Abstract
Background
Agaricus bisporus is commercially grown on compost, in which the available carbon sources consist mainly of plant-derived polysaccharides that are built out of various different constituent monosaccharides. The major constituent monosaccharides of these polysaccharides are glucose, xylose, and arabinose, while smaller amounts of galactose, glucuronic acid, rhamnose and mannose are also present.
Results
In this study, genes encoding putative enzymes from carbon metabolism were identified and their expression was studied in different growth stages of A. bisporus. We correlated the expression of genes encoding plant and fungal polysaccharide modifying enzymes identified in the A. bisporus genome to the soluble carbohydrates and the composition of mycelium grown compost, casing layer and fruiting bodies.
Conclusions
The compost grown vegetative mycelium of A. bisporus consumes a wide variety of monosaccharides. However, in fruiting bodies only hexose catabolism occurs, and no accumulation of other sugars was observed. This suggests that only hexoses or their conversion products are transported from the vegetative mycelium to the fruiting body, while the other sugars likely provide energy for growth and maintenance of the vegetative mycelium. Clear correlations were found between expression of the genes and composition of carbohydrates. Genes encoding plant cell wall polysaccharide degrading enzymes were mainly expressed in compost-grown mycelium, and largely absent in fruiting bodies. In contrast, genes encoding fungal cell wall polysaccharide modifying enzymes were expressed in both fruiting bodies and vegetative mycelium, but different gene sets were expressed in these samples.
Similar content being viewed by others
Background
Carbon catabolism serves fungi with energy in the form of reducing equivalents and ATP, as well as essential precursor metabolites for biosynthesis, such as glucose-6-phosphate and fructose-6-phosphate [1]. In nature plant biomass is the main carbon source for many fungal species. A. bisporus (the white button mushroom) is commercially cultivated on a composted mixture of lignocellulose-containing materials (mainly wheat straw and horse manure), which is highly selective for this fungus [2, 3].
The major constituents of the lignocellulose fraction of compost are cellulose and the hemicellulose xylan (70% of the biomass) [4] and lignin [5–7]. Due to their diverse and complex polymeric nature, degradation of plant cell wall polysaccharides to their monomeric constituent requires a large range of enzymes [8, 9]. Most of these enzymes have been divided into families in a classification system for Carbohydrate Active enZymes (CAZy, http://www.cazy.org) [10]. It has been shown that during mycelial growth and fruiting A. bisporus produces a range of extracellular enzymes, which are involved in the degradation of the lignocellulosic fraction in compost [11–14]. A shift in fungal metabolism takes place during development of the fruiting body of A. bisporus that is closely linked to an increased rate of cellulose and hemicellulose degradation [15]. The production of laccase and cellulase was suggested to be connected to the high rate and flow of carbon metabolism during fruiting body development [16, 17]. Lignin degradation by A. bisporus decreases towards the end of the mushroom production cycle [18–20].
The major monosaccharide constituents of lignocellulose are D-glucose, D-xylose, and L-arabinose, while smaller amounts of D-galactose, D-galacturonic acid, L-rhamnose and D-mannose are also present. These monosaccharides are taken up by the fungal cell and converted through specific pathways [21]. Both L-arabinose and D-xylose catabolism are part of the pentose catabolic pathway [22], which ends at D-xylulose-5-phosphate, an intermediate of the pentose phosphate pathway (PPP). D-Glucose can enter several biochemical pathways [9, 23, 24], but can also lead to the synthesis of mannitol, trehalose and other storage compounds, such as glycogen and fatty acids [25]. The minor components of polysaccharides present in compost are converted through the galacturonic acid catabolic pathway [26], the D-galactose catabolic pathways (the Leloir pathway, the oxido-reductive pathway and the DeLey Doudoroff pathway) [27] and the L-rhamnose catabolic pathway [28].
Studies on carbon metabolism in A. bisporus have mainly focused on mannitol and trehalose. Synthesis of mannitol in A. bisporus is mediated by an NADPH-dependent mannitol dehydrogenase using fructose as substrate [29]. Metabolism trehalose involves either the trehalose synthase complex, [30], or trehalose phosphorylase (EC 2.4.1.64), which catalyze the reversible hydrolysis of trehalose into glucose-1-phosphate and glucose [30]. Remarkable differences were found in carbon metabolism of fruiting body and vegetative mycelium [31–34]. Mannitol functions as an osmolyte, which accumulates to high levels during fruiting body growth while after sporulation the level of mannitol decreases rapidly [35]. It might also serve as a post-harvest reserve carbohydrate [31, 33, 36]. Trehalose also serves as a reserve carbohydrate, which is present at lower levels than mannitol that decline during fruiting body development. It has been suggested that trehalose is synthesized in the mycelium and translocated to the fruiting body [16, 32, 34].
Gene expression analysis of genes encoding enzymes for polysaccharide modification and sugar metabolism offers an improved understanding of carbohydrate utilization and the metabolic fate of monosaccharides in the litter degrading fungus A. bisporus. Here, we identified genes encoding enzymes involved in carbon metabolism using the recently sequenced A. bisporus genome [37]. The expression of these genes and genes encoding plant biomass degrading enzymes was analyzed during different stages of growth of A. bisporus, revealing significant differences between mycelium grown on plates, in compost or in casing-soil, and fruiting bodies.
Results
Identification and expression analysis of genes encoding enzymes of central metabolism
The two sequenced genomes of A. bisporus var. bisporus H97 and var. burnettii JB137-s8 were analyzed to identify genes involved in central carbon metabolism. Identification was performed using the confirmed pathway genes from other fungi (Additional file 1).
Gene expression was assessed in mycelium grown on defined medium, in casing layer and in compost, and in fruiting bodies, using specific custom 60-mer Agilent microarrays (see "Methods"). Only those genes with > 2-fold differences and P-value <0.05 in gene expression between compost/casing layer/fruiting body and culture-grown mycelium were considered to be differentially expressed (Additional file 2).
Glycolysis & gluconeogenesis
Most genes from glycolysis were moderately upregulated in compost and casing compared to undifferentiated mycelium grown on agar medium, while their levels were similar or downregulated in the fruiting bodies (Figure 1, Additional files 3 and 4). In contrast, the gluconeogenic gene encoding phosphoenolpyruvate carboxykinase (PEPCK) was 8-fold upregulated in fruiting bodies.
Schematic representation of the expression of genes of the different carbon metabolic pathways. Bars under the growth stages indicate the percentage of genes that are 2-fold upregulated (red), between 2-fold upregulated and 2-fold downregulated (green), and more than 2-fold downregulated (blue) in the sample compared to culture-grown mycelium.
Pentose phosphate pathway
Expression of most PPP genes is similar in casing, compost and fruiting bodies compared to plate grown mycelium, while only some genes are slightly up- (in compost and casing layer) or down-regulated (in fruiting bodies) (Figure 1, Additional files 3 and 4). There is no consistent effect on either the oxidative or the non-oxidative part of the PPP.
Pentose catabolic pathway
A significant increase in expression of most of the pentose catabolic pathway genes were detected in compost and to a lesser extent in the casing layer compared to plate grown mycelium, while their expression was reduced in fruiting bodies (Additional file 2). An exception was the putative L-xylulose reductase encoding gene that had reduced expression levels in compost and casing compared to plate-grown mycelium.
Catabolism of D-galactose, D-galacturonic acid, L-rhamnose and D-mannose
The putative A. bisporus genes of galacturonic acid catabolic pathway are strongly upregulated in compost and to a lesser extent in the casing layer, while they are down-regulated in fruiting bodies (Figure 2). Expression of genes from the D-galactose Leloir pathway was similar or elevated in all samples compared to plate-grown mycelium (Additional file 2). In contrast, nearly all genes of the D-galactose oxido-reductive pathway were upregulated in compost and downregulated in fruiting bodies (Additional file 2). Most genes from the rhamnose and mannose catabolic pathways (Additional file 1) [28] were similar or upregulated in compost, casing layer and fruiting bodies, compared to plate-grown mycelium (Additional file 2).
Mannitol and trehalose metabolism
The mannitol-1-phosphate dehydrogenase encoding gene was similarly expressed in compost, casing layer and fruiting bodies, while the mannitol dehydrogenase encoding gene was similar in compost and downregulated in casing layer and fruiting body (Figure 2).
Expression of most trehalose metabolism genes was similar or upregulated in samples from compost and casing layer in comparison to undifferentiated plate-grown mycelium (Additional file 2). The exception was the gene encoding the neutral trehalase (EC 3.2.1.28), which was downregulated in compost. In samples from fruiting bodies, a gene encoding a neutral trehalase was slightly upregulated.
Organic acid metabolism
Oxalic acid and citric acid are among the two most commonly produced organic acids by fungi [38]. No specific upregulation for oxalic acid metabolic genes was observed in any of the samples. In contrast, several of the citric acid metabolic genes were expressed at higher levels in fruiting bodies than in compost or the casing layer.
Comparison of the expression of carbon metabolic genes between A.bisporus and L. bicolor
Orthologs of A. bisporus carbon metabolic genes were identified in the genome of a mycorrhiza species L. bicolor S238N (Additional file 1), with the exception of genes for L-rhamnose utilization genes for which no homologs could be found in L. bicolor.
The gene expression differentiation pattern of fruiting body versus mycelium was calculated for both fungi. In contrast to the prevalent gene downregulation in glycolysis, PPP and PCP pathways in A. bisporus, most of the genes in these pathways showed constant expression in mature fruiting bodies and free-living mycelium in L. bicolor.
Expression of genes encoding plant cell wall polysaccharide degrading enzymes
Expression of genes encoding plant cell wall degrading enzymes from A. bisporus active against all the major plant cell wall polysaccharides was detected (Table 1). These genes are expressed at significantly higher levels in compost than in the other samples. For xylan and cellulose related genes, 90% and 64%, respectively, were expressed in compost while in casing layer and fruiting bodies less than 15% of these genes were expressed. In compost, expression of genes encoding enzymes targeting other polysaccharides (e.g. starch, pectin and xyloglucan) was also observed. Some genes of families GH5 and CE4, which contain enzymes acting on both plant and fungal cell wall polysaccharides, were upregulated in either compost or fruiting bodies.
Expression of genes encoding fungal cell wall degrading/modifying enzymes
Fungal cell wall degrading and modifying enzymes have received less attention than plant cell wall degrading enzymes, resulting in a less well defined assignment of function. During growth A. bisporus needs to synthesize and modify its cell wall. As growth occurs in compost, casing layer and fruiting bodies, genes encoding fungal cell wall modifying enzymes need to be expressed in all growth stages. However, as the morphology of these stages is not identical, different genes may be expressed in compost and fruiting bodies. A complete list of genes encoding putative fungal cell wall modifying enzymes can be found in Additional file 5, including their putative function. Of all genes encoding putative fungal cell wall modifying enzymes 36% were expressed in all three samples, indicating a basal set of fungal cell wall modifying enzymes. Only 20% of the genes were upregulated in the compost, while about 30% were upregulated in the fruiting bodies. None of the genes were specifically upregulated in the casing layer.
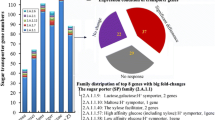
Some CAZy families related to fungal cell wall modification contain genes that were upregulated in compost as well as genes that were upregulated in fruiting bodies. This applies in particular to GH16 (endo-1,3(4)-β-glucanase), GH17 (endo-1,3-β-glucosidase) and GH18 (chitinases). Genes specifically expressed in compost were found in GH5, GH55 and GH72. Most of the genes of GH92 (α-mannosidase) are upregulated in compost. Genes specifically expressed in fruiting bodies were found in GH63 (α-glucosidase) and GT17 (glucan endo-1,3-β-glucosidase). Most genes from GT2 (chitin synthase), GT48 (1,3-β-glucan synthase), GT57 (α-1,3-glucosyltransferase) and GT15 were also upregulated in fruiting bodies.
Carbohydrate composition analysis of mycelium grown compost and casing layer and of fruiting bodies
Compost, casing layer and wheat straw were analysed for lignin, ash, protein, total carbohydrates and carbohydrate composition. Results are presented in Table 2. When the A. bisporus mushrooms have matured, compost consists of lignin (41% w/w) and ash (36% w/w), carbohydrates (17% w/w) and proteins (13%). Significant amounts of sandy particles and gravel are present in the compost and casing layer and due to the Klason lignin determination method we expect that some of this sandy inorganic material remained on the filter and is included in the calculated lignin amount [39]. The main monosaccharides released from compost by acid hydrolysis were xylose and glucose (4.4% w/w and 8.4% w/w, respectively). The composition of wheat straw was used as a reference for the composition of carbohydrates in raw compost as analysis showed that in raw compost the molar composition of carbohydrates is the same as in wheat straw (data not shown). The wheat straw composition determined in our study (Table 2) is in agreement with previously reported composition [40]. The molar composition of compost after mature mushrooms have been formed differs from that of wheat straw.
The casing layer is a mixture of calcium and peat that consists mainly of lignin (52% w/w) and ash (29% w/w). There are few carbohydrates present (14% w/w) and the main monosaccharides released after acid hydrolysis were xylose (1.4% w/w), mannose (0.6% w/w) and glucose (7.5% w/w) [41]. As mentioned above, the actual lignin amount is likely to be lower than measured due to calcium and sandy particles that remain on the filter after acid hydrolysis.
Aqueous extraction of compost, casing layer and fruiting bodies revealed that more than 95% of carbohydrates are insoluble. A high performance anion exchange (HPAEC) elution pattern of water extract from mycelium grown compost, casing layer and fruiting bodies was used to analyse the extract.
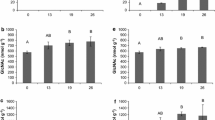
Changes in free soluble monosaccharides were observed in these samples. Concentrations of arabinose, galactose and xylose were high in compost, while only traces of these monosaccharides were found in casing layer and fruiting bodies (Table 3). High levels of glucose were observed in all samples. Mannitol and trehalose levels were significantly higher in fruiting bodies than in compost and casing layer (Table 3), as were the levels of citric acid (data not shown), while no oxalic acid was detected in the samples. The very high level of sorbitol in the compost samples could suggest a role as a transportable carbon compound from the vegetative mycelium to the fruiting body (Table 3).
Soluble oligosaccharides were detected in the compost, while none were detected in the casing layer or fruiting bodies (Figure 3). The peaks detected in the compost were compared to standards of xylo- and cello-dextran oligosaccharides (DP 2–6) and the elution pattern of the well described endoxylanase I digest of wheat arabinoxylan in order to identify them [42]. Mainly xylobiose (Figure 3 B), xylotriose (Figure 3 D), and presumably xylo-oligomers with attached glucuronic acid or its 4-O-methyl ether (Figure 3 F) were found. In addition to xylo-oligomers, cellobiose was detected. The small peaks that were detected are likely xylo- and cello-oligomers of higher degree of polymerisation and arabinose substituted xylo-oligomers.
HPAEC elution patterns of the water soluble fraction of compost, casing layer and fruiting bodies (xylobiose (A), cellobiose (B), xylotriose (C), single substituted xylo-oligomers (D), likely xylo-oligomers with GlcA substituent (E), double substituted xylo-oligomers (F), multiple substituted xylo-oligomers (G). Water extract of fruiting body was diluted 20 times. *WAX + EX1- digest of wheat arabinoxylan and endoxylanase I [42].
Discussion
In this study, genes encoding carbon metabolic genes were identified in the genome of A. bisporus and their expression in different growth stages was compared to the available carbohydrates and the expression of genes encoding carbohydrate modifying enzymes.
Compost is mainly focused on degrading plant biomass
Analysis of the expression of genes encoding plant and fungal polysaccharide modifying enzymes identified in the A. bisporus genome [37] revealed correlation between these genes and composition of carbohydrates. Expression analysis of CAZy-genes demonstrated that in compost the highest expressed genes are related to (hemi-) cellulose and pectin degradation, while also some genes related to β-1,3-glucan modification were expressed. A large decrease of carbohydrate content and, therefore, polysaccharides was revealed in the compost after growth of A. bisporus and fruiting body production. Expression data supports that the decrease in carbohydrates observed is partially caused by the growth of A. bisporus. About 90% of the genes encoding xylan degrading enzymes were upregulated in the compost. This correlates well with the detection of soluble xylo-oligosaccharides in compost. Higher proportions of arabinose and xylose in the water extracts of compost than in the water extracts of casing layer and fruiting bodies (Table 3) are in good agreement with the expression of genes encoding arabinofuranosidases, endoxylanases and β-xylosidases. The presence of xylo-oligomers in compost suggests that the β-xylosidase activity may be the limiting factor in xylose release. The pentose catabolic pathway was strongly upregulated in the compost and moderately upregulated in the casing layer, while it was downregulated in the fruiting bodies. This confirms the relevance of release and conversion of these pentoses as a main carbon source for A. bisporus during growth in compost.
Expression of genes encoding other plant polysaccharide degrading enzymes that are not normally associated with compost, e.g. starch, pectin and xyloglucan related genes, was also detected. In nature A. bisporus can grow on various substrates ranging from leaf litter and soil under cypress in coastal California to manured soil, composts of plant debris, and other horticultural and agricultural situations reported in Europe [43]. Growth on these different substrates is likely due to the ability of A. bisporus to produce a wide range of plant polysaccharide degrading enzymes and it may co-express genes aimed at different polysaccharides. Such a system is well described for the ascomycete Aspergillus niger, in which a single regulator (XlnR) activates the expression of genes related to cellulose, xylan and xyloglucan degradation [44, 45]. For this fungus six regulators involved in plant polysaccharide degradation have been described and they usually respond to the presence of the monomeric building blocks of the polysaccharides [44, 46–50]. While no homologs of these regulators have been found in basidiomycetes (Todd and de Vries, unpublished data), it is likely that basidiomycetes have developed similar systems using different regulators.
The casing layer serves as an intermediate phase
In the casing layer, which is a mixture of peat and lime, it is likely that the detected glucose and mannose at least partially drive from the mycelial cell wall, in the form of glucans and mannoproteins, respectively. While some genes encoding putative plant cell wall degrading enzymes were expressed in the casing layer, the level of up-regulation compared to plate-grown mycelium is much smaller than that in compost. In addition, expression of some chitinase encoding genes was detected. The casing layer seems to be an intermediate phase in which some genes related to plant biomass degradation are expressed, but also modification of the A. bisporus cell wall is an important process for the conversion to fruiting body morphology. The lack of soluble polysaccharides indicates that the role of the mycelium in the casing layer is mainly to supply carbohydrates to the fruiting body.
The fruiting body focuses on modification of fungal polysaccharides
For A. bisporus growth and development a basal set of fungal cell wall modifying enzymes is needed and about 36% of the genes encoding such enzymes were expressed in mycelium grown compost, casing layer and fruiting bodies. The other expressed genes encoding fungal cell wall modifying enzymes are upregulated during specific growth stages. This suggests that A. bisporus has specific genes for mycelium development and growth and others for fruiting body formation and modification. Some genes from GH16 (encoding endo-1,3(4)-β-glucanase), GH17 (encoding glucan endo-1,3-β-glucosidase) and GH18 (encoding chitinases) are upregulated in the compost while others from the same families are upregulated in the fruiting bodies. These results support the compositional and morphological differences found between mycelium and fruiting bodies [35]. Expression of different sets of genes encoding fungal cell wall modifying enzymes has also been described for other fungi. For example, in A. niger different sets of genes encoding chitinases, chitin synthases and alpha-1.3-glucan synthases were expressed in the centre and the periphery of plate grown cultures [51].
Enzymes from families GH5 and CE4 have several described activities, some of which are related to plant cell wall polysaccharides, while others are related to fungal cell wall polysaccharides (http://www.cazy.org). For some of the enzymes from these families upregulation in compost was observed, while others were upregulated in fruiting bodies. A strong correlation was observed between the putative function and the expression of genes from these families. While genes encoding putative plant biomass degrading enzymes were upregulated in compost, genes encoding putative fungal cell wall modifying enzymes were upregulated in fruiting bodies (Additional file 6).
Carbon metabolism is partially differentiated in A. bisporus
Expression analysis demonstrated that the pentose catabolic pathway and galacturonic acid pathway were strongly upregulated in compost and moderately upregulated in the casing layer, while they were downregulated in fruiting bodies. Most genes of the oxido-reductive galactose pathway were also higher expressed in compost than in fruiting bodies, which correlates with a higher galactose level in compost compared to fruiting bodies. In contrast to the pathways described above, the glycolytic pathway and PPP are active in all growth stages of A. bisporus. This correlates well with the presence of free glucose in all samples, suggesting that hexose catabolism is an important factor in all growth stages of A. bisporus. The PPP has been described as the major route of glucose catabolism in fruiting bodies of A. bisporus[35, 52, 53] as well as Lentinula edodes[54] as a greater proportion of glucose oxidation occurs via the PPP in the fruiting body than in vegetative mycelium, while glycolysis has been suggested to be the major pathway of sugar metabolism during fruiting body development in Pleurotus ostreatus, Coprinus cinereus and Schizophyllum commune[55–57].
The concentration of mannitol in fruiting bodies was six times higher than in compost. However, expression of mannitol pathway genes was significantly lower in fruiting bodies than in compost, suggesting that mannitol is synthesized in the vegetative mycelium and transported to the fruiting body. Earlier studies observed that mannitol functions as an osmoregulatory compound and facilitates a continuous influx of water from compost to the fruiting body to support turgor and fruiting body development [58, 59]. This would suggest that mannitol is unlikely to be transported by diffusion from the mycelium. Therefore, it should either be transferred by active transport or alternatively, be synthesized in the fruiting body. If the latter is the case, a possible explanation for the observed expression of the genes could be that the encoded enzymes are transported into the fruiting body.
Trehalase activity was reported to be highest during the peak of each flush, while low activity was detected during the interflush period [16], which correlates well with the highest expression of a putative trehalase encoding gene in fruiting bodies of our study. In contrast, trehalose phosphorylase was found to increase during the interflush period [34], which was also confirmed by the expression analysis in our study.
No significant differences were observed in the expression of genes related to oxalic acid metabolism in the different growth stages and the expression levels suggest that oxalic acid formation occurs in all stages. The high expression of one of the putative oxalate decarboxylase encoding genes could explain why no oxalic acid was detected in the samples as this could imply that degradation of oxalic acid occurs at least as fast as its synthesis. It should also be noted that only free oxalic acid was analysed in this study, while oxalic acid present in the form of calcium oxalate was not included.
In contrast, several of the genes involved in citric acid metabolism are higher expressed in the fruiting body than in compost and casing layer, which correlates well with the higher levels of citric acid that were detected in these samples. As citric acid is known to have preservative properties against bacteria in food [60], it is tempting to speculate that the accumulation of citric acid in fruiting bodies may also be involved in the defence mechanism of the mushroom against bacteria. Another explanation may be the high respiration rates of the fruiting bodies, which requires high expression of genes associated with the citric acid/Krebs cycle and mitochondria in general [54]. High expression of isocitrate lyase was also reported in brown-rot fungi, where this enzyme produced succinate and glyoxylate from isocitrate [61, 62]. Progressive downregulation of this gene was observed in the casing layer during the shift from vegetative mycelium to fruiting body [63].
The difference in carbon metabolism between A. bisporus and L. bicolor
Comparison of two basidiomycetes A. bisporus and L. bicolor didn’t show any correlation in expression of carbon metabolic genes. This could be explained by the difference in life styles of these two species. As a saprobe, A. bisporus is highly dependent on obtaining carbon from its surroundings. In contrast, the mycorrhizae L. bicolor obtains carbon from its symbiotic partner in the form of sucrose, placing a much lower demand on a versatile carbon metabolism.
Conclusions
The data from our study demonstrates that overall there is a clear correlation between expression of genes related to plant and fungal polysaccharides and the ability of A. bisporus to degrade these polysaccharides. We see a clear difference in genes expressed within mycelium grown compost and fruiting bodies supporting the hypothesis that different genes are expressed in A. bisporus mycelium and fruiting bodies. This supports previous results that this fungus produces different enzymes during its life cycle [64]. However, it should also be recognised that gene expression is likely to be dynamic and here we have examined it at the time point when first flush was harvested (approximately 34 days after compost was inoculated with spawn). Large oscillations of cellulase activity in the compost have been observed which co-ordinate with mushroom fruiting body production and oscillations of activities of fruiting body metabolic enzymes [16, 17, 65].
Moreover, our study demonstrates a clear correlation between the expression of genes encoding plant and fungal cell wall polysaccharides with the composition of carbohydrates in compost, casing layer and fruiting bodies. Genes encoding plant cell wall polysaccharide degrading enzymes were mainly expressed in compost-grown mycelium, and largely absent in fruiting bodies. In contrast, genes encoding fungal cell wall polysaccharide modifying enzymes were expressed in both fruiting bodies and vegetative mycelium in the compost, but different gene sets were expressed in these samples.
In the present study an in silico metabolic reconstruction of the central carbon metabolism in A. bisporus was performed and combined with expression analysis of the relevant genes in different growth stages of A. bisporus. The analysis of metabolic pathways in A. bisporus may provide information about the requirements of carbon source and energy metabolism during commercial growth of A. bisporus. We showed that during growth in compost and casing a much larger variety of carbon sources was used by A. bisporus than during growth on synthetic medium. In contrast, carbon metabolism in fruiting bodies appears to be mainly aimed at hexoses. This could indicate that only these sugars are transported towards the fruiting body from the vegetative mycelium, which implies that carbon transport to the fruiting bodies is a highly regulated and selective process.
Methods
Materials used
Compost, casing layer and fruiting bodies cultures were harvested at the first flush stage of A. bisporus strain A15 and were stored at -20°C. Samples (about 100 g) were collected, freeze dried and milled (<1 mm) (Retsch Mill MM 2000, Retsch, Haan, Germany). Duplicates were mixed in ratio 1:1. Wheat straw was collected as raw material and a representative sample was made by mixing 16 different freeze dried and milled samples of wheat straw in the same ratio. All chemicals, unless stated otherwise were obtained from Sigma, Merck or Fluka (Busch, Switzerland).
Water extraction
Milled compost, casing layer and fruiting bodies (0.4 g) were suspended in millipore water (20 mL) and boiled at 100°C for 10 min to inactivate enzyme activity, shaken vigorously and filtered (0.2 μm). The filtrate was used to analyse water soluble carbohydrates.
Analytical and spectrometric methods
Neutral carbohydrate composition
Neutral carbohydrate composition of wheat straw, compost and casing layer was analysed according to Englyst [66] using inositol as an internal standard. Samples were treated with 72% (w/w) H2SO4 (1h, 30°C) followed by hydrolysis with 1M H2SO4 for 3h at 100°C and the constituent sugars released were derivatised and analysed as their alditol acetates using gas chromatography (GC). The amount of neutral carbohydrates was corrected for mannitol, sorbitol and trehalose.
Uronic acid content
Uronic acids content of wheat straw, compost and casing layer was determined as anhydro-uronic acid by an automated m-hydroxydiphenyl assay [67] using an autoanalyser (Skalar Analytical BV, Breda, The Netherlands). Glucuronic acid was used as a reference.
Lignin content
Samples of wheat straw, compost and casing layer were analysed for acid insoluble (Klason) lignin. To each sample of 300 mg (dry matter) 3 ml of 72% (w/w) H2SO4 was added and samples were pre-hydrolysed for 1 h at 30°C. After this pre-hydrolysis, 37 ml of distilled water was added and samples were put in a boiling water bath for 3 h and shaken every half hour. Further, suspension was filtered over G4 glass filters (Duran Group GmbH, Mainz, Germany). The residual part was washed until it was free of acid and dried overnight at 105°C. The weight of the dried residual part was taken as a measure of the acid insoluble lignin content.
Protein content
Nitrogen content of wheat straw, compost and casing layer was analysed using the combustion (DUMAS) method on a Flash EA 1112 Nitrogen Analyser (Thermo Scientific, Rockford, IL, USA). Methionine (Acros Organics, New Jersey, USA) was used as a standard and protein content was calculated from the nitrogen content of the material, using a protein conversion factor of 6.25 [68].
Ash content
Samples of wheat straw, compost and casing layer (0.5 g) were dried in the oven overnight (105°C), weighed and put in the oven on 504°C overnight. Next day samples were weighed and difference between the mass at 105°C and 504°C was taken as ash content.
Chromatographic methods
Analysis of soluble carbohydrates, sorbitol, trehalose and mannitol
High-performance anion-exchange chromatography (HPAEC) was performed on an Ultimate 3000 system (Dionex, Sunnyvale, CA, USA) equipped with a CarboPac PA-1 column (2 mm x 250 mm ID) in combination with a CarboPac guard column (2 mm x 50 mm ID) and PAD detection. System was controlled by the Chromelion software (Dionex).
Separation and quantification of monosaccharides was done at a flow rate 0.4 ml/min, and the mobile phase consisted of (A) 0.1 M NaOH, (B) 1 M NaOAc in 0.1 M NaOH and (C) H2O. The elution profile was as follows: 0–40 min 100% C; 40.1-45.1 min from 100% A to 100% B, 45.1-50 min 100% B, 50.1-58 min 100% A, 58.1-73 min 100% C. From 0 to 40 min and from 58 to 73 min post column addition of 0.5 M NaOH at 0.1 ml/min was performed to detect and quantify the eluted saccharides.
Soluble carbohydrates. sorbitol, mannitol and trehalose were separated on the same system, including columns and detection. The flow rate used to separate sorbitol, mannitol and trehalose was 0.3 mL/min, and the mobile phase consisted of (A) 0.1 M NaOH, (B) 1 M NaOAc in 0.1 M NaOH and (C) H2O. The elution profile was as follows: 0–5 min 100% A, 5-25% 0-30% B, 25.1-30 min 100% B, 30–50 min 100% A.
Water soluble oligosaccharides were separated with a combination of linear gradients from two types of eluents, A: 0.1 M NaOH and B: 1 M NaOAc in 0.1 M NaOH. The elution profile was as following: 0–35 min: 0-38% B, cleaning step 3 min 100% B and equilibration step 12 min 100% A. As a reference for xylo-oligomers with substitution, elution pattern of wheat arabinoxylan (medium viscosity, Megazyme, Bray, Ireland) digest with a pure and well described endoxylanase I was used [42, 69], while as a standard for cellulose and xylan oligomers, cellodextrans and xylodextrans were used. Water extract of compost and casing layer were injected on the column without dilution and fruiting body water extract was diluted 20 times before injecting it on the column.
Organic acid analysis
Oxalic acid and citric acid were determined with an Ultimate system (Dionex, Sunnyvale, USA) equipped with a Shodex RI detector and an Aminex HPX 87H column (300 mm x 7.8 mm) (Bio-Rad, Hercules, CA, USA) plus pre-column [70]. Elution was performed by using 5 mM H2SO4 as eluent at a flow rate of 0.6 ml min-1 at 40°C.
Esterified acetic acid content
Samples of compost and casing layer (20 mg) were saponified with 1 mL of 0.4 M NaOH in isopropanol/H20 (1:1) for 3 h at room temperature. The acetic acid content was determined with an Ultimate system (Dionex) equipped with a Shodex RI detector and an Aminex HPX 87H column (300 mm x 7.8 mm) (Bio-Rad) plus pre-column [70]. Elution was performed by using 5 mM H2SO4 as eluent at a flow rate of 0.6 mL min-1 at 40°C. The level of acetic acid substituents was corrected for the free acetic acid in the sample.
Genome annotation and comparative genomics
A. bisporus var bisporus (http://genome.jgi.doe.gov/Agabi_varbisH97_2/Agabi_varbisH97_2.home.html), A. bisporus var burnetti (http://genome.jgi.doe.gov/Agabi_varbur_1/Agabi_varbur_1.home.html), Aspergillus niger, Aspergillus oryzae, Aspergillus nidulans, Phanerochaete chrysosporium or Postia placenta and Laccaria bicolor S238N genomes (http://genome.jgi-psf.org/Lacbi2/Lacbi2.home.html) were used to perform genomic comparisons. Full genome clusters of orthologous genes were created by OrthoMCL (http://www.ncbi.nlm.nih.gov/pubmed/12952885) with E-value 1e-5 and sequence matching coverage 60% as the cutoff (http://www.ncbi.nlm.nih.gov/pubmed/20152020). Carbon catabolic genes of Agaricus and Laccaria were identified by extracting the orthologous clusters containing known carbon catabolic genes from Aspergulli, P. chrysosporium or P. placenta.
Transcriptome analysis
Gene expression was profiled in the commercial (heterokaryon) strain A15. A. bisporus strain A15 was grown in compost made from wheat straw, chicken litter and gypsum in the proportions 10:6:0.5 w/w. The first phase of composting was with regular mixing and took approximately 25 days. At phase II of composting process compost was pasteurized with steam at 70°C for 7 days. Phase II compost was inoculated with 1–2% w/w A. bisporus mycelium spawn, placed in 50 kg growth trays, and incubated at 25°C, 95% relative humidity for 21 days. The colonised compost was covered by 5 cm peat-based casing layer and incubated for a further 7 days. The culture samples refer to axenic culture and the media used was compost extract medium [71]. Fresh pasteurised compost was oven dried for 48 h at 80°C. Dried compost was boiled in distilled water (7.5 g / l) for 1 h and cooled to room temperature. After centrifugation (5000 rpm, 20 min), the supernatant was used to make the medium [72]. Peptone (0.5% w/v) was added to the extract and the medium buffered to pH 7 using potassium phosphate buffer.
The fruiting body samples represent the mature mushroom stage 2 with a stretched, unbroken veil fruiting body (including the stipe, cap and pilei pellis (skin) tissues) [35]. The casing samples consisted of a mixture of mycelium aggregates, undifferentiated primordia (1–2 mm circular with no differentiation between stipe and cap tissues), differentiated primordia (~ 7 mm diameter, oval with some evidence of cap tissue differentiation). The compost samples represent the mycelium growing in wheat straw compost. The samples for RNA extraction were collected on separate occasions from separate mushroom houses. Four biological replicates of each developmental stage were analyzed [37].
RNA was prepared from fruiting body and culture samples using a standard Trizol protocol. RNA was extracted from compost and casing samples using a method based on the FastRNA Pro Soil-Direct kit (MP Biochemicals) [63]. RNA was quantified using a NanoDrop-1000 spectrophotometer and quality was monitored with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Custom arrays (Agilent ID 027120) were developed using 10,438 CDS (filtered model set) from the H97 v2 gene annotation; 5 x 60-mer oligos per CDS and the 8 x 60K randomised format were designed using the Agilent eArray software. Cyanine-3 (Cy3) labeled cRNA was prepared from 0.6 ug RNA using the Quick Amp Labelling kit (Agilent) according to the manufacturer’s instructions, followed by RNAeasy column purification (QIAGEN, Valencia, CA). Dye incorporation and cRNA yield were checked with the NanoDrop ND-1000 Spectrophotometer. 600 ng of Cy3-labelled cRNA (specific activity >10.0 pmol Cy3/ug cRNA) was fragmented at 60°C for 30 minutes in a reaction volume of 25 μl containing 1x Agilent fragmentation buffer and 2x Agilent blocking agent following the manufacturer’s instructions. On completion of the fragmentation reaction, 25 μl of 2x Agilent hybridization buffer was added to the fragmentation mixture and hybridized to Agilent arrays (ID 027120) for 17 hours at 65°C in a rotating Agilent hybridization oven. After hybridization, microarrays were washed 1 minute at room temperature with GE Wash Buffer 1 (Agilent) and 1 minute with 37°C GE Wash buffer 2 (Agilent) then 10 seconds in acetonitrile and 30 seconds in Stabilization and drying solution (Agilent). Slides were scanned immediately after washing on the Agilent’s High-Resolution C Scanner (G2505C US94100321) using one color scan setting for 8 x 60K array slides (Scan resolution 3um). The scanned images were analyzed with Feature Extraction Software (Agilent) using default parameters (protocol GE1_107_Sep09 and Grid: 027120_D_F_20100129) to obtain background subtracted and spatially detrended Processed Signal intensities. Features flagged in Feature Extraction as Feature Non-uniform outliers were excluded [37]. Only those genes with > 2-fold differences and P-value <0.05 in gene expression between compost/casing layer/fruiting body and culture-grown mycelium were considered to be differentially expressed. Comparison of ratios of compost/culture transcript profiles was used to identify the most highly upregulated transcripts found in mycelium grown on compost during vegetative growth. The comparison of compost/fruiting body transcript profiles highlights developmental stage differences during mushroom formation [37].
The Laccaria bicolor S238N transcriptomes of 2 weeks free-living mycelium (FLM) and mature fruiting bodies were extracted from Gene Expression Omnibus (GEO) by series number GSE9784. Gene expression profiles were extracted, normalized and analysed as described previously [73]. Only genes with 2-fold differences and P-value <0.05 were considered significantly differentially expressed.
Availability of supporting data
Micro array data from Agaricus bisporus and Laccaria bicolor used in this paper is available at GEO, accession number GSE39569 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39569) and GSE32559 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32559), respectively.
References
Chang ST, Miles PG: Mushrooms: cultivation, nutritional value, medicinal effect and environmental impact. 2004, Washington, D.C: CRC Press
Fermor TR, Randle PE, Smith JF: Compost as a substrate and its preparation. The biology and technology of the cultivated mushroom. Edited by: Flegg PB, Spencer DM, Wood DA. 1985, Chichester, UK: John Wiley and Sons, 81-110.
Tautorus TE, Townsley PM: Biotechnology in commercial mushroom fermentation. Bio/Technology. 1984, 2 (8): 696-701. 10.1038/nbt0884-696.
Jørgensen H, Vibe‒Pedersen J, Larsen J, Felby C: Liquefaction of lignocellulose at high‒solids concentrations. Biotechnol Bioeng. 2007, 96 (5): 862-870. 10.1002/bit.21115.
Lawther JM, Sun RC, Banks WB: Extraction, fractionation, and characterization of structural polysaccharides from wheat straw. J Agric Food Chem. 1995, 43 (3): 667-675. 10.1021/jf00051a021.
Vane CH, Abbott GD, Head IM: The effect of fungal decay (Agaricus bisporus) on wheat straw lignin using pyrolysis-GC-MS in the presence of tetramethylammonium hydroxide (TMAH). J Anal Appl Pyrolysis. 2001, 60 (1): 69-78. 10.1016/S0165-2370(00)00156-X.
Lynch J: Substrate availability in the production of composts. Science and engineering of composting: design environmental, microbiological and utilization aspects. Edited by: Hoitink HA, Keener HM. 1993, Columbus: The Ohio State University Press, 24-35.
Coutinho PM, Andersen MR, Kolenova K, van Kuyk PA, Benoit I, Gruben BS, Trejo-Aguilar B, Visser H, van Solingen P, Pakula T: Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009, 46: S161-S169. 10.1016/j.fgb.2008.07.020.
de Vries RP, Visser J: Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev. 2001, 65 (4): 497-522. 10.1128/MMBR.65.4.497-522.2001.
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B: The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009, 37 (suppl 1): D233-D238.
Gerrits J: Organic compost constituents and water utilized by the cultivated mushroom during spawn run and cropping. Mushroom Sci. 1969, 7 (11): 1-126.
Wood D, Thurston C, Griensven L: Progress in the molecular analysis of Agaricus enzymes. Genetics and breeding of Agaricus Proceedings of the First International Seminar on Mushroom Science, Mushroom Experimental Station, 4–17 May 1991. 1991, Horst, Netherlands: Pudoc, 81-86.
Fermor TR, Wood DA, Lincoln SP, Fenlon JS: Bacteriolysis by Agaricus bisporus. J Gen Appl Microbiol. 1991, 137: 15-22. 10.1099/00221287-137-1-15.
Yague E, Mehak-Zunic M, Morgan L, Wood DA, Thurston CF: Expression of CEL2 and CEL4, two proteins from Agaricus bisporus with similarity to fungal cellobiohydrolase I and beta-mannanase, respectively, is regulated by the carbon source. Microbiology. 1997, 143 (1): 239-244. 10.1099/00221287-143-1-239.
Wood DA, Goodenough PW: Fruiting of Agaricus bisporus changes in extracellular enzyme activities during growth and fruiting. Arch Microbiol. 1977, 114 (2): 161-165. 10.1007/BF00410778.
Wells TK, Hammond JBW, Dickerson AG: Variations in activities of glycogen phosphorylase and trehalase during the periodic fruiting of the edible mushroom Agaricus bisporus (Lange) Imbach. New Phytol. 1987, 105 (2): 273-280. 10.1111/j.1469-8137.1987.tb00864.x.
Claydon N, Allan M, Wood DA: Fruit body biomass regulated production of extracellular endocellulase during fruiting by Agaricus bisporus. T Brit Mycol Soc. 1988, 90: 85-90. 10.1016/S0007-1536(88)80183-9.
Wood DA, Leatham GF: Lignocellulose degradation during the life cycle of Agaricus bisporus. FEMS microbiology letters. 1983, 20 (3): 421-424. 10.1111/j.1574-6968.1983.tb00160.x.
Durrant AJ, Wood DA, Cain RB: Lignocellulose biodegradation by Agaricus bisporus during solid substrate fermentation. J Gen Appl Microbiol. 1991, 137: 751-755. 10.1099/00221287-137-4-751.
Waksman SA, Nissen W: On the nutrition of the cultivated mushroom, Agaricus campestris, and the chemical changes brought about by this organism in the manure compost. Am J Bot. 1932, 19 (6): 514-537. 10.2307/2436074.
Battaglia E, Benoit I, Gruben BS, de Vries RP: Plant cell wall derived sugars as substrates for fungi and industry. The sugar industry and cotton crops. Edited by: Jenkins PT. 2010, Hauppauge, NY: Nova science publishers, 65-94.
Witteveen CFB, Busink R, Vandevondervoort P, Dijkema C, Swart K, Visser J: L-arabinose and d-xylose catabolism in Aspergillus niger. J Gen Appl Microbiol. 1989, 135: 2163-2171.
Timell TE: Recent progress in the chemistry of wood hemicelluloses. Wood Sc Technol. 1967, 1: 45-70. 10.1007/BF00592255.
Hayashi T: Xyloglucans in the primary cell wall. Annu Rev Plant Physiol Plant Mol Biol. 1989, 40: 139-168. 10.1146/annurev.pp.40.060189.001035.
Martin F, Ramstedt M, Soderhall K, Canet D: Carbohydrate and amino acid metabolism in the ectomycorrhizal ascomycete Sphaerosporella brunnea during glucose utilization. Plant physiology. 1988, 86 (3): 935-940. 10.1104/pp.86.3.935.
Richard P, Hilditch S: d-Galacturonic acid catabolism in microorganisms and its biotechnological relevance. Appl Microbiol Biotechnol. 2009, 82 (4): 597-604. 10.1007/s00253-009-1870-6.
Mojzita D, Koivistoinen OM, Maaheimo H, Penttila M, Ruohonen L, Richard P: Identification of the galactitol dehydrogenase, LadB, that is part of the oxido-reductive D-galactose catabolic pathway in Aspergillus niger. Fungal Genet Biol. 2012, 49 (2): 152-159. 10.1016/j.fgb.2011.11.005.
Watanabe S, Saimura M, Makino K: Eukaryotic and bacterial gene clusters related to an alternative pathway of nonphosphorylated L-rhamnose metabolism. J Biol Chem. 2008, 283 (29): 20372-20382. 10.1074/jbc.M801065200.
Edmundowicz JM, Wriston JC: Mannitol dehydrogenase from Agaricus campestris. J Am Chem Soc. 1963, 238 (11): 3539-3541.
Wannet WJ, Op den Camp HJ, Wisselink HW, van der Drift C, Van Griensven LJ, Vogels GD: Purification and characterization of trehalose phosphorylase from the commercial mushroom Agaricus bisporus. Biochimica et biophysica acta. 1998, 1425 (1): 177-188. 10.1016/S0304-4165(98)00066-X.
Hammond JBW: Variations in enzyme activity during periodic fruiting of Agaricus bisporus. New Phytol. 1981, 89 (3): 419-428. 10.1111/j.1469-8137.1981.tb02323.x.
Hammond JBW: Sugar, sugar phosphate and NADP(H) levels in Agaricus bisporus fruit bodies. J Gen Appl Microbiol. 1985, 131: 329-333.
Beecher TM, Magan N, Burton KS: Water potentials and soluble carbohydrate concentrations in tissues of freshly harvested and stored mushrooms (Agaricus bisporus). Postharvest Biol Tec. 2001, 22 (2): 121-131. 10.1016/S0925-5214(00)00195-2.
Wannet WJ, Aben EM, van der Drift C, Van Griensven LJ, Vogels GD, Op den Camp HJ: Trehalose phosphorylase activity and carbohydrate levels during axenic fruiting in three Agaricus bisporus strains. Curr Microbiol. 1999, 39 (4): 205-210. 10.1007/s002849900446.
Hammond JB, Nichols R: Carbohydrate metabolism in Agaricus bisporus (Lange) Sing: changes in soluble carbohydrates during growth of mycelium and sporophore. J Gen Microbiol. 1976, 93 (2): 309-320. 10.1099/00221287-93-2-309.
Wood DA: Production and roles of extracellular enzymes during morphogenesis of basidiomycete fungi. Developmental biology of higher fungi. Edited by: Moore DM LAC, Wood DA, Frankland JC. 1985, Cambridge: Cambridge University Press, 375-387.
Morin E, Kohler A, Baker AR, Foulongne-Oriol M, Lombard V, Nagy LG, Ohm RA, Patyshakuliyeva A, Brun A, Aerts AL: Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc Natl Acad Sci USA. 2012, 109 (43): 17501-17506. 10.1073/pnas.1206847109.
Gadd GM: Fungal production of citric and oxalic acid: Importance in metal speciation, physiology and biogeochemical processes. Adv Microb Physiol. 1999, 41: 47-92.
Iiyama K, Stone BA, Macauley BJ: Compositional changes in compost during composting and growth of Agaricus bisporus. Appl Environ Microbiol. 1994, 60 (5): 1538-1546.
Kabel MA, Bos G, Zeevalking J, Voragen AGJ, Schols HA: Effect of pretreatment severity on xylan solubility and enzymatic breakdown of the remaining cellulose from wheat straw. Bioresour Technol. 2007, 98 (10): 2034-2042. 10.1016/j.biortech.2006.08.006.
Straatsma G, Gerrits JPG, Gerrits TM, Dencamp H, Vangriensven L: Growth-kinetics of Agaricus bisporus mycelium on solid substrate (mushroom compost). J Gen Microbiol. 1991, 137: 1471-1477. 10.1099/00221287-137-7-1471.
Kormelink FJM, Gruppen H, Vietor RJ, Voragen AGJ: Mode of action of the xylan-degrading enzymes from Aspergillus awamori on alkali extractable cereal arabinoxylan. Carbohydr Res. 1993, 249 (2): 355-367. 10.1016/0008-6215(93)84100-K.
Kerrigan RW, Carvalho DB, Horgen PA, Anderson JB: The indigenous coastal Californian population of the mushroom Agaricus bisporus, a cultivated species, may be at risk of extinction. Mol Ecol. 1998, 7 (1): 35-45. 10.1046/j.1365-294x.1998.00294.x.
van Peij N, Gielkens MMC, de Vries RP, Visser J, de Graaff LH: The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998, 64 (10): 3615-3619.
Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJI, de Graaff LH: EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl Environ Microbiol. 2002, 68 (4): 1556-1560. 10.1128/AEM.68.4.1556-1560.2002.
van Kuyk PA, Benen JA, Wosten HA, Visser J, de Vries RP: A broader role for AmyR in Aspergillus niger: regulation of the utilisation of D-glucose or D-galactose containing oligo- and polysaccharides. Appl Microbiol Biotechnol. 2012, 93 (1): 285-293. 10.1007/s00253-011-3550-6.
Yuan X-L, Roubos JA, van den Hondel CAMJJ, Ram AFJ: Identification of InuR, a new Zn(II)2Cys6 transcriptional activator involved in the regulation of inulinolytic genes in Aspergillus niger. Mol Genet Genomics. 2008, 279 (1): 11-26. 10.1007/s00438-007-0290-5.
Battaglia E, Visser L, Nijssen A, van Veluw G, Wösten H, de Vries R: Analysis of regulation of pentose utilisation in Aspergillus nige r reveals evolutionary adaptations in Eurotiales. Stud Mycol. 2011, 69 (1): 31-38. 10.3114/sim.2011.69.03.
Gruben BS, Zhou M, de Vries RP: GalX regulates the D-galactose oxido-reductive pathway in Aspergillus niger. FEBS letters. 2012, 586 (22): 3980-3985. 10.1016/j.febslet.2012.09.029.
Gruben BS: Novel transcriptional activators of Aspergillus involved in plant biomass utilization. 2012, Utrecht: Utrecht University
Levin AM, de Vries RP, Conesa A, de Bekker C, Talon M, Menke HH, van Peij NNME, Wosten HAB: Spatial differentiation in the vegetative mycelium of Aspergillus niger. Eukaryotic cell. 2007, 6 (12): 2311-2322. 10.1128/EC.00244-07.
Hammond JBW, Nichols R: Carbohydrate metabolism in Agaricus bisporus (Lange) Imbach.: Metabolism of C-14 labeled sugars by sporophores and mycelium. New Phytol. 1977, 79 (2): 315-325. 10.1111/j.1469-8137.1977.tb02211.x.
Hammond JBW: Carbohydrate metabolism in Agaricus bisporus - oxidative pathways in mycelium and sporophore. J Gen Microbiol. 1977, 102: 245-248. 10.1099/00221287-102-2-245.
Tan YH, Moore D: Glucose catabolic pathways in Lentinula edodes determined with radiorespirometry and enzymatic analysis. Mycol Res. 1995, 99: 859-866. 10.1016/S0953-7562(09)80742-9.
Moore D, Ewaze JO: Activities of some enzymes involved in metabolism of carbohydrate during sporophore development in Coprinus cinereus. J Gen Microbiol. 1976, 97: 313-322. 10.1099/00221287-97-2-313.
Schwalb MN: Changes in activity of enzymes metabolizing glucose 6-phosphate during development of the basidiomycete Schizophyllum. Dev Biol. 1974, 40 (1): 84-89. 10.1016/0012-1606(74)90110-9.
Chakraborty TK, Das N, Mukherjee M: Evidences of high carbon catabolic enzyme activities during sporulation of Pleurotus ostreatus (Florida). J Basic Microbiol. 2003, 43 (6): 462-467. 10.1002/jobm.200310275.
Kalberer PP: Influence of the water potential of the casing soil on crop yield and on dry-matter content, osmotic potential and mannitol content of the fruit bodies of Agaricus bisporus. J Hortic Sci. 1990, 65 (5): 573-581.
Stoop JM, Mooibroek H: Cloning and characterization of NADP-mannitol dehydrogenase cDNA from the button mushroom, Agaricus bisporus, and its expression in response to NaCl stress. Appl Environ Microbiol. 1998, 64 (12): 4689-4696.
Brul S, Coote P: Preservative agents in foods: Mode of action and microbial resistance mechanisms. Int J Food Microbiol. 1999, 50 (1–2): 1-17.
Munir E, Yoon JJ, Tokimatsu T, Hattori T, Shimada M: A physiological role for oxalic acid biosynthesis in the wood-rotting basidiomycete Fomitopsis palustris. Proc Natl Acad Sci USA. 2001, 98 (20): 11126-11130. 10.1073/pnas.191389598.
Martinez D, Challacombe J, Morgenstern I, Hibbett D, Schmoll M, Kubicek CP, Ferreira P, Ruiz-Duenas FJ, Martinez AT, Kersten P: Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc Natl Acad Sci USA. 2009, 106 (6): 1954-1959. 10.1073/pnas.0809575106.
Eastwood DC, Herman B, Noble R, Dobrovin-Pennington A, Sreenivasaprasad S, Burton KS: Environmental regulation of reproductive phase change in Agaricus bisporus by 1-octen-3-ol, temperature and CO2. Fungal Genet Biol. 2013, 55: 54-66.
Garcia Mendoza C, Perez Cabo A, Calonje M, Galan B, Novaes-Ledieu M: Chemical and structural differences in cell wall polysaccharides of two monokaryotic strains and their resulting dikaryon of Agaricus bisporus. Curr Microbiol. 1996, 33 (4): 211-215. 10.1007/s002849900101.
Burton KS, Hammond JBW, Minamide T: Protease activity in Agaricus bisporus during periodic fruiting (flushing) and sporophore development. Curr Microbiol. 1994, 28 (5): 275-278. 10.1007/BF01573205.
Englyst HN, Cummings JH: Simplified method for the measurement of total non-starch polysaccharides by gas–liquid chromatography of constituent sugars as alditol acetates. Analyst. 1984, 109 (7): 937-942. 10.1039/an9840900937.
Blumenkrantz N, Asboe-Hansen G: New method for quantitative determination of uronic acids. Anal Biochem. 1973, 54 (2): 484-489. 10.1016/0003-2697(73)90377-1.
Jones DB: Factors for converting percentages of nitrogen in foods and feeds into percentages of proteins. 1931, Washington, DC: US Department of Agriculture
Van Laere K, Voragen C, Kroef T, Van den Broek L, Beldman G, Voragen A: Purification and mode of action of two different arabinoxylan arabinofuranohydrolases from Bifidobacterium adolescentis DSM 20083. Appl Microbiol Biotechnol. 1999, 51 (5): 606-613. 10.1007/s002530051439.
Voragen AGJ, Schols HA, Pilnik W: Determination of the degree of methylation and acetylation of pectins by HPLC. Food hydrocolloids. 1986, 1 (1): 65-70. 10.1016/S0268-005X(86)80008-X.
Calvo-Bado L, Noble R, Challen M, Dobrovin-Pennington A, Elliott T: Sexuality and genetic identity in the Agaricus section Arvenses. Appl Environ Microbiol. 2000, 66 (2): 728-734. 10.1128/AEM.66.2.728-734.2000.
Sonnenberg AS, Wessels JG, Vangriensven LJ: An efficient protoplasting / regeneration system for A. bisporus and A. bitorquis. Curr Microbiol. 1988, 17 (5): 285-291. 10.1007/BF01571330.
Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V: The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008, 452 (7183): 88-92. 10.1038/nature06556.
Acknowledgements
AP and EJ were supported by grants of the Dutch Technology Foundation STW, Applied Science division of NWO and the Technology Program of the Ministry of Economic Affairs UGC 11108. MZ was supported by a grant from the Netherlands Organisation for Scientific Research (NWO) and the Netherlands Genomics Initiative 93511035 to RPdV. The comparative analysis of Agaricus and Lacaria genomes was carried out on the Dutch National e-infrastructure with the support of SURF Foundation. The authors thank Wim Arts and Geert Weijers for supplying the samples for the composition analysis. The authors also thank the Horticultural Development Company, UK that provided grants for Agaricus bisporus custom array development and transcriptomics.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AP and EJ analysed the data and drafted the manuscript. EJ, WdB, HG and MK performed and analysed carbohydrate compositions experiments. AB, KSB, MPC, DCE, AK and FM performed transcriptome analysis. PMC and BH performed comparative genomic analysis of CAZy genes. MZ performed the genomic and transcriptomic comparisons between A. bisporus and L. bicolor. AP, EB, BSG, MRM, MN, JvdB and AW analysed the A. bisporus genome for metabolic pathways. RPdV designed the study. All authors participated in data interpretation, and read and approved the final manuscript.
Aleksandrina Patyshakuliyeva, Edita Jurak contributed equally to this work.
Electronic supplementary material
12864_2013_5395_MOESM1_ESM.xlsx
Additional file 1:Genes encoding putative enzymes of carbon metabolism in A. bisporus and L. bicolor. Genes in A. bisporus var. bisporus were identified using orthologous clustering method based on best bi-derectional hits of all-vs-all blast to the genomes included in analysis. (XLSX 28 KB)
12864_2013_5395_MOESM2_ESM.xlsx
Additional file 2:Expression comparison of carbon metabolic genes in different growth stages of A. bisporus var. bisporus and L. bicolor . In the ratio between the values genes that are upregulated compared to culture-grown mycelium are in pink, while genes that are down regulated are in green. (XLSX 63 KB)
12864_2013_5395_MOESM3_ESM.pdf
Additional file 3:Proportion of upregulated genes of the different carbon metabolic pathways in compost, casing layer and fruiting bodies. Venn diagrams represent different carbon metabolic pathways indicating the percentage of genes that are 2-fold upregulated in the samples compared to culture-grown mycelium. (PDF 90 KB)
12864_2013_5395_MOESM4_ESM.pdf
Additional file 4:Proportion of downregulated genes of the different carbon metabolic pathways in compost, casing layer and fruiting bodies. Venn diagrams represent different carbon metabolic pathways indicating the percentage of genes that are 2-fold downregulated in the samples compared to culture-grown mycelium. (PDF 91 KB)
12864_2013_5395_MOESM5_ESM.xls
Additional file 5:Expression of genes encoding putative fungal and plant polysaccharide modifying enzymes. Putative functions are based on CAZy family assignment and homology to characterised enzymes. The activity on plant of fungal polysaccharides is putative and not always supported biochemically. The expression levels are the average of 4 biological replicates. (XLS 79 KB)
12864_2013_5395_MOESM6_ESM.pdf
Additional file 6:Maximum likelihood tree showing the correlation between plant biomass degrading and fungal cell wall modifying enzymes and upregulation of genes encoding these enzymes in compost or fruiting body. Phylogenetic tree of the members of CE4 (A) and GH5 (B) families together with characterized enzymes was based on maximum likelihood method with 1000 bootstraps replications and WAG substitution model. Text in pink boxes shows that genes encoding indicated enzymes are upregulated in compost/fruiting body. (PDF 114 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Patyshakuliyeva, A., Jurak, E., Kohler, A. et al. Carbohydrate utilization and metabolism is highly differentiated in Agaricus bisporus. BMC Genomics 14, 663 (2013). https://doi.org/10.1186/1471-2164-14-663
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-14-663