresponding LB plates. The competitive index (CI) was calculated as the ratio of the CFU of arcA mutant to the CFU of the WT strain recovered from the spleen, liver, and mesenteric lymph nodes (i.e.; CI = [arcA mutant/WT]output/[arcA mutant/WT]input).
Results
Bacterial growth kinetics
The growth kinetics of the WT and the arcA mutant strains were determined under anaerobic conditions in LB-MOPS-X. The arcA mutant strain grew at a slower rate than the WT strain. The doubling-times of the WT and arcA mutant were 37.0 ± 0.4 and 55.4 ± 0.1 min under anaerobic conditions. Due to the difference in the doubling-times of the two strains, cells used for RNA isolation and subsequent transcriptome profiling were allowed to grow for an equal number of generations (~five generations: OD600 = 0.30-0.35) instead of an equal length of time.
Anaerobic transcriptome profiling
Out of 4,579 genes, the two-tailed Student's t test, produced a set of 2,026 coding sequences showing a significant difference between the arcA mutant and the WT (p < 0.05). We restricted the analyses to only include highly affected genes (i.e., has a ratio ≥ 2.5-fold) as previously described [20]. Under this constraint, 392 genes were differentially expressed in the arcA mutant relative to the WT and, therefore, regulated directly or indirectly by ArcA. Of these, 147 genes were up-regulated and 245 genes were down-regulated (Additional file 1: Table S1). All the genes showing significant differential expression were classified into clusters of orthologous groups (COGs) [34–36] as defined by the National Center for Biotechnology Information (NCBI) http://www.ncbi.nlm.nih.gov/COG (Table 3). It should be noted that throughout the study we compared the levels of transcription in the arcA mutant to that in the WT strain. Thus, genes repressed by ArcA posses positive values (i.e., >1), while genes activated by ArcA have negative values (i.e., <1).
Microarray validation
Normalized mRNA levels from qRT-PCR are shown in Table 2. The microarray and qRT-PCR data were log2 transformed and plotted (Figure 1). The correlation between the two sets of data was 0.87 (p < 0.05).
Correlation between the microarray and the qRT-PCR data of 17 randomly selected genes. The ratios of changes in gene expression, from the microarray (each S. Typhimurium ORF was spotted in triplicate on the slide) and qRT-PCR experiments, for the arcA mutant relative to the WT were log2 transformed and linearly correlated. The genes selected and the primers used in qRT-PCR are listed in Table 2. Three amplifications of each of the 17 genes were made using 1:5:25 dilutions of the total RNA.
Logo graph and promoter analysis
To determine whether a binding site for ArcA might be present in the region upstream of the candidate ArcA-regulated genes, we searched the 5' regions of these highly affected genes (i.e., has a ratio ≥ ± 2.5-fold) for the presence of a putative ArcA-binding motif using a Salmonella logo graph (Figure 2) and found 155 genes contained potential ArcA binding sites. Furthermore, fifty-five out of the 147 ArcA-activated genes (37%), and 100 out of the 245 ArcA-repressed genes (41%) contained at least one putative ArcA-binding site (Additional file 1: Table S1).
Logo of the information matrix obtained from the alignment of ArcA sequences for S . Typhimurium. Sequences were obtained by searching the S. Typhimurium LT2 genome [Accession #: AE006468 (chromosome) and AE606471 (plasmid)] with known ArcA sequences derived from the corresponding ArcA-regulated genes in E. coli. A total of 20 E. coli sequences were used to obtain the logo shown. The total height of each column of characters represents the amount of information [measured in bits, which is the maximum entropy for the given sequence type (ex. Log2 4 = 2 bits for DNA/RNA and log2 20 = 4.3 bits for proteins)] for that specific position and the height of each individual character represents the frequency of each nucleotide.
ArcA as a repressor
Transcription of the genes required for aerobic metabolism, energy generation, amino acid transport, and fatty acid transport were anaerobically repressed by ArcA (Additional file 1: Table S1). In particular, the genes required for cytochrome-o-oxidase, succinyl-CoA synthetase, glutamate/aspartate transport, trehalose-6-phosphate biosynthesis, long-chain fatty acids transport, spermidine/putrescine transport, dipeptide transport, the genes encoding the two-component tricarboxylic transport system and the site-specific DNA factor for inversion stimulation (fis) were among the highest repressed by ArcA. Genes required for L-lactate transport and metabolism, phosphate transport, acetyl-CoA transferase, APC family/D-alanine/D-serine/glycine transport, putative cationic amino acid transporter, peptide methionine sulfoxide reductase, multiple antibiotic resistance operon, as well as many poorly characterized genes were also repressed by ArcA (Additional file 1: Table S1).
Additionally, some genes related to Salmonella virulence were repressed by ArcA. For example, the expression of the mgtCB operon (member of SPI-3) that is required for Mg2+ transport/growth in low-magnesium and involved in systemic infections in mice/intramacrophage survival [37–40], genes constituting the lambdoid prophage Gifsy-1 that contributes to the virulence of S. Typhimurium [41], and genes coding for a leucine-rich repeat protein (sspH2) that is translocated by and coordinately regulated with the SPI-2 TTSS [42] were highly repressed by ArcA (Figure 3A and Additional file 1: Table S1).
Organization of major genes for (A) SPI-3, (B) ethanolamine utilization, (C) propanediol utilization, and (D-F) flagellar biosynthesis and motility. The names of genes are listed to the left of the arrow, an (*) next to the gene indicates the presence of at least one ArcA motif in the 5' - region, the numbers listed to the right of the arrows indicates the ratio of gene expression in the arcA mutant relative to that in the WT, and the shading corresponds to the level of gene expression. NR = no expression ratio.
ArcA as an activator
Several of the genes involved in regulating flagellar biosynthesis, motility, chemotaxis, sugar transport, metabolism, and glycogen biosynthesis were found to be anaerobically activated by ArcA (Figure 3D-F and Additional file 1: Table S1). In particular, several of the middle (class 2) flagellar genes and late flagellar (class 3) genes had lower transcript levels in the arcA mutant than in the WT strain (Figure 3D-F). There was no significant difference in the transcript levels of the early flagellar genes (class 1) flhD and flhC, whose gene products FlhD/FlhC are the master regulators of flagellar biosynthesis (Figure 3E). Additionally, several newly identified flagellar genes [43] (i. e., mcpA, mcpC, and cheV) had lower expression levels in the arcA mutant than in the WT (Additional file 1: Table S1), while the expression of mcpB was not affected. Furthermore, genes coding for transcriptional repressor CytR, nitrite reductase, 2-dexoyribose-5-phosphate aldolase, thymidine phosphorylase, lysine/cadaverine transport protein, putrescine/ornithine antiporter, ornithine decarboxylase, ethanolamine operon, and propanediol operon as well as its transcriptional regulator PocR were activated by ArcA (Figure 3B and 3C, and Additional file 1: Table S1).
The expression of SPI-1 associated genes was not affected by a mutation in arcA. However, two SPI-3 genes, slsA, encoding a putative inner membrane protein required for colonization of chickens and calves [1, 44], and STM3784, a putative sugar phosphotransferase, were activated by ArcA as their expression levels were significantly lower in the mutant than in the WT (Figure 3A and Additional file 1: Table S1).
Phenotype of the arcAmutant
Next, we correlated some of the microarray findings with the corresponding phenotypes of the WT and the arcA mutant strains.
a. Flagellar biosynthesis and swarming motility
The microarray data showed that, in anaerobiosis, the expression of the flagellar biosynthesis, motility, and chemotaxis genes was lower in the arcA mutant than in the WT. Therefore, we compared the swarming motility of the WT and the arcA mutant in soft agar under anaerobic conditions (Table 4). The data indicated that the arcA mutant was ~100% non-motile compared to the WT and that the inclusion of parcA complemented (~57%) this phenotype. We also compared the WT and the arcA mutant under anaerobic conditions for the presence of flagella by using SEM (Figure 4A and 4C, left panel) and TEM (Figure 4B and 4D, right panel). The data (Table 4 and Figure 4) clearly showed that the arcA mutant Lacked flagella and was non-motile.
Comparison of the WT and the arcA mutant for surface appendages and flagella via microscopy. Scanning electron microscopy (SEM) was used to evaluate the WT (A) and the arcA mutant (C) for the presence/absence of surface appendages and negative staining followed by transmission electron microscopy (TEM) was used to evaluate the WT (B) and the arcA mutant (D) for the presence/absence of flagella. Cells were grown anaerobically in LB-MOPS-X media and the samples were prepared as described in Materials and Methods.
b. Virulence in mice
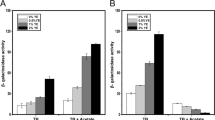
The microarray data (Additional file 1: Table S1) showed that ArcA does not significantly regulate the transcription of the virulence genes found in SPI-1, which are important for the ability of Salmonella to invade host epithelial cells [2, 3, 45–47]. However, few virulence genes related to SPI-2 (sspH2) and SPI-3 (mgtCB, slsA, STM3784) were affected by ArcA. Therefore, to evaluate these findings, we tested the virulence of the arcA mutant in a murine model of mucosal and acute infection using immunocompetent C57BL/6 mice. The arcA mutant was as virulent as the WT strain when 250 CFU/mouse were inoculated via i.p. (Figure 5A). Since intramacrophage survival and replication of Salmonella permits the colonization of the spleen and liver of mice [4, 48], a further virulence comparison of the WT and the arcA mutant was performed using a mixed infection assay. The data showed that the arcA mutant had a moderate competitive survival advantage in the reticuloendothelial system compared to the WT in all systemic organs examined following a p.o. or i.p. mixed infection (Figure 5B). In the majority of the mice, the arcA mutant was isolated in higher numbers than the WT, although these increases were not statistically significant (p > 0.05). The data generated with the competitive assays is in agreement with i.p. infection data, where the mice succumbed with similar kinetics after infection with arcA or WT bacteria.
Virulence comparison of the WT and the arcA mutant in 6-8 week old C57BL/6 mice. (A) Single infection assays, where two groups of five mice per strain (WT and arcA mutant) were challenged intraperitoneally using 250 CFU/mouse, as described in Materials and Methods. Percent survival is the number of mice surviving relative to the number of mice challenged at zero time; (B) Competitive infection assays, where groups of three 6-week-old mice were infected orally (p. o.) or i. p. with a 1:1 mixture of S. Typhimurium 14028 s and its isogenic arcA mutant. After 4 or 6 days following i.p. or p.o. infection, respectively, mice were euthanized and mesenteric lymph nodes (MLN), liver, and spleen were collected for enumeration of the WT and the mutant. The competitive index (CI) was calculated as described in the Materials and Methods.
Discussion
Although there are several reports on the regulation of specific genes by ArcA in non-virulent strains of E. coli [6–9, 13–17, 49–56] and in Salmonella spp. [57]. This is the first genome-wide study on the regulatory role of ArcA in S. Typhimurium (14028s) under anaerobic conditions. ArcA was found to directly or indirectly control the expression of at least 392 genes. In particular, we showed that ArcA is involved in energy metabolism, flagella biosynthesis, and motility. Additionally, the arcA mutant was as virulent as the WT, although it was non-motile. Furthermore, prior to the present report, none of the virulence genes (i. e., SPI-3 and Gifsy-1) had been identified as part of the Salmonella ArcA regulon. Finally, several genes involved in metabolism previously identified as being regulated by ArcA in E. coli [5–17, 49–52] were also identified in the present study (Additional file 1: Table S1).
Logo comparison
In a recent study, a logo was used to graphically compare multiple ArcA sequence alignments of Shewanella oneidensis [58] to that of E. coli [12]. The analysis revealed subtle changes in base pairs at each position between the sequences. Although the ArcA binding motifs of S. oneidensis and E. coli were similar, the arcA regulons and the physiological function of ArcA in these two organisms were different [58]. When comparing the ArcA logos of E. coli and S. oneidensis to the one generated herein for S. Typhimurium, we found that there is similarity between S. Typhimurium and both E. coli and S. oneidensis. However, while there is very little variation between the nucleotide sequences at each base pair of S. Typhimurium and E. coli, there is much more variation between S. Typhimurium and S. oneidensis. Therefore, when comparing the genes regulated by ArcA in these three organisms, it is evident that the ArcA regulons of E. coli and S. Typhimurium are more similar than that of S. oneidensis.
ArcA and carbon metabolism
Comparing our microarray data in S. Typhimurium to the published data of E. coli [5, 12], there are several aspects pertaining to metabolic regulation that are similar between these two organisms. Anaerobically, several ArcA-repressed genes identified in our microarray data are involved in metabolism and transport, while ArcA-activated genes included those coding for enzymes involved in glycogen synthesis and catabolism as well as those for gluconeogenesis. Expression of many of these genes was consistent with those reported in E. coli [5, 9, 11–14, 52], H. influenzae [59], and S. oneidensis [60]. The genes of the two-component tricarboxylic transport system (tctE, STM2786, STM2787, STM2788, and STM2789) were the most highly repressed by ArcA (Additional file 1: Table S1). This was not surprising since transport systems for substrates of aerobic pathways have been suggested to be candidates for regulation by ArcA [14]. A similar pattern of anaerobic regulation of these enzymes has also been seen in our previous global analysis of Fnr [20] (Additional file 1: Table S2).
In E. coli, the CsrA (carbon storage regulator) protein acts post-transcriptionally to balance carbon flow in the cell by activating genes involved in glycolysis and repressing genes participating in gluconeogenesis [61]. In the present study, we found that the transcription of csrA was not affected by a mutation in arcA, presumably CsrA remained fully functional in the mutant to provide the switch from glycolysis to gluconeogenesis by repressing the genes associated with glycolysis and activating those genes affiliated with gluconeogenesis. A mutation in arcA caused a 2.65-fold increase in the expression of ptsG, a glucose-specific IIB component of the PTS-system (STM1203), which is required for the first step in glucose metabolism. A similar 2-fold increase was noticed in E. coli and the binding of ArcA to the promoter of ptsG was demonstrated [54]. Under anaerobic conditions and in the absence of electron acceptors, where the reduced quinone carriers can activate ArcA, it seems to be more advantageous for S. Typhimurium and E. coli cells to control the rate of glucose metabolism in order to reduce the rate of production of acidic end-products. Thus, the adaptation to anaerobic environments requires the regulation of the rate of glycolysis, the utilization of the fermentation products, and the use of the tricarboxylic acid cycle and the glyoxylate shunt in order for the organism to compete with others during sudden changes in oxygen concentrations.
E. coli contains two oxidases in its respiratory chain. The first, which is known to decrease under anaerobic growth conditions and has a low affinity for oxygen, cytochrome o (encoded by the cyoABCDE) and the second, which is known to increase during anaerobic growth and has a high affinity for oxygen, cytochrome d (encoded by the cydAB) [62]. Our data show that, anaerobically, ArcA repressed the cyo operon (Additional file 1: Table S1), while the expression of cyd operon was slightly reduced in the arcA mutant relative to WT (i.e., ArcA is required for the activation of cyd). These results are in agreement with previous reports showing that a mutation in either arcA or arcB diminished cyd operon expression under aerobic and anaerobic conditions, while either mutation did not fully abolish repression of the cyo operon anaerobically [55].
Our data showed that the arcA mutant has a longer doubling time compared to the WT under anaerobiosis. This result is supported by our microarray data whereby several genes responsible for glycogen synthesis and catabolism as well as those for gluconeogenesis were down-regulated in the arcA mutant compared to the WT, while those genes regulating the tricarboxylic acid cycle (TCA), glyoxylate shunt, glycolysis, pentose phosphate shunt, and acetate metabolism were all up-regulated in the arcA mutant compared to the WT. Thus, under anaerobiosis, in arcA mutant cells, an energy imbalance is created, whereby the cells direct products through various metabolic pathways typically used during aerobiosis due to the de-repression of the TCA cycle, which subsequently yields a pool of reducing equivalents that can only be reduced in the presence of electron acceptors. Normally, during anaerobiosis, less energy in the form of ATP is generated. Thus, the arcA mutant cells appear to waste a vast amount of energy to express and maintain metabolic pathways that are not required under anaerobiosis, which may contribute to the slower growth rate of the culture. However, further work is required to determine NAD/NADH pools in the arcA mutant compared to the WT.
ArcA and hydrogenases
Hydrogen gas (H2) is an important energy source for the survival of pathogens in vivo [63] and is produced in the host via colonic bacterial fermentations [64]. Our results indicated that the hyb operon was activated in the arcA mutant, but these levels were not within our ± 2.5-fold threshold. Additionally, STM1538, STM1539, STM1786, STM1788, STM1790, and STM1791, which also code for hydrogenases were significantly repressed in the arcA mutant (Additional file 1: Table S1), in agreement with previous results [65].
ArcA regulation of cobalamine synthesis and metabolism
Propanediol (encoded by the pdu operon), a fermentation product of rhamnose or fucose [66, 67], and ethanolamine (encoded by the eut operon), an essential component of bacterial and eukaryotic cells, can be used by Salmonella as carbon and energy sources in the mammalian gastrointestinal tract [67]. Vitamin B12, its synthesis being encoded by the cob operon, is required for the metabolism of ethanolamine and propanediol, while anaerobic utilization of these substrates also requires the use of tetrathionate (ttr) as a terminal electron acceptor [68]. The positive regulatory protein, PocR, is necessary for the induction of the cob and pdu operons and is subject to global regulatory control via ArcA and/or Crp [69, 70].
In vivo expression technology (IVET) has shown that genes coding for cobalamine synthesis and 1,2-propanediol degradation are required for Salmonella replication in macrophages [71], that pdu genes may be necessary for intracellular proliferation within the host [72], and that pdu mutations, but not cob mutations can be attributed to a defect in virulence [73, 74]. Strains harboring mutations in ethanolamine utilization genes are attenuated in macrophages and in BALB/c mice when delivered orally, but not intraperitoneally [75].
Our data (Additional file 1: Table S1) show that pocR, the transcriptional regulator of propanediol utilization, was significantly activated by ArcA. Furthermore, all of the genes in the eut and pdu operons were activated by ArcA (Figure 3 and Additional file 1: Table S1). An arcA mutation in S. Typhimurium has been shown to cause reduced expression of the cob and pdu operons during anaerobic growth [69]. Interestingly, when comparing the data from the present study and our Fnr data [20], we found that ArcA and Fnr share in the regulation of the eut operon (Additional file 1: Table S2), while the pdu operon was more positively regulated by ArcA than by Fnr. Transcription of tetrathionate (ttr operon) was activated at equal levels by both Fnr and ArcA. Previous studies [68, 70] have shown that induction of the ttr operon is affected by Fnr, but not by ArcA. This may suggest that Fnr plays a more significant role in regulating the eut operon [70], while ArcA acts more significantly on regulating the genes associated with the pdu operon. Although, both the cob and pdu operons were both activated in the arcA mutant, this may be due to the effects of arcA on anaerobic pocR expression, which subsequently regulates the rest of each of these operons.
ArcA and flagellar biosynthesis/swarming motility/chemotaxis
Our data show that, anaerobically, ArcA positively regulates the expression of genes involved in flagellar biosynthesis, swarming motility, and chemotaxis (Figures 3 and 4; Table 3 and Additional file 1: Table S1) including many newly identified flagellar genes (i.e., mcpAC and cheV) [43]. Previously, we found that Fnr positively regulates many of the same the flagellar and chemotaxis genes under anaerobic conditions [20]; indeed the anaerobic motility phenotype of the arcA mutant was indistinguishable from that previously seen with the fnr mutant [20]. Furthermore, the expression of the flagellar biosynthesis, motility, and chemotaxis genes under anaerobiosis was more highly activated by Fnr than by ArcA (Additional file 1: Table S2).
A plethora of regulators affect the expression of flhDC and motility in E. coli and S. Typhimurium [20, 76–86]. Our data showed that ArcA activates class 2 and class 3 flagellar genes and we identified a potential ArcA binding site in filA, filZ, flgM, and flgN. ArcA seems to slightly repress flhDC (i. e., below our cut-off level of ±2.5-fold). In agreement with our work, ArcA was recently shown to be necessary for the expression of fliA in E. coli, but not for the master regulator, flhDC [56]. However, using in silico analysis, the authors did not identify ArcA binding sites in the promoter regions of fliA or other class 2 flagellar genes [56],
ArcA and antioxidant defenses
Under aerobic conditions, ArcA has been reported to be essential for the resistance of S. Enteritidis to RNS and ROS via an unknown mechanism [57]. In agreement with this report [57], we found that the arcA mutant of S. Typhimurium to be more sensitive to hydrogen peroxide (H2O2) under aerobic conditions (Additional file 1: Figure S2). Anaerobically, our data indicate that the expression of many of the antioxidant genes [i.e.: sodA, sodB, sodC1, and sodC2 (coding for superoxide dismutases) and katG and katE (coding for hydroperoxidases), and hmpA (coding for flavohemoglobin)] were not significantly affected by ArcA; however the expression of STM1731 (Mn-catalase, katN) was significantly increased in the arcA mutant compared to the WT (Additional file 1: Table S1). To date, the physiological role of Mn-catalase (KatN) in S. Typhimurium has not been thoroughly examined, although, its ability to scavenge H2O2 and work together with KatE, KatG, and the hydroperoxide reductases to protect the cell from oxidative stress has been demonstrated [87]. In addition, it has been proposed that the substitution of iron by manganese as a co-factor might be a way to circumvent iron restriction by the host during infection [88].
ArcA and pathogenesis
The majority of the virulence factors (~200 genes) of S. Typhimurium are chromosomally located within Salmonella pathogenicity islands (SPIs) [2, 89–93]. SPI-1 and SPI-2 both encode TTSSs [4, 45, 94]. SPI-1 effectors' proteins are required for epithelial cell invasion [95], while SPI-2 encodes secreted proteins, their specific chaperones [4], and a two-component regulatory system [96, 97], are all required for intracellular replication. Recently, SPI-1 invasion genes were found to be required for intramacrophage survival [98] and systemic infection in mice [99]. Our data have shown that most of the SPI-1 through SPI-5 genes were not significantly regulated by ArcA, with the exception of three genes contained within SPI-3 including, mgtC, mgtB, and slsA (Figure 3 and Additional file 1: Table S1). Thus, it is not surprising that our arcA mutant was determined to be as virulent as the WT strain following individual infection studies (Figure 5A), but was slightly more persistent than the WT following o. p. and i. p. competitive infection studies (Figure 5B), however, the difference was not statistically significant (p > 0.05).
Flagellar regulons have been shown to influence virulence gene expression in several pathogenic microorganisms [100–106]. Interestingly, data from our previous study [20], showed that the fnr mutant was non-motile and non-virulent, while in the present study, the arcA mutant was non-motile, but remained virulent. Clearly, the lack of motility does not necessarily correlate with the lack of virulence in S. Typhimurium.
Overlapping global regulation by ArcA and Fnr
ArcA and Fnr are two well known redox regulators in E. coli, S. Typhimurium, and other bacteria. We previously published the first report on the global role of Fnr in anaerobically grown S. Typhmurium [20]. The present study is the first report on the global regulatory role of ArcA in the same organism under the same experimental conditions and statistical constraints. Therefore, it is possible and reliable to compare genes/operons regulated by these two important transcriptional factors (i. e., ArcA and Fnr). The data indicated that ArcA and Fnr shared in the regulation of 120 genes; while the numbers of genes solely regulated by either ArcA or Fnr were 272 and 191, respectively. The 120 genes that were regulated by either ArcA or Fnr are listed (Additional file 1: Table S2). These include genes involved in cytochrome c oxidase (cyoABCDE), glutamate/aspartate transport (gltlJKL), dipeptide transport (dppABCDF), succinyl-CoA synthetase - α & β subunits (sucDC), tricarboxylic transport (STM2786, STM2787, STM2788), L-lactate transport and metabolism (lldPRD), nitrite reductase (nrfAB), 2-dexoyribose-5-phosphate aldolase (deoC), thymidine phosphorylase (deoA), aerotaxis sensor receptor (aer), ethanolamine utilization (eut operon - STM2454-2470), and several genes involving flagellar biosynthesis, motility, and chemotaxis. Interestingly, most of the 120 genes were regulated by ArcA and Fnr in the same fashion (i.e., repressed or activated) except for yneB (putative fructose-1,6-bisphosphate aldolase - STM4078), which was activated by ArcA, but repressed by Fnr (Additional file 1: Table S2). The opposing regulation of yneB by ArcA and Fnr indeed warrant further studies.
Conclusion(s)
Herein, we report on the role of the two-component regulator, ArcA, in the genome-wide response to oxygen in Salmonella. Our data clearly demonstrate that ArcA serves, directly or indirectly, as a regulator/modulator of genes involved in aerobic/anaerobic energy metabolism and motility. In a recent study [20], we demonstrated that the oxygen sensing, global regulator, Fnr participates in coordinating anaerobic metabolism, flagellar biosynthesis, motility, chemotaxis, and virulence in S. Typhimurium. In the present study, we identified a set of 120 genes whose regulation is shared between ArcA and Fnr. We also demonstrated that Fnr plays a more hierarchical role than ArcA in pathogenesis. Furthermore, under our experimental conditions, we demonstrated that the lack of motility does not necessarily correspond to the lack of virulence in S. Typhimurium.
References
Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, et al: Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2004, 54: 994-1010. 10.1111/j.1365-2958.2004.04323.x.
Galan JE: Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol. 2001, 17: 53-86. 10.1146/annurev.cellbio.17.1.53.
Wallis TS, Galyov EE: Molecular basis of Salmonella induced enteritis. Mol Microbiol. 2000, 36: 997-1005. 10.1046/j.1365-2958.2000.01892.x.
Cirillo DM, Valdivia RH, Monack DM, Falkow S: Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998, 30: 175-188. 10.1046/j.1365-2958.1998.01048.x.
Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, et al: Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J Biol Chem. 2005, 280: 15084-15096. 10.1074/jbc.M414030200.
Chao GL, Shen J, Tseng CP, Park SJ, Gunsalus RP: Aerobic regulation of isocitrate dehydrogenase gene (icd) expression in Escherichia coli by the arcA and fnr gene products. J Bacteriol. 1997, 179: 4299-4304.
Park S-J, Chao G, Gunsalus RP: Aerobic regulation of the sucABCD genes of Escherichia coli, which encode alpha-ketoglutarate dehydrogenase and succinyl coenzyme A synthetase: roles of ArcA, Fnr, and the upstream sdhCDAB promoter. J Bacteriol. 1997, 179: 4138-4142.
Gunsalus RP, Park S-J: Aerobic-anaerobic regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994, 145: 437-450. 10.1016/0923-2508(94)90092-2.
Nystrom T, Larsson C, Gustafsson L: Bacterial defense against aging: role of the Escherichia coli ArcA regulator in gene expression, readjusted energy flux and survival during stasis. EMBO J. 1996, 15: 3219-3228.
Nunn WD: A molecular view of fatty-acid catabolism in Escherichia coli. Microbiol Rev. 1986, 50: 179-192.
Lin ECC, Iuchi S: Regulation of gene expression in fermentative and respiratory systems in Escherichia coli and related bacteria. Annu Rev Genet. 1991, 25: 361-387. 10.1146/annurev.ge.25.120191.002045.
Liu XQ, De Wulf P: Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem. 2004, 279: 12588-12597. 10.1074/jbc.M313454200.
Shalel-Levanon S, San KY, Bennett GN: Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and glycolysis pathway in Escherichia coli under growth conditions. Biotechnol Bioeng. 2005, 92: 147-159. 10.1002/bit.20583.
Iuchi S, Lin EC: arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988, 85: 1888-1892. 10.1073/pnas.85.6.1888.
Iuchi S, Chepuri V, Fu HA, Gennis RB, Lin ECC: Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol. 1990, 172: 6020-6025.
Iuchi S, Aristarkhov A, Dong JM, Taylor JS, Lin ECC: Effects of nitrate respiration on expression of the Arc-controlled operons encoding succinate dehydrogenase and flavin-linked L-lactate dehydrogenase. J Bacteriol. 1994, 176: 1695-1701.
Cho BK, Knight EM, Palsson BO: Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA. Microbiology SGM. 2006, 152: 2207-2219. 10.1099/mic.0.28912-0.
Oshima T, Aiba H, Masuda Y, Kanaya S, Sugiura M, Wanner BL, et al: Transcriptome analysis of all two-component regulatory system mutants of Escherichia coli K-12. Mol Microbiol. 2002, 46: 281-291. 10.1046/j.1365-2958.2002.03170.x.
McClelland M, Florea L, Sanderson K, Clifton SW, Parkhill J, Churcher C, et al: Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 2000, 28: 4974-4986. 10.1093/nar/28.24.4974.
Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, et al: FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J Bacteriol. 2007, 189: 2262-2273. 10.1128/JB.00726-06.
Archer CD, Elliott T: Transcriptional control of the nuo operon which encodes the energy-conserving NADH dehydrogenase of Salmonella typhimurium. J Bacteriol. 1995, 177: 2335-2342.
Iuchi S, Matsuda Z, Fujiwara T, Lin EC: The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990, 4: 715-727. 10.1111/j.1365-2958.1990.tb00642.x.
Sambrook J, Russell DW: Molecular Cloning: A Laboratory Manual. 2001, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory
Porwollik S, Wong RMY, Sims SH, Schaaper RM, DeMarini DM, McClelland M: The delta uvrB mutations in the Ames strains of Salmonella span 15 to 119 genes. Mutat Res. 2001, 483: 1-11.
Satterthwaite FE: An approximate distribution of estimates of variance components. Biometrics Bull. 1946, 2: 110-114. 10.2307/3002019.
Higuchi R, Fockler C, Dollinger G, Watson R: Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY). 1993, 11: 1026-1030. 10.1038/nbt0993-1026.
Miron M, Woody OZ, Marcil A, Murie C, Sladek R, Nadon R: A methodology for global validation of microarray experiments. Bmc Bioinformatics. 2006, 7: 333-10.1186/1471-2105-7-333.
Robison K, McGuire AM, Church GM: A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J Mol Biol. 1998, 284: 241-254. 10.1006/jmbi.1998.2160.
Lee HS, Lee YS, Kim HS, Choi JY, Hassan HM, Chung MH: Mechanism of regulation of 8-hydroxyguanine endonuclease by oxidative stress: roles of fnr, arcA, and fur. Free Radic Biol Med. 1998, 24: 1193-1201. 10.1016/S0891-5849(97)00427-9.
Hassan HM, Schrum LW: Roles of manganese and iron in the regulation of the biosynthesis of manganese superoxide dismutase in Escherichia coli. FEMS Microbiol Rev. 1994, 14: 315-323. 10.1111/j.1574-6976.1994.tb00105.x.
van Helden J: Regulatory sequence analysis tools. Nucleic Acids Res. 2003, 31: 3593-3596. 10.1093/nar/gkg567.
Crooks GE, Hon G, Chandonia JM, Brenner SE: WebLogo: a sequence logo generator. Genome Res. 2004, 14: 1188-1190. 10.1101/gr.849004.
Allen CA, Fedorka-Cray PJ, Vazquez-Torres A, Suyemoto M, Altier C, Reeni Ryder L, et al: In vitro and in vivo assessment of Salmonella enterica serovar Typhimurium DT104 virulence. Infect Immun. 2001, 69: 4673-4677. 10.1128/IAI.69.7.4673-4677.2001.
Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, et al: Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005, 33: D39-D45. 10.1093/nar/gki062.
Tatusov RL, Koonin EV, Lipman DJ: A genomic perspective on protein families. Science. 1997, 278: 631-637. 10.1126/science.278.5338.631.
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al: The COG database: an updated version includes eukaryotes. Bmc Bioinformatics. 2003, 4: 41-10.1186/1471-2105-4-41.
Snavely MD, Miller CG, Maguire ME: The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J Biol Chem. 1991, 266: 815-823.
Groisman EA: The ins and outs of virulence gene expression: Mg2+ as a regulatory signal. Bioessays. 1998, 20: 96-101. 10.1002/(SICI)1521-1878(199801)20:1<96::AID-BIES13>3.0.CO;2-3.
Blanc-Potard AB, Groisman EA: The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997, 16: 5376-5385. 10.1093/emboj/16.17.5376.
Adkins JN, Mottaz HM, Norbeck AD, Gustin JK, Rue J, Clauss TRW, et al: Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol Cell Proteomics. 2006, 5: 1450-1461. 10.1074/mcp.M600139-MCP200.
Figueroa-Bossi N, Bossi L: Inducible prophages contribute to Salmonella virulence in mice. Mol Microbiol. 1999, 33: 167-176. 10.1046/j.1365-2958.1999.01461.x.
Miao EA, Scherer CA, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ, et al: Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999, 34: 850-864. 10.1046/j.1365-2958.1999.01651.x.
Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, et al: Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006, 188: 2233-2243. 10.1128/JB.188.6.2233-2243.2006.
Blanc-Potard AB, Solomon F, Kayser J, Groisman EA: The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol. 1999, 181: 998-1004.
Collazo CM, Galan JE: The invasion-associated type-III protein secretion system in Salmonella: a review. Gene. 1997, 192: 51-59. 10.1016/S0378-1119(96)00825-6.
Zhou D, Galan J: Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 2001, 3: 1293-1298. 10.1016/S1286-4579(01)01489-7.
Zhang SP, Kingsley RA, Santos RL, Andrews-Polymenis H, Raffatellu M, Figueiredo J, et al: Molecular pathogenesis of Salmonella enterica serotype typhimurium-induced diarrhea. Infect Immun. 2003, 71: 1-12. 10.1128/IAI.71.1.1-12.2003.
Shea JE, Beuzon CR, Gleeson C, Mundy R, Holden DW: Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999, 67: 213-219.
Silverman PM, Rother S, Gaudin H: Arc and Sfr functions of the Escherichia coli K-12 arcA gene product are genetically and physiologically separable. J Bacteriol. 1991, 173: 5648-5652.
Silverman PM, Wickersham E, Rainwater S, Harris R: Regulation of the F-plasmid tray promoter in Escherichia coli K-12 as a function of sequence context. J Mol Biol. 1991, 220: 271-279. 10.1016/0022-2836(91)90012-U.
Six S, Andrews SC, Unden G, Guest JR: Escherichia coli possesses two homologous anaerobic C4-dicarboxylate membrane transporters (DcuA and DcuB) distinct from the aerobic dicarboxylate transport-system (Dct). J Bacteriol. 1994, 176: 6470-6478.
Cunningham L, Gruer MJ, Guest JR: Transcriptional regulation of the aconitase genes (acnA and acnB) of Escherichia coli. Microbiology UK. 1997, 143: 3795-3805. 10.1099/00221287-143-12-3795.
Levanon SS, San KY, Bennett GN: Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol Bioeng. 2005, 89: 556-564. 10.1002/bit.20381.
Jeong JY, Kim YJ, Cho N, Shin D, Nam TW, Ryu S, et al: Expression of ptsG encoding the major glucose transporter is regulated by ArcA in Escherichia coli. J Biol Chem. 2004, 279: 38513-38518. 10.1074/jbc.M406667200.
Cotter PA, Gunsalus RP: Contribution of the Fnr and ArcA gene-products in coordinate regulation of cytochrome-o and cytochrome-d oxidase (cyoABCDE and cydAB) genes in Escherichia coli. FEMS Microbiol Lett. 1992, 91: 31-36. 10.1111/j.1574-6968.1992.tb05179.x.
Kato Y, Sugiura M, Mizuno T, Aiba H: Effect of the arcA mutation on the expression of flagella genes in Escherichia coli. Biosci Biotechnol Biochem. 2007, 71: 77-83. 10.1271/bbb.60375.
Lu S, Killoran PB, Fang FC, Riley LW: The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun. 2002, 70: 451-461. 10.1128/IAI.70.2.451-461.2002.
Gao H, Wang X, Yang ZK, Palzkill T, Zhou J: Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analysis. Bmc Genomics. 2008, 9: 42-10.1186/1471-2164-9-42.
Wong SMS, Alugupalli KR, Ram S, Akerley BJ: The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol Microbiol. 2007, 64: 1375-1390. 10.1111/j.1365-2958.2007.05747.x.
Gralnick JA, Brown CT, Newman DK: Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol Microbiol. 2005, 56: 1347-1357. 10.1111/j.1365-2958.2005.04628.x.
Romeo T: Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998, 29: 1321-1330. 10.1046/j.1365-2958.1998.01021.x.
Cotter PA, Chepuri V, Gennis RB, Gunsalus RP: Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J Bacteriol. 1990, 172: 6333-6338.
Olson JW, Maier RJ: Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002, 298: 1788-1790. 10.1126/science.1077123.
Maier RJ: Availability and use of molecular hydrogen as an energy substrate for Helicobacter species. Microbes Infect. 2003, 5: 1159-1163. 10.1016/j.micinf.2003.08.002.
Zbell AL, Benoit SL, Maier RJ: Differential expression of NiFe uptake-type hydrogenase genes in Salmonella enterica serovar Typhimurium. Microbiology. 2007, 153: 3508-3516. 10.1099/mic.0.2007/009027-0.
Obradors N, Badia J, Baldoma L, Aguilar J: Anaerobic metabolism of the L-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1988, 170: 2159-2162.
Badia J, Ros J, Aguilar J: Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae. J Bacteriol. 1985, 161: 435-437.
Price-Carter M, Tingey J, Bobik TA, Roth JR: The alternative electron acceptor tetrathionate supports B-12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J Bacteriol. 2001, 183: 2463-2475. 10.1128/JB.183.8.2463-2475.2001.
Chen P, Ailion M, Bobik T, Stormo G, Roth J: Five promoters integrate control of the cob/pdu regulon in Salmonella typhimurium. J Bacteriol. 1995, 177: 5401-5410.
Ailion M, Bobik TA, Roth JR: Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol. 1993, 175: 7200-7208.
Klumpp J, Fuchs TM: Identification of novel genes in genornic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology SGM. 2007, 153: 1207-1220. 10.1099/mic.0. 2006/004747-0.
Heithoff DM, Conner CP, Hentschel U, Govantes F, Hanna PC, Mahan MJ: Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999, 181: 799-807.
Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ: Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998, 95: 4641-4645. 10.1073/pnas.95.8.4641.
Bjorkman J, Rhen M, Andersson DI: Salmonella typhimurium cob mutants are not hyper-virulent. FEMS Microbiol Lett. 1996, 139: 121-126. 10.1016/0378-1097(96)00129-2.
Stojiljkovic I, Baumler AJ, Heffron F: Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995, 177: 1357-1366.
Sperandio V, Torres AG, Kaper JB: Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002, 43: 809-821. 10.1046/j.1365-2958.2002.02803.x.
Goodier RI, Ahmer BMM: SirA orthologs affect both motility and virulence. J Bacteriol. 2001, 183: 2249-2258. 10.1128/JB.183.7.2249-2258.2001.
Teplitski M, Goodier RI, Ahmer BMM: Pathways leading from BarA/SirA to motility and virulence gene expression in Salmonella. J Bacteriol. 2003, 185: 7257-7265. 10.1128/JB.185.24.7257-7265.2003.
Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, et al: Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999, 181: 7500-7508.
Shin S, Park C: Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995, 177: 4696-4702.
Shi WY, Zhou YN, Wild J, Adler J, Gross CA: DnaK, DnaJ, and GrpE are required for flagellum synthesis in Escherichia coli. J Bacteriol. 1992, 174: 6256-6263.
Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G: LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol. 2002, 45: 521-532. 10.1046/j.1365-2958.2002.03032.x.
Francez-Charlot A, Laugel B, Van Gemert A, Dubarry N, Wiorowski F, Castanie-Cornet MP, et al: RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol. 2003, 49: 823-832. 10.1046/j.1365-2958.2003.03601.x.
Ellermeier CD, Slauch JM: RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J Bacteriol. 2003, 185: 5096-5108. 10.1128/JB.185.17.5096-5108.2003.
Bertin P, Terao E, Lee EH, Lejeune P, Colson C, Danchin A, et al: The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J Bacteriol. 1994, 176: 5537-5540.
Altier C, Suyemoto M, Ruiz AI, Burnham KD, Maurer R: Characterization of two novel regulatory genes affecting Salmonella invasion gene expression. Mol Microbiol. 2000, 35: 635-646. 10.1046/j.1365-2958.2000.01734.x.
Hebrard M, Viala JPM, Meresse P, Barras F, Aussel L: Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009, 191: 4605-4614. 10.1128/JB.00144-09.
Horsburgh MJ, Wharton SJ, Karavolos M, Foster SJ: Manganese: elemental defence for a life with oxygen?. Trends Microbiol. 2002, 10: 496-501. 10.1016/S0966-842X(02)02462-9.
Hacker J, Kaper J: The concept of pathogenicity islands. Pathogenicity Islands and Other Mobile Virulence Elements. Edited by: Hacker J, Kaper J. 1999, Washington, DC: American Society for Microbiology, 1-11.
Bowe F, Lipps CJ, Tsolis RM, Groisman E, Heffron F, Kusters JG: At least four percent of the Salmonella typhimurium genome is required for fatal infection of mice. Infect Immun. 1998, 66: 3372-3377.
Hensel M, Nikolaus T, Egelseer C: Molecular and functional analysis indicates a mosaic structure of Salmonella pathogenicity island 2. Mol Microbiol. 1999, 31: 489-498. 10.1046/j.1365-2958.1999.01190.x.
Hensel M: Salmonella pathogenicity island 2. Mol Microbiol. 2000, 36: 1015-1023. 10.1046/j.1365-2958.2000.01935.x.
Groisman EA, Ochman H: Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993, 12: 3779-3787.
Shea JE, Hensel M, Gleeson C, Holden DW: Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996, 93: 2593-2597. 10.1073/pnas.93.6.2593.
Collazo CM, Galan JE: The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997, 24: 747-756. 10.1046/j.1365-2958.1997.3781740.x.
Ochman H, Groisman EA: Distribution of pathogenicity islands in Salmonella spp. Infect Immun. 1996, 64: 5410-5412.
Deiwick J, Nikolaus T, Erdogan S, Hensel M: Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999, 31: 1759-1773. 10.1046/j.1365-2958.1999.01312.x.
Chan K, Kim CC, Falkow S: Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect Immun. 2005, 73: 5438-5449. 10.1128/IAI.73.9.5438-5449.2005.
Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM: Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2006, 2: e11-10.1371/journal.ppat.0020011.
Yoon H, Lim S, Heu S, Choi S, Ryu S: Proteome analysis of Salmonella enterica serovar Typhimurium fis mutant. FEMS Microbiol Lett. 2003, 226: 391-396. 10.1016/S0378-1097(03)00641-4.
Wilson RL, Libby SJ, Freet AM, Boddicker JD, Fahlen TF, Jones BD: Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol Microbiol. 2001, 39: 79-88. 10.1046/j.1365-2958.2001.02192.x.
Sheikh J, Hicks S, Dall'Agnol M, Phillips AD, Nataro JP: Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol. 2001, 41: 983-997. 10.1046/j.1365-2958.2001.02512.x.
Schmitt CK, Ikeda JS, Darnell SC, Watson PR, Bispham J, Wallis TS, et al: Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect Immun. 2001, 69: 5619-5625. 10.1128/IAI.69.9.5619-5625.2001.
Schechter LM, Jain S, Akbar S, Lee CA: The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect Immun. 2003, 71: 5432-5435. 10.1128/IAI.71.9.5432-5435.2003.
Goldberg MD, Johnson M, Hinton JCD, Williams PH: Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol Microbiol. 2001, 41: 549-559. 10.1046/j.1365-2958.2001.02526.x.
Falconi M, Prosseda G, Giangrossi M, Beghetto E, Colonna B: Involvement of Fis in the H-NS-mediated regulation of virF gene of Shigella and enteroinvasive Escherichia coli. Mol Microbiol. 2001, 42: 439-452. 10.1046/j.1365-2958.2001.02646.x.
Acknowledgements
This work was supported in part by the North Carolina Agricultural Research Services (to HMH), and by NIH grants R01AI034829, R01AI022933, R21AI057733, and R01AI52237 and generous gifts from Mr. Sidney Kimmel and Mr. Ira Lechner (MM and SP), NIH grants AI054959 and RR16082 (AV-T and JJ-C). We appreciate the donation of the anti-ArcA antibodies from Dr. Philip Silverman and Ms. Robin Harris at the Department of Botany and Microbiology, University of Oklahoma. We would also like to thank Valerie Knowlton for her assistance with the microscopy. We are grateful to Drs. Gabriele Gusmini and Russell Wolfinger for their assistance with the statistical analyses/SAS software.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
MRE transduced and confirmed the arcA mutant, constructed and confirmed the complement plasmid (parcA), performed the Western blotting, performed the growth studies in aerobic and anaerobic conditions, compared our microarray data to that in other studies, conducted the qRT-PCR and lethality studies in the presence of hydrogen peroxide, participated in the motility studies, and contributed to the writing/editing of the manuscript. RCF conducted the microarrays and performed the analysis, constructed the logo, participated in motility studies, and contributed to the editing of the manuscript. AV-T and JJ-C carried-out all of the mice studies. MM and SP constructed and provided the microarray slides. HMH conceived the idea, directed the research, and contributed to the writing/editing of the manuscript. All authors have read and approved the final manuscript.
Electronic supplementary material
12866_2010_1345_MOESM1_ESM.DOC
Additional file 1: Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv . Typhimurium. Identification of ArcA by Western blot; Effects of H2O2 on viability of the ArcA mutant; List of genes differentially regulated by ArcA; and List of genes shared with the Fnr regulon. A. Supplemental Methods: Western blot analysis of ArcA. H2O2 survival assays. B. Supplemental Figures: Figure S1. Western blot of total proteins of the WT, arcA mutant, and arcA-/parcA complement strains. Figure S2. Effects of hydrogen peroxide on viability of the WT and the arcA mutant under anerobiosis. C. Supplemental Tables: Table S1. Differentially expressed genes and the presence/absence of putative ArcA-binding motifs in their 5' regions. Table S2. Comparison of the 120 genes shared between the ArcA and the Fnr regulons of S. Typhimurium under anaerobiosis. (DOC 1014 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Evans, M.R., Fink, R.C., Vazquez-Torres, A. et al. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol 11, 58 (2011). https://doi.org/10.1186/1471-2180-11-58
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-11-58