Abstract
Background
Immunological and genetic findings implicate Th17 effector cytokines in the pathogenesis of inflammatory bowel disease (IBD). Expression of Th17 pathway-associated genes is mainly studied in colonic disease. The present study assessed the mRNA expression levels of Th17 effector cytokines (IL17A, IL17F, IL21, IL22 and IL26) and genes involved in differentiation (IL6, IL1B, TGFB1, IL23A and STAT3) and recruitment of Th17 cells (CCR6 and CCL20) by quantitative real-time PCR analysis of colonic and ileal biopsies from 22 healthy control subjects, 26 patients with Crohn's disease (CD) and 12 patients with ulcerative colitis (UC). Inflammation was quantified by measuring expression of the inflammatory mediators IL8 and TNF.
Results
Evaluation of mRNA expression levels in colonic and ileal control samples revealed that TNF, TGFB1, STAT3 and CCR6 were expressed at higher levels in the ileum than in the colon. Expression of all the Th17 pathway-associated genes was increased in inflamed colonic samples. The increased expression of these genes was predominantly observed in samples from UC patients and was associated with more intense inflammation. Although increased expression of IL17A, IL17F, IL21 and IL26 was detected in inflamed ileal samples, expression of the indispensable Th17 cell differentiation factors TGFB1 and IL23A, the signaling molecule STAT3 and the Th17 recruitment factors CCR6 and CCL20 were unchanged.
Conclusions
Our findings suggest that immune regulation is different in colonic and ileal disease, which might have important consequences for therapeutic intervention.
Similar content being viewed by others
Background
The location and pattern of inflammation in inflammatory bowel disease (IBD) are variable. Whereas ulcerative colitis (UC) is limited to the colon with a sharp delineation between the involved and non-involved mucosa, Crohn's disease (CD) can affect any part of the gastrointestinal tract and is associated with patchy distribution of mucosal lesions. Ileal localization occurs in about 80% of CD patients, and about 30% of CD patients have isolated ileal disease. Although it is generally accepted that IBD develops as a result of an altered immune response to luminal content in a genetically susceptible host, the mechanism by which the site of disease is selected remains unknown. Important differences in function, architecture and bacterial distribution between the ileum and colon have been described. Peyer's patches, which consist of aggregated lymphoid tissue and play a central role in the induction of mucosal immune responses, are a hallmark of the terminal ileum. Increased numbers of mucosa-associated E. coli are observed in IBD, and adherent invasive E. coli (AIEC) strains were highly associated with the ileal mucosa in CD patients. Moreover, the reduced number of goblet cells in the ileum results in decreased mucus secretion and increased contact between the mucosa and luminal content [1–3].
Several subsets of T helper (Th) cells contribute to defensive responses at inflammatory sites [4]. Dendritic cell-derived cytokines skew the differentiation of naïve CD4+ T cells into Th1, Th2, Th17 or regulatory T cell (Treg) subsets. For many years, CD was believed to be mediated by Th1 cytokines, while UC was believed to be mediated by Th2 cytokines; however, recent data have implicated Th17 cells in the pathogenesis of IBD [5–9]. The invasion of extracellular bacteria into the intestinal mucosa triggers the expression of IL-23A, driving Th17 cells to release IL-17A, IL-17F, IL-21, IL-22 and IL-26, which in turn exert a number of proinflammatory effects on intestinal epithelial cells, endothelial cells, macrophages and fibroblasts [10]. In addition to their proinflammatory functions, IL-17A, IL-17F and IL-22 have been reported to induce increased expression of epithelial barrier protective genes such as defensins, mucins, tight junction proteins and lipopolysaccharide-binding proteins [11–14].
The differentiation of Th17 cells depends on the activation of janus kinase 2 (JAK2), signal transducer and activator of transcription 3 (STAT3) and the transcription factor RAR-related organ receptor C2 (RORC2) and is regulated by a combination of cytokines, including IL-6, IL-1B (IL-1β), TGFB1 (TGFβ), IL-23A, and the autocrine activity of IL-21 [4, 15–20]. Chemokine (C-C motif) receptor 6 (CCR6) which is expressed on the surface of Th17 cells, contributes to their recruitment to chemokine (C-C motif) ligand 20 (CCL20) produced at the inflamed mucosa [21].
The important role of Th17 cells in the pathogenesis of IBD is also supported by genome-wide association studies, which have demonstrated that CCR6, STAT3, JAK2, IL23R and IL12B are CD susceptibility genes [22–24]. Interestingly, single nucleotide polymorphisms (SNPs) within IL23R, IL12B, STAT3, JAK2, the IL22/IL26 and the IL2/IL21 gene cluster have also been found to be associated with UC [22, 24–27].
Although the expression of Th17-related genes has been studied previously, most studies included only colonic samples and were focused on a limited number of genes. Increased expression of IL17A, IL17F, IL22, IL26, IL21, CCL20 and CCR6 has been found in inflamed colonic tissues of IBD patients [4, 14, 28–33]. In only one study, IL17A and IL23 were mildly increased in active ileal CD samples [34].
To examine the possible differences in the expression levels of genes involved in the Th17 pathway, we assessed the mRNA levels of the Th17 effector cytokines and genes involved in the differentiation and recruitment of Th17 cells in both colonic and ileal biopsies of healthy controls, UC patients and CD patients.
Results
The mRNA expression levels of inflammatory cytokines and Th17-related genes in colonic and ileal samples of healthy controls
The expression level of the proinflammatory cytokine IL8 was equal in colonic and ileal controls, while the expression level of TNF (TNFα) was slightly higher in ileal control samples than in colonic control samples (P = 0.037) (Figure 1A).
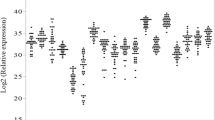
mRNA expression levels of proinflammatory and Th17-pathway-associated genes in colon and ileum of healthy controls. The relative transcription levels are determined by SYBR Green qPCR using the geometric mean of GAPDH, HPRT and SDHA as an endogenous control in colonic and ileal samples from healthy controls. The data are expressed as geometric means. The geometric mean of the colonic controls was set as 1. Data are presented on a log scale. (a) proinflammatory cytokines, (b) Th17 effector cytokines, (c) Th17 differentiation factors, (d) Th17 recruitment factors. *P < 0.05, **P < 0.01, ***P < 0.001
The expression levels of the Th17 effector cytokines IL17A, IL17F, IL21, IL22 and IL26 were comparable between colonic and ileal control samples (Figure 1B); however, the expression of these cytokines in each individual sample was not consistent. In one ileal sample and none of the colonic samples, all five cytokines were expressed. In one colonic sample and four ileal samples, three to four cytokines were expressed, while in eight colonic samples and seven ileal samples, one to two cytokines were expressed. In four colonic samples and three ileal samples, none of the five cytokines were detected.
We next examined the expression of genes involved in the differentiation of Th17 cells. TGFβ (P = 0.0005) and STAT3 (P = 0.007) were expressed at higher levels in ileal control samples than in colonic control samples, while IL6, IL1β and IL23A expression levels were similar in colonic and ileal control samples (Figure 1C).
Expression of genes involved in the recruitment of Th17 cells was also assessed. CCR6 expression was higher in ileal control samples than in colonic control samples (P = 0.0008), while CCL20 expression was similar between colonic and ileal controls (Figure 1D).
The mRNA expression levels of inflammatory mediators in inflamed colonic and ileal samples of IBD patients
Although all samples were taken from endoscopically inflamed mucosa, we quantified inflammation by measuring the expression of the proinflammatory cytokines IL8 and TNFα. The expression level of IL8 has been shown to be associated with the grade of inflammation [35, 36]. IL8 was strongly induced in inflamed colonic samples from UC (P < 0.0001) and CD patients (P = 0.0004) and in inflamed ileal samples from CD patients (P < 0.0001) (Figure 2A). The expression level of IL8 was significantly higher in UC samples than in colonic CD samples (P = 0.017). Expression of TNFα was only significantly increased in UC samples (P = 0.0002); however, a tendency for increase was observed in ileal CD samples (P = 0.052) (Figure 2A). Expression levels of TNF α were significantly higher in UC samples than in colonic CD samples (P = 0.028).
mRNA expression levels of proinflammatory and Th17-pathway-associated genes in inflamed colonic and ileal samples of IBD patients. Fold changes in transcription levels are determined by SYBR Green qPCR using the geometric mean of GAPDH, HPRT and SDHA as an endogenous control in inflamed colonic UC, colonic CD and ileal CD samples. The fold changes shown are relative to control samples. The data are presented as box plots with medians and interquartile ranges. (a) proinflammatory cytokines, (b) Th17 effector cytokines, (c) Th17 differentiation factors, (d) Th17 recruitment factors. *P < 0.05, **P < 0.01, ***P < 0.001, #P < 0.0001
The mRNA expression levels of Th17 effector cytokines in inflamed colonic and ileal samples of IBD patients
In UC samples, IL17A (P = 0.0002), IL21 (P = 0.0021), IL22 (P = 0.0003) and IL26 (P = 0.0017) were strongly induced, and IL17F (P = 0.046) was weakly induced, while in colonic CD samples, IL17A (P = 0.0084) and IL22 (P = 0.001) were strongly induced, and IL21 (P = 0.047), IL26 (P = 0.048) and IL17F (P = 0.011) were weakly induced (Figure 2B). In ileal CD samples, IL22 (P = 0.0015) was strongly induced, while IL17A (P = 0.013), IL26 (P = 0.029) and IL17F (P = 0.019) were weakly induced, and expression levels of IL21 were similar to those in ileal controls. Furthermore, in the colon, expression levels of IL17A (P = 0.005) and IL21 (P = 0.036) were significantly higher in UC samples than in CD samples. In CD samples, expression of IL21 (P = 0.0097) was significantly higher in the colon than in the ileum.
The mRNA expression levels of genes involved in the differentiation of Th17 cells in inflamed colonic and ileal samples of IBD patients
IL1β (UC, P≤0.0001; CD, P = 0.0009), IL6 (UC, P≤0.0001; CD, P = 0.0007), TGFβ (UC, P = 0.0002; CD, P = 0.0001) and IL23A (UC, P = 0.0006; CD, P = 0.003) were strongly induced in both UC and colonic CD (Figure 2C). STAT3 was only slightly induced in colonic CD (P = 0.015) and UC (P = 0.018). Although a strong induction of the Th17 differentiation factor IL1β (P = 0.0007) and a weak induction of IL6 (P = 0.015) were observed in ileal samples, TGFβ, IL23A and STAT3 were not induced. In the colon, IL23A was expressed at higher levels in UC samples than in CD samples (P = 0.027). In CD samples, TGFβ (P = 0.0008) and IL23A (P = 0.010) were expressed at higher levels in the colon than in the ileum.
The mRNA expression levels of genes involved in the recruitment of Th17 cells in inflamed colonic and ileal samples of IBD patients
Expression levels of both CCR6 (UC, P = 0.0004; CD, P = 0.011) and CCL20 (UC; P = 0.0003, CD; P = 0.0009) were significantly increased in colonic CD and UC samples but not in ileal CD samples (Figure 2D).
The mRNA expression levels of the master transcription factor RORC in inflamed colonic and ileal samples of IBD patients
Two sets of primers for RORC were developed. The RORC primers detect both the full-length transcript and the shorter T cell-specific isoform, while the RORC1 primers detect only the full-length transcript. A strong correlation was found between RORC1 and RORC expression levels (R = 0.776, P < 0.001). When comparing the RORC1 and RORC levels between healthy controls and UC and CD patients, higher significance levels were achieved using the primer detecting both transcripts. This suggests a stronger role for the T cell-specific RORC2 isoform.
In colonic samples from CD and UC patients, RORC mRNA expression levels were comparable to those in healthy controls, while in ileal CD samples, expression levels of RORC were lower than those in control samples (P = 0.0019) (Figure 3).
mRNA expression levels of RORC in inflamed colonic and ileal samples of IBD patients. The relative transcription levels of RORC are determined by SYBR Green qPCR using the geometric mean of GAPDH, HPRT and SDHA as an endogenous control in (a) colonic and (b) ileal samples from healthy controls, UC patients and CD patients. The geometric mean of the control samples was set as 1. Data are presented on a log scale. **P < 0.01
Discussion
A growing body of human studies and studies in mouse models has shown that Th17 effector cytokines promote chronic intestinal inflammation through the induction of multiple proinflammatory mediators. The role of Th17 cells has predominantly been studied in CD; these cells have rarely been studied in UC and have never been studied in healthy controls. Although it is generally accepted that IBD develops as a result of an altered immune response to luminal content in a genetically susceptible host, the factors influencing the selection of the disease site remain unknown. At present, even the mechanisms controlling development of ileitis and/or colitis in transgenic or gene-targeted mouse models are unclear.
In the present screening study the mRNA expression signatures of Th17 pathway-associated genes were evaluated in colonic and ileal samples of healthy controls, UC and CD patients. We first compared the expression levels in healthy colons to those in healthy ilea. Evaluation of the Th17 effector cytokines revealed no significant differences in expression levels between the colon and the ileum. Notably, these cytokines were not detectable in the majority of control samples, although expression was more frequently observed in ileal samples than in colonic samples. In addition, the expression of one Th17 effector cytokine was not necessarily linked to the expression of the other effector cytokines in a single mucosal specimen. The relative stasis of luminal content in the terminal ileum supports our observation that Th17 effector cytokines were more often present in the ileum than in the colon because IL-17A, IL-17F and IL-22 enhance the production of antimicrobial peptides, which protect the intestinal mucosa against bacterial invasion [11–14]. The diversity of bacteria present among individuals might explain why not all individuals express Th17 effector cytokines.
Recently, a subset of CD4+ T cells that provide help to B cells for antibody production in germinal centers (GC), termed follicular helper T cells (Tfh), were identified [37, 38]. Tfh cells produce IL-21, which is necessary for GC formation. The increased number of ileal controls expressing IL21 could reflect the presence of Peyer's patches in the terminal ileum, where B lymphocytes predominate in the GC. The associated increased ileal expression of the downstream signaling molecule STAT3 supports this assumption.
CCR6, which is present not only on Th17 cells but also on Treg cells, B cells, neutrophils and immature dendritic cells, plays a critical role in the migration of these cells to its ligand CCL20, which is produced at inflammatory sites [14, 20, 33]. CCR6-positive cells have been detected in lymphoid organs like Peyer's patches and isolated lymphoid follicles and seem to be more common in the ileum than in the colon. TGFβ was shown to be the main factor for induction of CCR6 mRNA expression in Th17 cells and dendritic cells [39, 40]. In our study, the increased expression of CCR6 in ileal controls was correlated with increased ileal TGFβ (r = 0.785, P = 0.0003).
The increased expression of the proinflammatory cytokine IL8 in the inflamed colonic and ileal IBD samples confirmed the endoscopic inflammatory state of our samples. The relationship between the expression of proinflammatory cytokines and the grade of inflammation or disease activity index has been described before and is supported by our observation that IL8 expression levels in samples from patients in remission are similar to those of healthy controls (data not shown) [35, 36]. TNFα is less useful as parameter of inflammation because reports about its expression and secretion in inflamed samples are contradictory [41–43]. In support of this view, expression of TNFα was increased in UC patients and not increased in CD patients.
Except for ileal IL21, significant induction of all Th17 effector cytokines was observed in inflamed colonic and ileal IBD samples. Moreover, induction of IL17A and IL21 was significantly more pronounced in UC than in colonic CD, and this induction was associated with more intense inflammation as defined by increased induction of IL8. In contrast, this association was not found in ileal CD where only marginal induction of Th17 cytokines was detected in ileal CD samples with IL8 expression levels similar to those observed in UC samples. Marked induction of IL1β, IL6, TGFβ and IL23A, genes involved in the differentiation of Th17 cells, was observed in colonic inflammation. The downstream signaling molecule STAT3 was only moderately increased in colonic samples. In ileal CD samples, except for a strong induction of IL1β, only a weak induction of IL6 and no increase in TGFβ, IL23A and STAT3 was detected. In parallel, significant increases in CCR6 and CCL20, genes involved in the recruitment of Th17 cells, were only observed in colonic samples, supporting a less pronounced infiltration of Th17 cells in ileal inflammation.
The observed increase in expression levels of IL17A, IL17F, IL22 and IL26 and the downstream proinflammatory cytokines IL6 and IL1β in the inflamed ileum could originate from cells other than Th17 cells. Lymphoid tissue inducer-like cells, which are important in the development of lymphoid organs, are an innate source of IL-17A and IL-22 [44]. Paneth cells, which are common in the ileum, also express IL-17A [45]. Natural killer cells, natural killer T cells and a newly identified T helper cell, Th22 cells, which are involved in inflammatory skin disorders, are sources of IL-22 [46]. Importantly, IL-17A, IL-17F and IL-22 mediate protective effects through the induction of defensins [11–14].
Although mRNA expression levels are not the optimal way to study the activity of transcription factors, the significantly reduced expression of ileal RORC support a defect in the Th17 pathway in ileal disease.
We should consider the fact that gene expression could be affected by the use of anti-inflammatory drugs. Although 42% of the included patients were on medication, statistical analysis did not show an effect.
Given that SNPs within CCR6, STAT3, JAK2, IL23R, IL12B, the IL22/IL26 and the IL2/IL21 gene cluster have been found to be associated with CD and or UC, we should take in mind the influence of these SNPs on Th17 cytokine profiles. Recently, response to anti-TNF therapy was demonstrated to be modulated by IL23R variants, linking Th17 function to biologicals [47]. Twenty-two percent of the samples in this study were included in the GWAS of Barrett and thus genotyped for CCR6, STAT3, JAK2, IL23R and IL12B[23]. Unfortunately, due to a low frequency of patients heterozygous or homozygous for the different risk alleles, such comparisons were not conclusive [47].
A lot of factors inherent to the heterogeneous nature of biopsies may influence RNA levels. Although we have to be cautious with the extrapolation of mRNA expression data to functional immunological conclusions, biological replicates and the previously demonstrated association between RNA and protein levels of most genes included in this study contribute to reliable conclusions.
Conclusions
In conclusion, this study demonstrated important differences in the expression of Th17-associated genes in colonic and ileal disease. Inflammation of the colon of both CD and UC patients is clearly associated with increased expression levels of Th17 effector cytokines and genes involved in the differentiation, amplification and recruitment of Th17 cells, whereas in the inflamed ileum, essential factors for differentiation and recruitment of Th17 cells are missing. The differential expression of Th17-related genes between the colon and ileum could reflect different immune regulation in the colon and ileum, suggesting different therapeutic approaches for CD patients with colonic versus ileal disease. Future clinical trials of agents blocking Th17-related genes should take into account the disease location in CD patients. In addition, the difference in expression profiles between the colon and ileum also provide the potential of identifying diagnostic biomarkers.
Methods
Patients and samples
A total of 72 macrodissected intestinal tissue samples from 22 healthy controls, 12 UC patients and 26 CD patients were obtained during colonoscopy with a Single-Use Biopsy Forceps Radial Jaw3 (Boston Scientific, El Coyol, Costa Rica) (Table 1). The size of a biopsy specimen was usually between 2-4 mm2 with an estimated average weight of 6.4 mg. UC and CD patients were diagnosed based on clinical, endoscopic and histological criteria. Patients characteristics, medication intake and the Montreal classification which gives an overview of the subclassification of IBD patients is shown in Table 1[48]. Mucosal inflammation was defined as the presence of endoscopic signs of disease activity. Samples from healthy controls were taken from the ileum and sigmoid of patients who underwent colonoscopy to screen for cancer or polyps. All biopsies collected during colonoscopy were immediately stored in RNALater (Ambion, Cambridgeshire, UK) at -80°C. The study was in accordance with the guidelines of the Helsinki Declaration (1964 and amended in 1975, 1983, 1989, 1996 and 2000) of the World Medical Association. Informed consent was obtained from all patients, and the protocol was approved by the local Ethics Committee of Ghent University Hospital (EC UZG 2004/242).
RNA extraction, cDNA synthesis and amplification
Total RNA was extracted from 2-3 pooled mucosal samples using an RNeasy Mini Kit (Qiagen, Westburg BV, The Netherlands) with on-column DNAse treatment (Qiagen). Needle homogenization was performed. The total RNA was quantified using spectrophotometry (Nanodrop; Thermo Scientific, Wilmington, USA) and ranged from 150 ng to 16.8 μg with an average of 5.9 μg total RNA. The quality of the RNA, expressed as RNA quality indicator (RQI), was checked by automated electrophoresis (Experion, Bio-Rad, Hercules, California) and ranged from 7.4 to 10 with an average of 8.6. Starting from 20 ng of total RNA, the WT-Ovation RNA Amplification System (Nugen Technologies Inc., San Carlos, USA) was used to the letter of the manufacturer's instructions, generating approximately 6 μg of cDNA. First strand cDNA is prepared from total RNA using both oligo-dT and random hexamer primers and reverse transcriptase. After the generation of double strand cDNA, a DNA amplification step developed by NuGEN was performed. The cDNA was diluted to 50 μl.
Quantitative real-time PCR
PCR amplification reactions were carried out in a total volume of 8 μl containing 2× SYBR Green I Master Mix (Eurogentec, Seraing, Belgium), 3 μl 1/100 cDNA (~3.75 ng) and 250 nM forward and reverse primers (BioLegio, Nijmegen, The Netherlands). All reactions were performed in 384-well plates (LightCycler 480 Multiwell Plates 384, white and LightCycler 480 Sealing Foils from Roche) on the CFX384 real-time PCR detection system (Bio-Rad, Hercules, California), followed by a regression Cq value determination method. Cycling conditions were as follows: 95°C for 10 min followed by 45 cycles of 95°C for 10 s and 60°C for 30 s, followed by a dissociation curve analysis from 60 to 95°C. Because instrument and liquid handling variations were shown to be minimal using the Tecan Freedom Evo robot for pippeting (< 6% CV in 0.5 μl and <3% CV in 2 μl), and a large number of biological replicates were used, no PCR replicates were carried out. Primers containing neither SNPs nor secondary structures were designed for Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), Succinate dehydrogenase complex subunit A (SDHA), Hypoxanthine-guanine phosphoribosyltransferase (HPRT), IL8, TNF, TGFB1, IL1B, IL6, IL23A, CCR6, STAT3, IL17A, IL17F, IL21, IL22, IL26, CCL20 and RORC (Table 2). For RORC, two sets of primers were designed, RORC, which detects the mRNA levels of both isoforms, and RORC1 which detects only the full-length transcript. BLAST searches confirmed that only the target genes were 100% covered. A 6 point 4-fold standard dilution series (highest concentration; 32 ng/μl) of a cDNA mixture of all samples included in the study diluted in 5 ng/μl tRNA (Roche) was used to test the PCR efficiency of the primers. The dynamic range had to cover at least 3 orders of dilution. Only primers with an efficiency between 88% and 112% were retained (Table 2). Correlation coefficients of the targets were between 0.9843<R2 < 1, with a mean of 0.9942 (Table 2). The PCR efficiency for each gene was calculated according to the equation E = 10 (-1/slope). Each sample has been revised for a melting-curve with a single sharp peak with a high correlation between the observed and the expected Tm (mean variation of 0.9°C). Samples with other patterns than a single sharp peak at the expected Tm, defined as multiple peaks, a single broader peak or a shoulder peak, were omitted. Cq values of samples with flattened melting-curves were set as 45. An amplification signal in the no template control (NTC) was ignored as long as the difference in Cq value between the NTC and the highest Cq >5. Although the pre-amplification method of NuGEN does not amplify genomic DNA, possible gDNA contamination was assessed using intronic primers. We confirmed that gDNA was undetectable in a dilution of up to 32 ng/μl cDNA [49]. The mRNA expression level of each gene was determined in Excel by using the comparative 2-(delta delta Cq) method and normalized to the geometric mean of the stably-expressed reference genes GAPDH, SDHA and HPRT as determined by geNorm [50].
According to the MIQE guidelines, the minimum information for publication of quantitative real-time PCR experiments was provided [51].
Statistical analysis
Statistical differences were assessed using a non-parametrical Mann-Whitney U test (two tailed probabilities). The correlations were analyzed with Spearman's correlation coefficient. P-values less than 0.05 were considered significant. Statistical analysis was performed using SPSS software 11.5 (SPSS, Chicago, USA).
References
Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF: Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterol. 1998, 115: 1405-1413. 10.1016/S0016-5085(98)70019-8.
Kotlowski R, Bernstein CN, Sepehri S, Krause DO: High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007, 56: 669-675. 10.1136/gut.2006.099796.
Burke DA, Axon ATR: Adhesive Escherichia coli in inflammatory bowel disease and infective diarrhea. BMJ. 1988, 297: 102-104. 10.1136/bmj.297.6641.102.
Brand S: Crohn's disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn's disease. Gut. 2009, 58: 1152-1167. 10.1136/gut.2008.163667.
Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D: IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006, 116: 1310-1316. 10.1172/JCI21404.
Parronchi P, Romagnani P, Annunziato F, Sampognaro S, Becchio A, Giannarini L, Maggi E, Pupilli C, Tonelli F, Romagnani S: Type 1 T-helper cell predominance and interleukin-12 expression in the gut of patients with Crohn's disease. Am J Pathol. 1997, 150: 823-832.
Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, Powrie F, Maloy KJ: Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006, 203: 2473-2483. 10.1084/jem.20061099.
Hue S, Maloy K, McKensie B, Cua D, Powrie F: IL-23 and not IL-12 is essential for the development of IBD. Inflamm Bowel Dis. 2006, 12: S25-10.1097/00054725-200604002-00059.
Heller F, Florian P, Bojarski C, Richter J, Christ M, Hillenbrand B, Mankertz J, Gitter AH, Burgel N, Fromm M, Zeitz M, Fuss I, Strober W, Schulzke JD: Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterol. 2005, 129: 550-564.
Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F, Lecron JC, Kastelein RA, Cua DJ, McClanahan TK, Bowman EP, Malefyt RD: Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007, 8: 950-957. 10.1038/ni1497.
Wolk K, Kunz S, Witte E, Friedrich M, Asadullah K, Sabat R: IL-22 increases the innate immunity of tissues. Immunity. 2004, 21: 241-254. 10.1016/j.immuni.2004.07.007.
Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, Fouser LA: Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med. 2006, 203: 2271-2279. 10.1084/jem.20061308.
Kinugasa T, Sakaguchi T, Gu XB, Reinecker HC: Claudins regulate the intestinal barrier in response to immune mediators. Gastroenterol. 2000, 118: 1001-1011. 10.1016/S0016-5085(00)70351-9.
Brand S, Beigel F, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Diepolder H, Marquardt A, Jagla W, Popp A, Leclair S, Herrmann K, Seiderer J, Ochsenkuhn T, Goke B, Auernhammer CJ, Dambacher J: IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006, 290: G827-G838. 10.1152/ajpgi.00513.2005.
Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C: T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008, 28: 29-39. 10.1016/j.immuni.2007.11.016.
Yang L, Anderson DE, Baecher-Allan C, Hastings WD, Bettelli E, Oukka M, Kuchroo VK, Hafler DA: IL-21 and TGF-beta are required for differentiation of human T(H)17 cells. Nature. 2008, 454: 350-U55. 10.1038/nature07021.
Pene J, Chevalier S, Preisser L, Venereau E, Guilleux MH, Ghannam S, Moles JP, Danger Y, Ravon E, Lesaux S, Yssel H, Gascan H: Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008, 180: 7423-7430.
Manel N, Unutmaz D, Littman DR: The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor ROR gammat. Nat Immunol. 2008, 9: 641-649. 10.1038/ni.1610.
Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR: The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006, 126: 1121-1133. 10.1016/j.cell.2006.07.035.
Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, Giudici F, Romagnani P, Parronchi P, Tonelli F, Maggi E: Phenotypic and functional features of human Th17 cells. J Exp Med. 2007, 204: 1849-1861. 10.1084/jem.20070663.
Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM: Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008, 180: 214-221.
Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang HY, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH: A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006, 314: 1461-1463. 10.1126/science.1135245.
Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwilliam R, Tremelling M, Deloukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ: Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature Genetics. 2008, 40: 955-962. 10.1038/ng.175.
Anderson CA, Massey DCO, Barrett JC, Prescott NJ, Tremelling M, Fisher SA, Gwilliam R, Jacob J, Nimmo ER, Drummond H, Lees CW, Onnie CM, Hanson C, Blaszczyk K, Ravindrarajah R, Hunt S, Varma D, Hammond N, Lewis G, Attlesey H, Watkins N, Ouwehand W, Strachan D, McArdle W, Lewis CM, Lobo A, Sanderson J, Jewell DP, Deloukas P, Mansfield JC, Mathew CG, Satsangi J, Parkes M: Investigation of Crohn's disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterol. 2009, 136: 523-529. 10.1053/j.gastro.2008.10.032.
Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M, Schreiber S: Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nature Genetics. 2008, 40: 713-715. 10.1038/ng.148.
Silverberg MS, Cho JH, Rioux JD, McGovern DPB, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, Barmada MM, Klei L, Daly MJ, Abraham C, Bayless TM, Bossa F, Griffiths AM, Ippoliti AF, Lahaie RG, Latiano A, Pare P, Proctor DD, Regueiro MD, Steinhart AH, Targan SR, Schumm LP, Kistner EO, Lee AT, Gregersen PK, Rotter JI, Brant SR, Taylor KD, Roeder K, Duerr RH: Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nature Genetics. 2009, 41: 216-220. 10.1038/ng.275.
Glas JN, Stallhofer J, Ripke S, Wetzke M, Pfennig S, Klein W, Epplen JT, Griga T, Schiemann U, Lacher M, Koletzko S, Folwaczny M, Lohse P, Goke B, Ochsenkuhn T, Muller-Myhsok B, Brand S: Novel genetic risk markers for ulcerative colitis in the IL2/IL21 region are in epistasis with IL23R and suggest a common genetic background for ulcerative colitis and celiac disease. Am J Gastroenterol. 2009, 104: 1737-1744. 10.1038/ajg.2009.163.
Schmechel S, Konrad A, Diegelmann J, Glas J, Wetzke M, Paschos E, Lohse P, Goke B, Brand S: Linking genetic susceptibility to Crohn's disease with Th17 cell function: IL-22 serum levels are increased in Crohn's disease and correlate with disease activity and IL23R genotype status. Inflamm Bowel Dis. 2008, 14: 204-212. 10.1002/ibd.20315.
Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y: Increased expression of interleukin-17 in inflammatory bowel disease. Gut. 2003, 52: 65-70. 10.1136/gut.52.1.65.
Dambacher J, Beigel F, Golluscio R, Olszak T, Zitzmann K, Eichhorst ST, Otte JM, Seiderer J, Diepolder H, Auemhammer CJ, Ochsenkuehn T, Goeke B, Brand S: The novel IL-10 related cytokine IL-26 is increased in active inflammatory bowel disease and intestinal epithelial cells express the functional IL-26 receptor complex. Gastroenterol. 2006, 130: A696-
Andoh A, Zhang ZB, Inatomi O, Fujino S, Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S, Takayanagi A, Shimizu N, Fujiyama Y: Interleukin-22, a member of the IL-10 subfamily, induces inflammatory responses in colonic subepithelial myofibroblasts. Gastroenterol. 2005, 129: 969-984. 10.1053/j.gastro.2005.06.071.
Seiderer J, Elben I, Diegelmann J, Glas J, Stallhofer J, Tillack C, Pfennig S, Jurgens M, Schmechel S, Konrad A, Goke B, Ochsenkuhn T, Muller-Myhsok B, Lohse P, Brand S: Role of the novel th17 cytokine IL-17F in inflammatory bowel disease (IBD): Upregulated colonic IL-17F expression in active Crohn's disease and analysis of the IL17F p. Hisl6lArg polymorphism in IBD. Inflamm Bowel Dis. 2008, 14: 437-445. 10.1002/ibd.20339.
Brand S, Olszak T, Beigel F, Diebold J, Otte JM, Eichhorst ST, Goke B, Dambacher J: Cell differentiation dependent expressed CCR6 mediates ERK-1/2, SAPK/JNK, and Akt signaling resulting in proliferation and migration of colorectal cancer cells. J Cell Biochem. 2006, 97: 709-723. 10.1002/jcb.20672.
Holtta V, Klemetti P, Sipponen T, Kociubinski G, Westerholm-Ormio M, Solo H, Rasanen L, Kolho KL, Farkkila M, Savilahti E, Vaarala O: IL-23/IL-17 immunity as a hallmark of Crohn's disease. Inflamm Bowel Dis. 2008, 14: 1175-1184. 10.1002/ibd.20475.
Mazzucchelli L, Hauser C, Zgraggen K, Wagner H, Hess M, Laissue JA, Mueller C: Expression of interleukin-8 gene in inflammatory bowel disease is related to the histological grade of active inflammation. Am J Pathol. 1994, 144: 997-1007.
Ishiguro Y: Mucosal proinflammatory cytokine production correlates with endoscopic activity of ulcerative colitis. J Gastroenterol. 1999, 34: 66-74. 10.1007/s005350050218.
Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C: A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008, 29: 127-137. 10.1016/j.immuni.2008.06.001.
Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, Dong C: Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008, 29: 138-149. 10.1016/j.immuni.2008.05.009.
Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C: CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008, 181: 8391-8401.
Sato K, Kawasaki H, Nagayama H, Enomoto M, Morimoto C, Tadokoro K, Juji T, Takahashi TA: TGF-beta 1 reciprocally controls chemotaxis of human peripheral blood monocyte-derived dendritic cells via chemokine receptors. J Immunol. 2000, 164: 2285-2295.
Stevens C, Walz G, Singaram C, Lipman ML, Zanker B, Muggia A, Antonioli D, Peppercorn MA, Strom TB: Tumor necrosis factor-alpha, interleukin-1 beta, and interleukin-6 expression in inflammatory bowel disease. Dig Dis Sci. 1992, 37: 818-826. 10.1007/BF01300378.
Dionne S, Hiscott J, D'Agata I, Duhaime A, Seidman EG: Quantitative PCR analysis of TNF-alpha and IL-1 beta mRNA levels in pediatric IBD mucosal biopsies. Dig Dis Sci. 1997, 42: 1557-1566. 10.1023/A:1018895500721.
Cui G, Olsen T, Christiansen I, Vonen B, Florholmen J, Goll R: Improvement of real-time polymerase chain reaction for quantifying TNF-alpha mRNA expression in inflamed colorectal mucosa: an approach to optimize procedures for clinical use. Scand J Clin Lab Invest. 2006, 66: 249-259. 10.1080/00365510600590472.
Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, Littman DR, O'Shea JJ: Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009, 206: 35-41. 10.1084/jem.20072713.
Takahashi N, Vanlaere I, de Rycke R, Cauwels A, Joosten LAB, Lubberts E, van den Berg WB, Libert C: IL-17 produced by Paneth cells drives TNF-induced shock. J Exp Med. 2008, 205: 1755-1761. 10.1084/jem.20080588.
Eyerich K, Foerster S, Pennino D, Carbone T, Cianferani F, Odorisio T, Traidl-Hoffmann C, Durham S, Schmidt-Weber C, Cavani A: Th22 cells represent a distinct human T lymphocyte subset of tissue-signaling leukocytes. J Invest Dermatol. 2009, 129: S2-
Jurgens M, Laubender RP, Hartl F, Weidinger M, Seiderer J, Wagner J, Wetzke M, Beigel F, Pfennig S, Stallhofer J, Schnitzler F, Tillack C, Lohse P, Goke B, Glas J, Ochsenkuhn T, Brand S: Disease activity, ANCA, and IL23R genotype status determine early response to infliximab in patients with ulcerative colitis. Am J Gastroenterol. 2010, 105: 1811-1819. 10.1038/ajg.2010.95.
Satsangi J, Silverberg MS, Vermeire S, Colombel JF: The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006, 55: 749-753. 10.1136/gut.2005.082909.
Vermeulen J, Derveaux S, Lefever S, De SE, De PK, Yigit N, De Paepe A, Pattyn F, Speleman F, Vandesompele J: RNA pre-amplification enables large-scale RT-qPCR gene-expression studies on limiting sample amounts. BMC Res Notes. 2009, 2: 235-10.1186/1756-0500-2-235.
Vandesompele J, De PK, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F: Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3: RESEARCH0034-10.1186/gb-2002-3-7-research0034.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT: The MIQE guidelines: Minimum Information for publication of Quantitative real-time PCR Experiments. Clin Chem. 2009, 55: 611-622. 10.1373/clinchem.2008.112797.
Acknowledgements
This study was supported by a concerted action grant GOA2001/12051501 of Ghent University, Belgium.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SB had substantial contributions to the conception, design, execution and analysis of the study, and drafted the manuscript; DL participated in the design and interpretation of the data; HP and MDV carried out the sampling of gut specimens and contributed to the interpretation of the data, LM and KO contributed to the RNA extraction, RNA quality determination, cDNA synthesis and qPCR analysis; JV participated in designing and organizing the qPCR analysis; JV, NB, GV and DE participated in critically revising the manuscript; MDV carefully revised and edited the manuscript with important intellectual contributions and coordinated the research group. All authors read and approved the final version of the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Bogaert, S., Laukens, D., Peeters, H. et al. Differential mucosal expression of Th17-related genes between the inflamed colon and ileum of patients with inflammatory bowel disease. BMC Immunol 11, 61 (2010). https://doi.org/10.1186/1471-2172-11-61
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2172-11-61