Abstract
Background
RpoS is a conserved stress regulator that plays a critical role in survival under stress conditions in Escherichia coli and other γ-proteobacteria. RpoS is also involved in virulence of many pathogens including Salmonella and Vibrio species. Though well characterized in non-pathogenic E. coli K12 strains, the effect of RpoS on transcriptome expression has not been examined in pathogenic isolates. E. coli O157:H7 is a serious human enteropathogen, possessing a genome 20% larger than that of E. coli K12, and many of the additional genes are required for virulence. The genomic difference may result in substantial changes in RpoS-regulated gene expression. To test this, we compared the transcriptional profile of wild type and rpoS mutants of the E. coli O157:H7 EDL933 type strain.
Results
The rpoS mutation had a pronounced effect on gene expression in stationary phase, and more than 1,000 genes were differentially expressed (twofold, P < 0.05). By contrast, we found 11 genes expressed differently in exponential phase. Western blot analysis revealed that, as expected, RpoS level was low in exponential phase and substantially increased in stationary phase. The defect in rpoS resulted in impaired expression of genes responsible for stress response (e.g., gadA, katE and osmY), arginine degradation (astCADBE), putrescine degradation (puuABCD), fatty acid oxidation (fadBA and fadE), and virulence (ler, espI and cesF). For EDL933-specific genes on O-islands, we found 50 genes expressed higher in wild type EDL933 and 49 genes expressed higher in the rpoS mutants. The protein levels of Tir and EspA, two LEE-encoded virulence factors, were elevated in the rpoS mutants under LEE induction conditions.
Conclusion
Our results show that RpoS has a profound effect on global gene expression in the pathogenic strain O157:H7 EDL933, and the identified RpoS regulon, including many EDL933-specific genes, differs substantially from that of laboratory K12 strains.
Similar content being viewed by others
Background
Enterohemorrhagic Escherichia coli O157:H7 is a serious human pathogen that is responsible for many food-borne epidemic outbreaks, and the infection of E. coli O157:H7 can cause bloody diarrhea, hemorrhagic colitis and the hemolytic uremic syndrome [1, 2]. The pathogenesis caused by E. coli O157:H7 is a complex process that requires a coordinated expression of virulence factors and regulators [1]. Known virulence factors in E. coli include the type III secretion factors encoded on the LEE pathogenicity island [3] and Shiga toxins (StxI and StxII) (reviewed in [4]). Many regulators are involved in mediating expression of these virulence factors. For example, genes on the LEE island are under control of H-NS [5], IHF [5], ClpXP [6] and three LEE-encoded regulators Ler, GrlA, and GrlR [7].
In E. coli and many other gamma-proteobacteria, the global stress response is controlled by the stationary phase sigma factor RpoS [8, 9]. RpoS is induced in many stress conditions, including near-UV exposure [10], acid shock [11], heat shock [12], oxidative stress [10], and starvation [13], many of which E. coli may experience during growth and survival in natural environments. RpoS controls a large regulon consisting of 10% of the genome in E. coli K12 strains in stationary phase and stress conditions [14–17]. Even in exponential phase when RpoS is expressed at low levels, mutation in rpoS affects the expression of a large set of genes as well [18, 19], and RpoS is important for DNA damage response in early exponential phase cells [20]. Though there is an identifiable core set of RpoS-regulated genes, the RpoS-dependence of many genes within the RpoS regulon varies depending on experimental conditions and strain backgrounds [16, 18, 19].
The effect of RpoS on virulence has been examined in many pathogens, and results differ depending on species. RpoS is critical for virulence of Salmonella [21] and Vibrio cholerae [22]. By contrast, RpoS does not appear to be required for virulence in P. aeruginosa [23] and Y. enterocolitica [24]. How RpoS is involved in enteropathogenesis of E. coli remains elusive, primarily because of the lack of a proper animal model since mice are not susceptible to infection of E. coli pathogens [25]. To overcome this problem, a model of using Citrobacter rodentium, a natural mouse enteropathogen closely related to E. coli has been widely used to simulate E. coli infection [25]. We have found that RpoS is important for full virulence of C. rodentium [26], suggesting an important role of RpoS in E. coli infection. Consistently, there are a few virulence traits regulated by RpoS. For example, curli production, important for virulence of Salmonella and E. coli, is positively regulated by RpoS [26–29]. The effect of RpoS on expression of the LEE virulence genes appears to vary depending on strain backgrounds and experimental conditions. For example, Sperandio et al. (1999) reported that the LEE3 operon and tir are positively regulated by RpoS in EHEC strain 86-24 [30]. However, in EHEC O157:H7 Sakai strain, LEE expression is enhanced in rpoS mutants [6, 31]. It is likely that the expression of LEE genes is modulated differently depending on strain backgrounds. Surprisingly, expression of LEE genes appears to differ between O157:H7 Sakai and EDL933 strains as well (see Fig. 1 in [32]). The role of RpoS in strain EDL933 has not been tested. Furthermore, there has been no genomic profiling specifically investigating the involvement of RpoS in regulation of virulence genes in enteropathogenic E. coli and other related pathogens.
The genomes of E. coli K12 reference strain MG1655 and O157:H7 strain EDL933 differ considerably [33]. EDL933 and MG1655 possess 5.5 Mb and 4.6 Mb genome sizes, respectively, sharing 4.1 Mb backbone DNA [33]. DNA segments that are unique to one or the other strain and scattered within each genome are termed "O-islands" in O157:H7 and "K-islands" in K12 [33]. O-islands consist of 1.34 Mb DNA sequence encoding 26% of all EDL933 genes, while K-islands consist of 0.53 Mb harboring 12% of the genes in MG1655 genome [33]. Many genes on the O-islands are important in pathogenicity (e.g., genes on the LEE islands) [33]. In addition, gene polymorphisms on the backbone are common, since 75% of the backbone genes encode proteins that differ by at least one amino acid in these two strains [33]. Some genes are extremely divergent. In the case of yadC, the protein sequence in K12 and O157:H7 is only 34% identical [33]. The genome divergence between O157:H7 and K12 may have a substantial effect on gene regulation.
E. coli O157:H7 diverged from K12 strain about 4.5 million years ago [34], and genes on O-islands have been acquired through horizontal gene transfer [33–35]. How O-island genes are integrated into preexisting regulatory circuits controlled by RpoS is still unknown. Given that RpoS is known to regulate genes of nonessential functions [8, 9, 15, 16], it is possible these O-island genes are preferentially under control of RpoS rather than RpoD, the housekeeping sigma factor. This has yet to be tested.
To examine RpoS-regulated gene expression in a pathogenic strain, we employed the E. coli O157:H7 strain EDL933 since this strain can cause serious human health problems and its genome is fully sequenced [33]. To compare with our previous results [15, 18], we sampled wild type and isogenic rpoS mutants of EDL933 under the same growth conditions and compared their transcriptome expression in exponential phase (OD600 = 0.3) and early stationary phase (OD600 = 1.5). Herein we report that rpoS mutation had a profound effect on transcriptome expression. Genes under control of RpoS included many EDL933-specific genes on the O-islands. Besides stress response genes, RpoS also regulated the expression of genes involved in metabolic pathways, transcription, and virulence.
Results
Expression of RpoS during growth in LB media
Although RpoS controls the expression of a large set of genes, mutation of rpoS has little effect on growth rate of E. coli K12 strain MG1655 [17, 18]. To test whether this is applicable to pathogenic E. coli EDL933, we compared the growth of rpoS mutants with wild type EDL933 grown in LB. Both the growth rate and the time to enter stationary phase were similar between wild type and rpoS mutants of EDL933 (Figure 1). The generation time in exponential phase was approximately 26 min. This equivalence is important for comparison of genomic expression since the expression of many genes is affected by growth rate [36]. As expected, the protein level of RpoS was found to be low in early exponential phase, followed by a substantial increase during entry of stationary phase (Figure 1).
Growth of EDL933 in LB media. Cultures were inoculated from overnight cultures to a starting OD600 = 0.0001 and incubated aerobically at 37°C at 200 rpm. RNA samples were isolated at OD600 = 0.3 and 1.5 as indicated. RpoS (ðS) protein levels were tested by Western blot analyses using monoclonal anti-RpoS antiserum as described in Materials and Methods. This experiment was performed in triplicate using independent isolates. Averaged values were used for construction of the growth curve.
Expression of genes under control of RpoS
The mutation in rpoS had a pronounced effect on genomic expression of EDL933 in stationary phase but a minor effect in exponential phase (Figure 2). In exponential phase when RpoS protein level was low, we found that 11 genes were differentially expressed in the rpoS mutants (Table 1), while in stationary phase, more than 1,000 genes were expressed differently as a result of rpoS mutation (twofold, P < 0.05) (Table 2 and Additional file 1). The false discovery rate was 1.4%. Among these stationary phase genes, 596 genes were expressed higher in the wild type EDL933, including 105 previously known RpoS-dependent genes in K12 strains. In addition, a mutation in rpoS led to increased expression of 536 genes (Table 3 and Additional file 1), indicating that the negative effect of RpoS on gene expression is also extensive. For genes on O-islands that are specific to EDL933, 50 genes showed higher expression in wild type and the expression of 49 genes was elevated in the rpoS mutants.
Transcriptome profile of WT EDL933 and rpoS mutants. Scatterplot was used to examine the effects of RpoS on gene expression in exponential (A) and stationary (B) phase. Probe sets (including genes and intergenic regions) are outlined by two parallel lines into three different groups: probe sets expressed at least twofold higher in the WT (red), those expressed more than twofold higher in rpoS mutants (green), and those not differentially expressed (black). LI: log2-transformed expression intensity.
RpoS-regulated functions in exponential phase
The expression of 11 genes was impaired in rpoS mutants in exponential phase (Table 1). Three genes, motAB and yhjH, are involved in the motor function of flagella. The gene yciF, encoding a putative structural protein, is RpoS-dependent in K12 strains [16]. There were seven EDL933-specific unknown genes under control of RpoS, two of which, Z3023 and Z3026, encode putative secreted proteins and play a role in colonization of E. coli O157:H7 in the bovine GI tract [37]. By contrast, the rpoS mutation had a much larger impact on gene expression in stationary phase. We thus focused on the analysis of the RpoS regulon in stationary phase.
RpoS-regulated functions in stationary phase
Stress response
As expected, many of the identified RpoS up-regulated genes were those that are important for stress response. For example, the rpoS mutation resulted in decreased expression of stress response genes yhiO (uspB), yhbO, gadAXW, gadB, gadE, osmY, csiD, and katE that are known be RpoS-dependent in K12 strains [38]. The genes gadAXW, gadB, and gadE are important for acid resistance [39], osmY for hyperosmotic resistance [40], yhiO (uspB) for ethanol tolerance [41], katE for oxidative response [42, 43], and yhbO for survival under oxidative, heat, UV, and pH stresses [16, 44]. Consistently, survival of rpoS mutants under low pH, oxidative stress, and heat exposure was severely impaired in comparison with wild type EDL933 strain (Figure 3).
Two starvation-induced genes, csiD (for carbon) and psiF (for phosphate) were also expressed higher in EDL933 wild type than in the rpoS mutants. Unlike in K12, the genes that encode universal stress proteins uspA, yecG (uspC), yiiT (uspD), ydaA (uspE) showed attenuated expression in rpoS mutants (this study) while their expression is not dependent on RpoS in K12 [45, 46].
Transporter and Membrane proteins
The expression of many genes for nutrient transport was affected by the rpoS mutation (Figure 4). Most of these genes encode proteins belonging to the ATP-Binding Cassette (ABC) transporter family. RpoS positively regulated ABC transporter genes included those for transport of oligopeptide (encoded by oppABCDF), dipeptide (dppABDF), putrescine (potFGH), maltose (malEFGK), glutamate/aspartate (gltIJKL), D-xylose (xylFHG) and sn-glycerol-3-P (ugpABCE). The expression of genes yehWXYZ, encoding a predicted ABC transporter, was also highly dependent on RpoS. Transporter genes expressed higher in the rpoS mutants included those for spermidine/putrescine (potABCD), glycine/proline (proWXY), and Zinc (znuABC). Besides ABC transporters, the tnaB gene encoding a tryptophan transporter and the dcuB gene encoding a transporter for C4-dicarboxylates (e.g., fumarate and malate) uptake were expressed at a lower level in the rpoS mutants compared with that in wild type EDL933. The gene cstA, encoding a peptide transporter that is induced under carbon starvation, has been shown to be negatively regulated by RpoS in a K12 strain [47], while we found that the expression of cstA was attenuated in the rpoS mutants of EDL933.
Effect of RpoS on expression of transporter genes. The mean expression ratio (MER/RpoS-dependence level) is given after each gene. Genes highlighted in red were expressed higher in wild type, those in blue were expressed higher in the rpoS mutant, and those in grey were not found to be significantly different (P > 0.05).
Metabolism
RpoS had a substantial effect on expression of metabolic genes, primarily for utilization of amino acids and carbohydrates (Figure 5). LB medium is rich in amino acids that can be utilized by E. coli as nutrient sources [48]. We found that the expression of genes for utilization of serine (tdcB), proline (putA), glutamine (ybaS), aspartate (asnB), arginine (astCABDE), tryptophan (tnaA), threonine (ilvBCDEMG), and alanine (dadAX) was expressed higher in the wild type EDL933 than in the rpoS mutants. The genes yneH and alr, encoding isoenzymes of YbaS and DadX, respectively, were expressed higher in the rpoS mutants (Figure 5). Pyruvate and glutamate appeared to be two common intermediate metabolites in RpoS-regulated amino acid utilization (Figure 5). For carbohydrate utilization, genes whose expression is positively regulated by RpoS included those encoding for putrescine degradation (puuABCD), fatty acid beta-oxidation (fadBA, fadD, fadE, and fadIJ), fucose utilization (fucAO, fucIK, lldD, and aldA), glucarate degradation (garDLR), glyoxylate cycle (aceBA, acnA, and gltA), and synthesis of trehalose (otsBA) and glycogen (glgABC) (Figure 5). The cdd and udp genes for pyrimidine degradation were reduced in expression in the rpoS mutant, while the expression of genes udk, cmk, upp, and codA that are involved in the pyrimidine biosynthesis pathway was enhanced.
Metabolic pathways that are regulated by RpoS in stationary phase. Genes expressed higher in wild type are colored red and those expressed higher in rpoS mutants are blue. Genes whose differential expression was not significant (P > 0.05) are in black. The mean expression ratio (MER: WT/rpoS) is indicated after each gene.
Some of these metabolic genes may play an important role in colonization and pathogenesis of E. coli in vivo in host environments. For example, the expression of fucAO is important for colonization of E. coli in mouse intestine [49]. Mutants defected in metabolism of maltose and glycogen are also impaired in colonization of EDL933 in mouse intestine [50].
Transcription Regulation
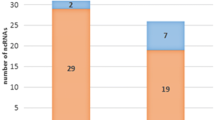
The expression of 29 genes encoding known transcriptional regulators was affected by the rpoS mutation. Sixteen genes (lsrR, mhpR, prpR, putA, lldR, hcaR, galS, gadXWE, fucR, dgsA, csgD, cdaR, bolA, and xylR) were expressed higher in the wild type EDL933 while 13 genes (dicA, deoR, birA, uhpA, marR, metJ, pdhR, purR, rcsA, arsR, asnC, cspA, and fis) were expressed higher in the rpoS mutants (Additional file 1). The observed differential expression of many genes in the rpoS mutants may be an indirect effect of RpoS through these intermediate regulators. Some regulatory genes are known to be RpoS-controlled, such as bloA [51], gadE [52], and csgD [28]. Expression of the hcaR gene, encoding the hydrocinnamic acid regulator, is stationary phase dependent but RpoS-independent in E. coli K12 strain [53]. Here we found that expression of hcaR was induced in stationary phase in both wild type EDL933 and rpoS mutants. However, the induction level was significantly higher in wild type, indicating that RpoS is important for full expression of hcaR.
Virulence and O-island genes
We found that 10% of the identified RpoS-regulated genes are located on O-islands. Among them, 50 genes were expressed higher in wild type EDL933 in stationary phase (Table 4) while 49 genes expressed higher in the rpoS mutants (Table 5). The functions of most of these genes are still unknown. On the LEE island (located on the O-island 148), three genes, ler, cesF and Z5139, were expressed significantly higher in wild type EDL933 than in the rpoS mutants (Table 4), while the eae gene, encoding the outer membrane intimin protein essential for colonization and virulence, was expressed twofold higher in rpoS mutants (Table 5). The expression of other genes on the LEE islands was not significantly affected by the rpoS mutation. The espI gene, though not located on the LEE island, encodes a secreted protein whose secretion requires the LEE-encoded type III secretion system [54]. The expression of espI was 78 fold higher in the wild type EDL933. The nlpA gene, encoding an inner membrane protein that is required for virulence in Haemophilus influenzae [55], was impaired in its expression in the rpoS mutants. The dppA operon, required for colonization by uropathogenic E. coli [56], was expressed much higher in the wild type EDL933 than rpoS mutants.
Western blot analysis of LEE proteins under LEE-induction conditions
Growth condition plays a considerable effect on LEE gene expression [57, 58]. The expression of LEE genes is low in LB media and is induced in LB supplemented with sodium bicarbonate or DMEM media in 5% CO2 [57, 58]. To determine whether the expression of LEE genes was controlled by RpoS under these LEE-induction conditions, we examined the expression of one gene from each of the five LEE islands by qPCR using cultures grown in LB supplemented with 44 mM sodium bicarbonate media [57]. All genes tested were expressed higher in the rpoS mutants. The ratio of expression in rpoS mutants verse wild type EDL933 for ler (LEE1), sepZ (LEE2), escV (LEE3), tir (LEE4), sepL (LEE5), grlR and grlA (LEE regulator) was 2.8 ± 0.5, 1.3 ± 0.4, 5.5 ± 0.4, 4.8 ± 0.4, 6.4 ± 0.4, 4.7 ± 0.4, and 7.6 ± 0.4, respectively. Western blot analysis revealed that the expression of Tir and EspA was enhanced in the rpoS mutants of EDL933 (Figure 6). Similar results were obtained in cultures grown in DMEM media, another LEE induction condition (Figure 6). Consistent with previous results, neither Tir nor EspA was detected in LB without sodium bicarbonate (data not shown).
Western blot analysis of Tir and EspA expression in wild type and rpoS mutants. Cultures were grown aerobically at 37°C in LB media supplemented with 44 mM NaHCO3 to OD600 = 1.5 or in DMEM media in 5% CO2 (two known LEE-induction conditions). Cell pellets were resuspended in SDS loading buffer and boiled for 5 min. Resultant cell extracts were resolved on a 10% SDS-PAGE gel. Proteins were transferred to a PVDF membrane by electrophoresis, followed by incubation of the membrane with anti-Tir or anti-EspA specific antibody. Signals were detected using ECL solution and Hyperfilm-ECL film (Amersham).
Negative regulation by RpoS
As mentioned above, we found 536 genes expressed higher in rpoS mutants in stationary phase (Table 3 and Additional file 1). These genes are involved in many cellular functions, including metabolism (e.g., thiI and guaBA), nutrient transport (e.g., ampG, cmr and uraA), and DNA modification (e.g., endA and nth). The expression of almost all genes in the purine biosynthesis pathway was enhanced in the rpoS mutant (Figure 7). The rsxABCDGE operon that is required for the reduction of SoxR was also expressed higher in the rpoS mutants (Table 3). Interestingly, the flagellar genes and the TCA cycle genes, whose expression is negatively regulated by RpoS in E. coli K12 strains [15], were not differentially expressed in the rpoS mutant of EDL933. The flagellar sigma factor FliA, was expressed similarly in wild type EDL933 and rpoS mutants (Figure 8).
RpoS-regulation of genes required for de novo biosynthesis of purine nucleotides pathway I in stationary phase. RpoS-dependence (MER) is indicated in parentheses. A negative value (-) denotes RpoS-negative regulation. The pathway map is adapted from the EcoCyc database. Genes that were significantly differentially expressed (P < 0.05) are highlighted in bold.
Expression of FliA in WT and rpoS mutants of EDL933 in LB. Western blot analyses of the expression of the flagella sigma factor FliA were performed using monoclonal antibody to FliA as described in Material and Methods. To confirm equal protein loading, another protein gel run in parallel was stained by Coomassie blue R250.
Verification of microarray results
To validate the microarray results, we determined the expression level and RpoS dependence of candidate genes by qPCR (Figure 9). The RpoS-dependence levels of all 12 genes tested were in good correlation between results of microarray and qPCR. Because the rpoS sequence is absent in the rpoS null mutant tested in this study, the signal difference for rpoS between wild type EDL933 and rpoS mutant strains serves as an internal control for the sensitivity of microarray data. We found the expression difference of the two rpoS probe sets was about 5,000 fold between wild type and rpoS mutants. As expected, we also found many known RpoS-regulated genes (e.g., osmY, katE and astC) were identified as RpoS-controlled genes in this study.
Discussion
In this study, we have characterized the RpoS regulon of the important pathogenic E. coli O157:H7 strain EDL933. Comparison with previous data obtained using laboratory K12 strains reveals substantial differences between the composition of RpoS regulon in K12 and O157:H7 EDL933. As might be expected, the RpoS-regulon identified in EDL933 is much larger than that of K12, which is partly attributable to the larger number of genes present in the pathogenic strain. Another factor may be different levels of the expression of RpoS itself. Indeed, we found that the level of RpoS was higher in EDL933 than in MG1655 in early stationary phase (Additional file 2), consistent with previous results that RpoS levels vary among E. coli isolates [59]. Though there is a core set of genes regulated by RpoS in both K12 and EDL933 strains, the RpoS-dependence of a large number of genes (~80% of RpoS-dependent genes in EDL933) is strain-specific, including a group of RpoS-dependent genes on O-islands and several virulence determinant genes. RpoS has a larger effect on exponential phase gene expression in K12 strain than in EDL933 [18, 19]. These results suggest that RpoS regulation may be strongly dependent on strain background. Consistent with this, there are many known phenotypic differences between K12 and EDL933. For example, MG1655 and EDL933 differ in utilization of nutrients and location of colonization during in vivo growth in mouse intestine [50, 60, 61].
The expression of a large number of genes was higher in the rpoS mutants, indicating negative control of RpoS on gene expression. As a sigma factor, negative control exerted by RpoS is likely an indirect effect, probably resulting from sigma factor competition [45]. Because the number of sigma factors exceeds that of core RNA polymerase, different sigma factors compete for binding to the core enzyme [62]. Deletion of RpoS, a major sigma factor in stationary phase, may thus result in increased amount of core enzyme associated with other sigma factors and their-directed gene expression. In E. coli K12 strain, there is also a large number of genes negatively regulated by RpoS [15]. For example, expression of genes for chemotaxis and flagella is negatively regulated by RpoS in K12 [15, 17]. However, this was not the case in EDL933 (this study), suggesting the negative regulation of RpoS was also strain-specific. In other pathogens, the effect of RpoS on flagella expression is variable (Table 6) [15, 17, 63–71]. In P. aeruginosa, expression of the flagellar gene fliF as well as genes for chemotaxis is positively regulated by RpoS [64]. In Vibrio cholerae, RpoS positively controls the expression of chemotaxis and flagellar genes during pathogenesis [68]. In Legionella pneumophila and S. typhimurium, RpoS is important for expression of flagella [63, 65]. However, flagella gene expression is independent of RpoS in S. typhimurium strain LT2 [66], which has a mutant allele of RpoS [72].
The intestinal growth environment inhabited by EHEC E. coli is complex. Utilization of glycogen [50], maltose [50], L-fucose [49], galactose [61], arabinose [61], and ribose [61] is important for colonization by E. coli. We found that an rpoS mutation attenuates the expression of genes involved in metabolism of these sugars (Figure 5), suggesting a role of RpoS in regulation of bacterial colonization. This is consistent with our previous findings in an animal model that wild type C. rodentium colonizes mouse colon better than rpoS mutants [26]. The contribution of RpoS-regulated metabolism to in vivo colonization needs to be further evaluated through construction of mutations in relevant pathways to identify specific causal factors.
The expression of most genes on the LEE island is under control of Ler, a LEE-encoded regulator [73, 74], and thus LEE genes is expected to be expressed similarly. However, previous results have shown that this is not the case [75, 76]. Consistent with this, our results show that RpoS had an opposing effect on LEE gene expression, suggesting that LEE genes are under differential control for expression. The difference in expression of LEE genes may be due to the lack of induction signals for LEE expression in LB. Under induction conditions, all LEE genes tested were expressed higher in the rpoS mutants (this study).
A recent microarray study reviewed differences in the heat shock response of E. coli O157:H7 EDL933 and K12 strains, and attributed discrepancies to experimental conditions and/or genomic compositions [77]. About 30 EDL933 specific genes are differentially expressed during heat shock [77]. Only four of the top 25 heat shock response genes were RpoS-dependent (this study), suggesting that other regulators (e.g., the heat shock sigma factor RpoH) are required for the full heat shock response. Again, differences in methodology (e.g., array platforms and experimental conditions) make it difficult to directly compare results.
Gene expression profiling has greatly improved our knowledge of the role of RpoS in regulation of genes and many cellular functions. However, we are still far from fully understanding the physiological role of RpoS. For example, a large portion of RpoS-regulated genes are those with unknown or putative functions. Factors responsible for strain-specific effects also remain elusive. Furthermore, the regulation of RpoS itself is not fully understood. Recent studies have identified two anti-adaptor proteins, IraM (previously known as YcgW) [78] and IraD (YjiD) [20], which stabilize RpoS through inhibition of RssB-ClpXP directed proteolysis. RpoS activity has also been found to be transiently inhibited by FliZ in post exponential phase [79]. It is likely that there are other unidentified factors involved in the regulatory network of RpoS.
Conclusion
Our results reveal the first snapshot overview of RpoS-regulated transcriptome expression in non-K12 strains. This, together with previous results regarding RpoS control in laboratory strains, provides a useful database for understanding how global regulators (e.g., RpoS) can gain additional functions in pathogenic E. coli strains.
Methods
Strains, media and growth conditions
E. coli strain O157:H7 EDL933 and its rpoS mutant derivative were employed in this study. Cultures were grown aerobically at 37°C with shaking at 200 rpm in Luria-Bertani media, and growth was monitored spectrophotometrically at OD600. Antibiotics were used at the following concentrations: ampicillin (100 μg/ml) and chloramphenicol (25 μg/ml).
Construction of EDL933 rpoS deletion mutant
An rpoS non-polar deletion mutant was constructed by homologous recombination as described previously [80, 81]. Briefly, a linear DNA fragment, harboring the chloramphenicol resistant gene cat and homologous rpoS-flanking sequences, was amplified using pKD3 plasmid (template) and primers FP1 (CCTCGCTTGAGACTGGCCTTTCTGACAGTGCTTACGTGTAGGCTGGAGCTGCTTC) and RP1 (ATGTTCCGTCAAGGGATCACGGGTAGGAGCCACCTTCATATGAATATCCT CCTTAG) and introduced into EDL933 competent cells by electroporation. Transformants were selected on LB chloramphenicol plates. The cat gene was further removed by recombination with the FLP recombinase. The loss of rpoS was confirmed by PCR using flanking primers and by sequencing.
RNA preparation
RNA samples were prepared as previously described [18]. Overnight cultures were diluted into fresh media at a starting OD600 of 0.0001 to allow cells to grow at least ten generations prior to RNA isolation in exponential phase. Cultures grown in triplicate were sampled at OD600 = 0.3 (exponential phase) and OD600 = 1.5 (stationary phase), conditions used in our previous studies for comparison [15, 18]. RNA samples were prepared using hot acidic phenol (pH 4.3, Sigma-Aldrich), and the quality of RNA was examined using a Bioanalyzer 2100 (Agilent Technologies).
Microarray analysis
The Affymetrix GeneChip E. coli Genome 2.0 Array was employed in this study. This array chip contains more than 10,000 probe sets that cover all genes in the genomes of four type E. coli strains, K12 MG1655, O157:H7 EDL933, O157:H7 Sakai, and the uropathogen, CFT073. A gene that is present in all genomes with high similarity in sequence is represented by a single probe set. Although this is an effective approach to minimize the total number of probe sets used to cover all four genomes, some homologous genes with low sequence similarity in the four strains may be represented by more than one probe set. For example, there are two probe sets in the array representing rpoS (probe set IDs: 1761030_s_at and 1767783_s_at) because the rpoS sequence in the strain CFT073 harbors an internal mutation that results in two truncated genes, c3306 (519 bp probing to 3' end of rpoS) and c3307 (435 bp probing to 5' end of rpoS). Both probe sets hybridized to rpoS transcripts and the resultant signals in wild type samples were 4,939 and 7,643 time higher than those in the knockout rpoS mutants, respectively (this study). Though both probe sets are representative of rpoS, this leads to duplication. To avoid this problem, microarray data were curated to remove redundant probe sets in our analysis. Microarray samples were analyzed using dChip [82] and BRB Arraytools [83], as described previously [17]. Samples were log2 transformed and normalized using the GCRMA method [84]. RpoS dependence of genes is represented by the mean expression ratio (MER) of WT and rpoS mutants. The significance of expression difference was tested using Student's t-tests. Genes with MER value ≥2 or ≤0.5 and P value < 0.05 were considered to be controlled by RpoS [17]. The false discovery rate (FDR) was estimated by 1,000 time random permutations as previously described [17]. Microarray data can be accessed in the Gene Expression Omnibus database at the National Center for Biotechnology Information under the accession number GSE17420.
Quantitative real-time PCR (qPCR)
To confirm microarray results, we tested gene transcription by qPCR as previously described [17]. Primers were designed using the PerlPrimer program [85] and synthesized by the MOBIX laboratory at McMaster University. RNA samples were prepared as for microarray analysis. First strand cDNA was synthesized using a cDNA synthesis kit (New England Biolabs). Gene amplification was detected using SYBR green (Clontech) in a MX3000P qPCR system (Stratagene). The expression level of genes was determined by constructing a standard curve using serial dilutions of EDL933 genome DNA with known concentrations. The 16S RNA gene, rrsA, was used as a reference control to normalize differences in total RNA quantity among samples [86].
Western blot analyses
Cultures were grown in LB media aerobically at 37°C and sampled periodically. Samples were immediately mixed with chloramphenicol (150 μg/ml) and placed on ice to stop protein synthesis, followed by centrifugation at 15,000 × g for 2 min. Cell pellets were flash frozen in liquid nitrogen prior to use. Cell pellets were thawed on ice, resuspended to OD600 = 1.0 with SDS loading buffer, and boiled for 5 min. Samples of 10 μl were resolved on 10% SDS-PAGE and transferred to PVDF membrane [17]. The PVDF membrane was then blocked with 5% milk solution, incubated with mouse monoclonal antibodies for RpoS (NeoClone, Madison, WI), Tir or EspA (a gift from B. Coombes), and HRP-conjugated Goat anti-mouse secondary antibody (Bio-Rad, Hercules, CA). The signal was detected using the ECL solution (Amersham, Pittsburgh, PA) and Hyperfilm-ECL film (Amersham, Pittsburgh, PA). To ensure that equal amounts of protein were loaded, another SDS-PAGE gel was run in parallel and stained with Coomassie Blue R-250.
Survival of mutants upon exposure to stress conditions
Stationary phase cultures were washed and diluted in 0.9% NaCl before exposure to stress. A total number of 1.0 × 108 cells were exposed to 1 ml of acidic LB (pH2.5, adjusted with HCl) and 15 mM H2O2, respectively, while 5.0 × 103 cells were treated at 55°C for heat exposure. Viable cells were enumerated by serial plating on LB media, and survival expressed as a percentage determined by dividing the number of viable cells by the number of cells before treatment.
References
Paton JC, Paton AW: Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin Microbiol Rev. 1998, 11: 450-479.
Karmali MA: Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989, 2: 15-38.
McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB: A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995, 92: 1664-1668. 10.1073/pnas.92.5.1664.
O'Brien AD, Holmes RK: Shiga and Shiga-like toxins. Microbiol Rev. 1987, 51: 206-220.
Barba J, Bustamante VH, Flores-Valdez MA, Deng W, Finlay BB, Puente JL: A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol. 2005, 187: 7918-7930. 10.1128/JB.187.23.7918-7930.2005.
Tomoyasu T, Takaya A, Handa Y, Karata K, Yamamoto T: ClpXP controls the expression of LEE genes in enterohaemorrhagic Escherichia coli. FEMS Microbiol Lett. 2005, 253: 59-66.
Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vazquez A, Barba J, Ibarra JA, O'Donnell P, Metalnikov P, et al: Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci USA. 2004, 101: 3597-3602. 10.1073/pnas.0400326101.
Dong T, Joyce C, Schellhorn HE: The Role of RpoS in Bacterial Adaptation. Bacterial Physiology – A Molecular Approach. Edited by: Walid M El-Sharoud. 2008, Springer, Berlin, Germany, 313-337.
Hengge-Aronis R: The general stress response in Escherichia coli. Bacterial Stress Response. Edited by: Storz G, Hengge-Aronis R. 2000, ASM press, Washington, DC, 161-178.
Sammartano LJ, Tuveson RW, Davenport R: Control of sensitivity to inactivation by H2O2 and broad-spectrum near-UV radiation by the Escherichia coli katF (rpoS) locus. J Bacteriol. 1986, 168: 13-21.
Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL: Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994, 176: 1729-1737.
Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W: Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J Bacteriol. 1991, 173: 7918-7924.
Lange R, Hengge-Aronis R: Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991, 5: 49-59. 10.1111/j.1365-2958.1991.tb01825.x.
Lacour S, Landini P: SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J Bacteriol. 2004, 186: 7186-7195. 10.1128/JB.186.21.7186-7195.2004.
Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE: Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics. 2004, 272: 580-591. 10.1007/s00438-004-1089-2.
Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R: Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005, 187: 1591-1603. 10.1128/JB.187.5.1591-1603.2005.
Dong T, Schellhorn HE: Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol Genet Genomics. 2009, 281: 19-33. 10.1007/s00438-008-0389-3.
Dong T, Kirchhof MG, Schellhorn HE: RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol Genet Genomics. 2008, 279: 267-277. 10.1007/s00438-007-0311-4.
Rahman M, Hasan MR, Oba T, Shimizu K: Effect of rpoS gene knockout on the metabolism of Escherichia coli during exponential growth phase and early stationary phase based on gene expressions, enzyme activities and intracellular metabolite concentrations. Biotechnol Bioeng. 2006, 94: 585-595. 10.1002/bit.20858.
Merrikh H, Ferrazzoli AE, Bougdour A, Olivier-Mason A, Lovett ST: A DNA damage response in Escherichia coli involving the alternative sigma factor, RpoS. Proc Natl Acad Sci USA. 2009, 106: 611-616. 10.1073/pnas.0803665106.
Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG: The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992, 89: 11978-11982. 10.1073/pnas.89.24.11978.
Merrell DS, Tischler AD, Lee SH, Camilli A: Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect Immun. 2000, 68: 6691-6696. 10.1128/IAI.68.12.6691-6696.2000.
Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE: Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999, 181: 3890-3897.
Iriarte M, Stainier I, Cornelis GR: The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995, 63: 1840-1847.
Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S: Citrobacter rodentium of mice and man. Cell Microbiol. 2005, 7: 1697-1706. 10.1111/j.1462-5822.2005.00625.x.
Dong T, Coombes BK, Schellhorn HE: Role of RpoS in the virulence of Citrobacter rodentium. Infect Immun. 2009, 77: 501-507. 10.1128/IAI.00850-08.
Romling U: Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005, 62: 1234-1246. 10.1007/s00018-005-4557-x.
Romling U, Bian Z, Hammar M, Sierralta WD, Normark S: Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998, 180: 722-731.
Uhlich GA, Cooke PH, Solomon EB: Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol. 2006, 72: 2564-2572. 10.1128/AEM.72.4.2564-2572.2006.
Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB: Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci USA. 1999, 96: 15196-15201. 10.1073/pnas.96.26.15196.
Iyoda S, Watanabe H: ClpXP protease controls expression of the type III protein secretion system through regulation of RpoS and GrlR levels in enterohemorrhagic Escherichia coli. J Bacteriol. 2005, 187: 4086-4094. 10.1128/JB.187.12.4086-4094.2005.
Laaberki MH, Janabi N, Oswald E, Repoila F: Concert of regulators to switch on LEE expression in enterohemorrhagic Escherichia coli O157:H7: interplay between Ler, GrlA, HNS and RpoS. Int J Med Microbiol. 2006, 296: 197-210. 10.1016/j.ijmm.2006.02.017.
Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al: Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001, 409: 529-533. 10.1038/35054089.
Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS: Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000, 406: 64-67. 10.1038/35017546.
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, et al: Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001, 8: 11-22. 10.1093/dnares/8.1.11.
Tao H, Bausch C, Richmond C, Blattner FR, Conway T: Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999, 181: 6425-6440.
Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS: Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology. 2004, 150: 3631-3645. 10.1099/mic.0.27448-0.
Karp PD, Keseler IM, Shearer A, Latendresse M, Krummenacker M, Paley SM, Paulsen I, Collado-Vides J, Gama-Castro S, Peralta-Gil M, et al: Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007, 35: 7577-7590. 10.1093/nar/gkm740.
Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW: GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol. 2003, 49: 1309-1320. 10.1046/j.1365-2958.2003.03633.x.
Yim HH, Villarejo M: osmY, a new hyperosmotically inducible gene, encodes a periplasmic protein in Escherichia coli. J Bacteriol. 1992, 174: 3637-3644.
Farewell A, Kvint K, Nystrom T: uspB, a new sigmaS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J Bacteriol. 1998, 180: 6140-6147.
Imlay JA, Linn S: Mutagenesis and stress responses induced in Escherichia coli by hydrogen peroxide. J Bacteriol. 1987, 169: 2967-2976.
Sak BD, Eisenstark A, Touati D: Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci USA. 1989, 86: 3271-3275. 10.1073/pnas.86.9.3271.
Abdallah J, Caldas T, Kthiri F, Kern R, Richarme G: YhbO protects cells against multiple stresses. J Bacteriol. 2007, 189: 9140-9144. 10.1128/JB.01208-07.
Farewell A, Kvint K, Nystrom T: Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998, 29: 1039-1051. 10.1046/j.1365-2958.1998.00990.x.
Gustavsson N, Diez A, Nystrom T: The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol Microbiol. 2002, 43: 107-117. 10.1046/j.1365-2958.2002.02720.x.
Dubey AK, Baker CS, Suzuki K, Jones AD, Pandit P, Romeo T, Babitzke P: CsrA regulates translation of the Escherichia coli carbon starvation gene, cstA, by blocking ribosome access to the cstA transcript. J Bacteriol. 2003, 185: 4450-4460. 10.1128/JB.185.15.4450-4460.2003.
Sezonov G, Joseleau-Petit D, D'Ari R: Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007, 189: 8746-8749. 10.1128/JB.01368-07.
Autieri SM, Lins JJ, Leatham MP, Laux DC, Conway T, Cohen PS: L-fucose stimulates utilization of D-ribose by Escherichia coli MG1655 DeltafucAO and E. coli Nissle 1917 DeltafucAO mutants in the mouse intestine and in M9 minimal medium. Infect Immun. 2007, 75: 5465-5475. 10.1128/IAI.00822-07.
Jones SA, Jorgensen M, Chowdhury FZ, Rodgers R, Hartline J, Leatham MP, Struve C, Krogfelt KA, Cohen PS, Conway T: Glycogen and maltose utilization by Escherichia coli O157:H7 in the mouse intestine. Infect Immun. 2008, 76: 2531-2540. 10.1128/IAI.00096-08.
Lange R, Hengge-Aronis R: Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor sigma S. J Bacteriol. 1991, 173: 4474-4481.
Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW: GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol. 2003, 49: 1309-1320. 10.1046/j.1365-2958.2003.03633.x.
Turlin E, Perrotte-Piquemal M, Danchin A, Biville F: Regulation of the early steps of 3-phenylpropionate catabolism in Escherichia coli. J Mol Microbiol Biotechnol. 2001, 3: 127-133.
Mundy R, Petrovska L, Smollett K, Simpson N, Wilson RK, Yu J, Tu X, Rosenshine I, Clare S, Dougan G, et al: Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect Immun. 2004, 72: 2288-2302. 10.1128/IAI.72.4.2288-2302.2004.
Chanyangam M, Smith AL, Moseley SL, Kuehn M, Jenny P: Contribution of a 28-kilodalton membrane protein to the virulence of Haemophilus influenzae. Infect Immun. 1991, 59: 600-608.
Haugen BJ, Pellett S, Redford P, Hamilton HL, Roesch PL, Welch RA: In vivo gene expression analysis identifies genes required for enhanced colonization of the mouse urinary tract by uropathogenic Escherichia coli strain CFT073 dsdA. Infect Immun. 2007, 75: 278-289. 10.1128/IAI.01319-06.
Abe H, Tatsuno I, Tobe T, Okutani A, Sasakawa C: Bicarbonate ion stimulates the expression of locus of enterocyte effacement-encoded genes in enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2002, 70: 3500-3509. 10.1128/IAI.70.7.3500-3509.2002.
Kenny B, Abe A, Stein M, Finlay BB: Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997, 65: 2606-2612.
King T, Ishihama A, Kori A, Ferenci T: A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol. 2004, 186: 5614-5620. 10.1128/JB.186.17.5614-5620.2004.
Miranda RL, Conway T, Leatham MP, Chang DE, Norris WE, Allen JH, Stevenson SJ, Laux DC, Cohen PS: Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004, 72: 1666-1676. 10.1128/IAI.72.3.1666-1676.2004.
Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, et al: Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008, 76: 1143-1152. 10.1128/IAI.01386-07.
Ishihama A: Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000, 54: 499-518. 10.1146/annurev.micro.54.1.499.
Hammer BK, Tateda ES, Swanson MS: A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol. 2002, 44: 107-118. 10.1046/j.1365-2958.2002.02884.x.
Schuster M, Hawkins AC, Harwood CS, Greenberg EP: The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 2004, 51: 973-985. 10.1046/j.1365-2958.2003.03886.x.
Cogan TA, Jorgensen F, Lappin-Scott HM, Benson CE, Woodward MJ, Humphrey TJ: Flagella and curli fimbriae are important for the growth of Salmonella enterica serovars in hen eggs. Microbiology. 2004, 150: 1063-1071. 10.1099/mic.0.26791-0.
Kutsukake K: Autogenous and global control of the flagellar master operon, flhD, in Salmonella typhimurium. Mol Gen Genet. 1997, 254: 440-448. 10.1007/s004380050437.
Adams P, Fowler R, Kinsella N, Howell G, Farris M, Coote P, O'Connor CD: Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics. 2001, 1: 597-607. 10.1002/1615-9861(200104)1:4<597::AID-PROT597>3.0.CO;2-P.
Nielsen AT, Dolganov NA, Otto G, Miller MC, Wu CY, Schoolnik GK: RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006, 2: e109-10.1371/journal.ppat.0020109.
Hulsmann A, Rosche TM, Kong IS, Hassan HM, Beam DM, Oliver JD: RpoS-dependent stress response and exoenzyme production in Vibrio vulnificus. Appl Environ Microbiol. 2003, 69: 6114-6120. 10.1128/AEM.69.10.6114-6120.2003.
Ito A, May T, Kawata K, Okabe S: Significance of rpoS during maturation of Escherichia coli biofilms. Biotechnol Bioeng. 2008, 99: 1462-1471. 10.1002/bit.21695.
Makinoshima H, Aizawa S, Hayashi H, Miki T, Nishimura A, Ishihama A: Growth phase-coupled alterations in cell structure and function of Escherichia coli. J Bacteriol. 2003, 185: 1338-1345. 10.1128/JB.185.4.1338-1345.2003.
Swords WE, Cannon BM, Benjamin WH: Avirulence of LT2 strains of Salmonella typhimurium results from a defective rpoS gene. Infect Immun. 1997, 65: 2451-2453.
Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB: The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol Microbiol. 1999, 33: 296-306. 10.1046/j.1365-2958.1999.01473.x.
Kaper JB, Nataro JP, Mobley HL: Pathogenic Escherichia coli. Nat Rev Microbiol. 2004, 2: 123-140. 10.1038/nrmicro818.
Kendall MM, Rasko DA, Sperandio V: Global effects of the cell-to-cell signaling molecules autoinducer-2, autoinducer-3, and epinephrine in a luxS mutant of enterohemorrhagic Escherichia coli. Infect Immun. 2007, 75: 4875-4884. 10.1128/IAI.00550-07.
Bergholz TM, Wick LM, Qi W, Riordan JT, Ouellette LM, Whittam TS: Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol. 2007, 7: 97-10.1186/1471-2180-7-97.
Carruthers MD, Minion C: Transcriptome analysis of Escherichia coli O157:H7 EDL933 during heat shock. FEMS Microbiol Lett. 2009, 295: 96-102. 10.1111/j.1574-6968.2009.01587.x.
Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S: Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008, 68: 298-313. 10.1111/j.1365-2958.2008.06146.x.
Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R: Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008, 22: 2434-2446. 10.1101/gad.475808.
Datsenko KA, Wanner BL: One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000, 97: 6640-6645. 10.1073/pnas.120163297.
Dong T, Chiang SM, Joyce C, Yu R, Schellhorn HE: Polymorphism and selection of rpoS in pathogenic Escherichia coli. BMC Microbiol. 2009, 9: 118-10.1186/1471-2180-9-118.
Li C, Wong WH: Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001, 98: 31-36. 10.1073/pnas.011404098.
Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y: Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007, 3: 11-17.
Wu Z, Irizarry RA: Preprocessing of oligonucleotide array data. Nat Biotechnol. 2004, 22: 656-658. 10.1038/nbt0604-656b.
Marshall OJ: PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004, 20: 2471-2472. 10.1093/bioinformatics/bth254.
Kobayashi A, Hirakawa H, Hirata T, Nishino K, Yamaguchi A: Growth phase-dependent expression of drug exporters in Escherichia coli and its contribution to drug tolerance. J Bacteriol. 2006, 188: 5693-5703. 10.1128/JB.00217-06.
Acknowledgements
This study was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) and Canadian Institutes of Health Research (CIHR) to HES. TD is a recipient of an Ontario Graduate Scholarship. We thank B. Coombes for providing antibodies of Tir and EspA. We also thank C. Joyce for reviewing the manuscript and R. Yu and X. Liang for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
TD performed the experiments and wrote the manuscript. HES is the principal investigator who supervised the project and revised the manuscript. Both authors read and approved the final manuscript.
Electronic supplementary material
12864_2009_2233_MOESM1_ESM.xls
Additional file 1: Expression of RpoS-regulated genes. The data show the expression of RpoS-regulated genes (twofold, P < 0.05) in E. coli O157:H7 EDL933 wild type and rpoS mutants in LB stationary phase (OD600 = 1.5) (XLS 361 KB)
12864_2009_2233_MOESM2_ESM.pdf
Additional file 2: Western blot analysis of RpoS expression in Escherichia coli O157:H7 strain EDL933 and E. coli K12 strain MG1655. Cultures of EDL933 and MG1655 were grown in LB media at 37°C with vigorous shaking at 200 rpm and sampled at OD600 = 0.3 in exponential phase, OD600 = 1.5 in stationary phase, and overnight. The rpoS mutant of EDL933 was included as a negative control (lane 7). The beta subunit of RNA polymerase RpoB serves as an internal loading control. The expression level of RpoS was higher in EDL933 than in MG1655 in exponential (OD600 = 0.3) and early stationary phase (OD600 = 1.5). However, the level of RpoS reached a higher level in MG1655 than EDL933 in overnight samples. This is consistent with previous reports that RpoS expression varies depending on strain background (King et al., 2004; See the manuscript for reference). (PDF 21 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Dong, T., Schellhorn, H.E. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157:H7 strain EDL933. BMC Genomics 10, 349 (2009). https://doi.org/10.1186/1471-2164-10-349
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-10-349