Abstract
Background
Indian populations endowed with unparalleled genetic complexity have received a great deal of attention from scientists world over. However, the fundamental question over their ancestry, whether they are all genetically similar or do exhibit differences attributable to ethnicity, language, geography or socio-cultural affiliation is still unresolved. In order to decipher their underlying genetic structure, we undertook a study on 3522 individuals belonging to 54 endogamous Indian populations representing all major ethnic, linguistic and geographic groups and assessed the genetic variation using autosomal microsatellite markers.
Results
The distribution of the most frequent allele was uniform across populations, revealing an underlying genetic similarity. Patterns of allele distribution suggestive of ethnic or geographic propinquity were discernible only in a few of the populations and was not applicable to the entire dataset while a number of the populations exhibited distinct identities evident from the occurrence of unique alleles in them. Genetic substructuring was detected among populations originating from northeastern and southern India reflective of their migrational histories and genetic isolation respectively.
Conclusion
Our analyses based on autosomal microsatellite markers detected no evidence of general clustering of population groups based on ethnic, linguistic, geographic or socio-cultural affiliations. The existence of substructuring in populations from northeastern and southern India has notable implications for population genetic studies and forensic databases where broad grouping of populations based on such affiliations are frequently employed.
Similar content being viewed by others
Background
Human diversity in India is defined by 4693 different, documented population groups that include 2205 major communities, 589 segments and 1900 territorial units spread across the country [1]. Anthropologically, the populations are grouped into four major ethnic categories, which include the Australoid, Indo-Caucasoid, Indo-Mongoloid and Negrito populations and linguistically broadly classified as Indo-European, Dravidian, Austro-Asiatic and Sino-Tibetan speakers. The complex structure of the Indian population is attributed to incessant, historical waves of migrations into India, the earliest, by the Austric speakers around 70,000 years ago, followed by the Dravidian speakers from middle-east Asia and the Sino-Tibetan speakers from China and southeast Asia around 8000 to 10,000 years ago. The last major migration is believed to have occurred around 4000 years ago by several waves of Indo-European speakers [2]. Earlier genetic studies to understand the prevailing diversity among extant Indian populations analyzing populations that were predefined either based on ethnicity, language, culture or geography have interpreted existence of different levels of genetic relationships among population groups [3–6] that broadly attest the theories of migration and assimilation of different populations. However, recent molecular analyses have also asserted genetic similarity across populations spread over diverse geographic regions of the country, revealing a gradation of genetic lineages underscoring the genetic correlation amongst populations [7, 8].
The striking social attribute of the Indian populations is their strict practice of endogamy across all social ranks that has resulted in emergence of diverse population-specific social traditions and formation of distinct linguistic dialects due to subsequent isolation of populations. Although uniparental, biallelic markers have deciphered the common major Paleolithic contributions [9], resolution of many sub-lineages is still awaited in order to decipher finer genetic signatures defining populations that have resisted admixture for centuries. Patterns of variation across recently diverged populations can be successfully characterized with fast-evolving microsatellite markers [10][11][12]. Genetic drift among isolated, small populations manifests as characteristic allele frequency patterns that have been recently effectively characterized to identify genetic clusters that corresponded well with predefined geographically or linguistically similar populations [13].
With these rationales, we have analyzed 15 highly polymorphic autosomal microsatellite markers including 13 core forensic loci, which have been extensively used to reveal the ethnological and anthropological affinity of diverse populations ([10][11][12], [14][15][16][17][18][19]). In order to decipher if geographic proximity, linguistic, ethnic and socio-cultural affiliations have played a role in genetic differentiation of extant Indian populations these markers were analyzed in over 3522 individuals drawn from 54 endogamous populations representing major ethnic and linguistic groups spread across diverse geographic regions of the country (Table 1). Distribution of alleles across populations was evaluated to ascertain presence of group-specific patterns if any. Extent of molecular variance evident among pre-defined groups based on ethnicity, language, geography and socio-cultural hierarchy was evaluated to determine if such classifications were supported genetically. In addition, a model-based clustering algorithm was applied to infer population groups differentiated by their characteristic allele frequencies and to detect presence of cryptic population subdivisions.
Results
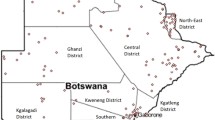
A number of alleles of the different microsatellite loci analyzed were found to be present unique to specific populations with discernable distribution along geographic and ethnic affiliations evident only among few of the populations. Populations like the Gond (a tribal population) from Chattisgarh; Irular, Chakkiliyar, Gounder and Pallar (Australoid populations) from the southern state of Tamil Nadu; showed genetic isolation, evident from the presence of alleles confined within these populations (Figure 1). On the contrary, allele 15.2 of the D3S1358 locus was found to be prevalent among the Gowda and Muslims in the state of Karnataka and allele 18.2 of the FGA locus was present among the Thakur and Kurmi of Uttar Pradesh exhibiting a regional distribution. Sharing of allele 24.2 of the FGA locus was also observed between Lepcha and the Nepali of Sikkim, who share similar ethnic and geographic origins.
Significantly, most frequent alleles were shared among some ethnically and linguistically related populations. The populations of Sikkim, Lai and Lusei of Mizoram that shared Mongol ancestry had a high frequency of allele 12 of the D7S820 locus. Analogous results were obtained for allele 13 of the D5S818 locus, which was in high incidence amongst the Bhutia of Sikkim and Mara of Mizoram. The Indo-Caucasoids, Lingayat of Karnataka; Yadav and Baniya of Bihar, and the geographically proximate Australoid, Kurmi had allele 7 of the Penta E locus in high frequency. Allele 18 of the same locus was present in high frequencies among the Dravidian speaking Australoids, Gowda of Karnataka; Irular of Tamil Nadu as well as among the Indo-European speaking Indo-Caucasoids, Khandayat and Gope of Orissa.
Analysis of molecular variance (Table 2) failed to support the geographic, ethnic, linguistic or socio-cultural grouping of Indian populations suggesting little variation between the different groups. We then employed a cluster-based algorithm to ascertain the extent to which the observed discrete patterns of allele distributions would delineate populations. In order to maintain uniformity of estimated probabilities across runs for a given value of K with large datasets [20], we initially used small K to analyze the 54 populations in this study and then subdivided the dataset into smaller groups to dissect the regional diversity.
In the countrywide dataset, at K = 5, associated with maximum posterior probability (Table 3), individuals displayed partial membership to multiple clusters with some populations exhibiting distinctive identities that did not correspond to geographic, linguistic or ethnic affiliation (Figure 2). Populations such as Thakur and Khatri from Uttar Pradesh and Baniya from Bihar showed similarity with southern populations such as Naikpod Gond and Chenchu from Andhra Pradesh and with a few individuals from Maharashtra and Lepcha of Sikkim. Populations from the northeastern state of Mizoram exhibited a distinct clustering, different from populations of similar ethnicity from Sikkim, while some individuals from Saora and Gope from the eastern state of Orissa shared a similar degree of membership as the Mizoram populations. Of the southern populations, those from Karnataka and Andhra Pradesh were differentiated into two groups with populations from Tamil Nadu exhibiting split membership to both groups.
Bar plot of estimation of the membership coefficient (Q) for each individual of the Indian population grouped on geographic distribution. Each individual is represented by a thin vertical line, which is partitioned into K colored segments that represent the individual's estimated membership fractions in K clusters. Black lines separate individuals of different population groups based on geography. Population groups are labeled below the figure, with their geographical affiliations above it. The figure shown for K = 5 is based on the highest probability run at that K.
At the regional level (Figure 3), amongst northern Indian populations, at K = 5, where the highest posterior probability was associated, Thakur were identified to be distinct from Jat and Uttar Pradesh Kurmi. The Khatri were found substructured with few individuals exhibiting membership similar to the Thakur.
In the East, Bihar Brahmin, Bhumihar, Kayasth, Rajput, Yadav, Bihar Kurmi, Orissa Brahmin, Khandayat, Karan, Juang and Paroja shared similar membership to multiple clusters revealing a common genetic structure. Baniya of Bihar were found similar to two of the northern Indian populations while Gope and few individuals from Saora of Orissa shared similar identities as the populations from Mizoram of northeastern India.
The northeastern populations from Mizoram were identified to be distinct from those of Sikkim. Three clusters were evident with Hmar, Mara, Lai and Lusei of Mizoram all representing one group while Lepcha of Sikkim were distinct representing the second group and the third group comprised Nepali and Bhutia of Sikkim.
In the south, Lingayat, Gowda, Brahmin and Muslim of Karnataka along with Vanniyar, Gounder and Pallar of Tamil Nadu separated from rest of the populations. Irular of Tamil Nadu and Yerukula of Andhra Pradesh presented distinct identities while Chenchu and Naikpod Gond of Andhra Pradesh exhibited similar affinities. Rest of the populations from Tamil Nadu; Chakkiliyar, Paraiyar, Tanjore Kallar and from Andhra Pradesh; Brahmin, Raju, Komati, Kamma Chaudhury, Kapu Naidu, Reddy and Lambadi displayed mixed membership to multiple clusters.
Populations from western and central India showed absence of any distinct grouping with individuals having symmetrical membership across inferred clusters. The above results reveal genetic similarity across populations with a few presenting distinct identities that did not follow traditional groupings of geography, language or ethnicity. Populations from southern India and northeastern India largely exhibited structuring while most Indian populations shared similar membership in multiple clusters.
Discussion
Contemporary molecular studies on Indian populations were focused to uncover the genetic relationship among geographically, linguistically or ethnically related populations [21–25]. Recently, few studies involving a larger number of populations have correlated the genetic relatedness of the populations with linguistic [6] or socio-cultural affinities, [3, 5] though genetic uniformity across populations has also been largely observed [7, 8]. The current study employs microsatellite markers to decipher allele frequency changes that would effectively detect recently isolated populations whose times of divergences were shorter than those detectable by uniparental markers. Distribution of alleles across the microsatellite loci studied among the populations predominantly demonstrates the occurrence of alleles unique only to a few populations (Figure 1). This pattern is probably due to the result of genetic isolation and drift experienced by the populations that follow strict endogamous practices. The distribution of the most frequent allele was in general, uniform across populations suggesting their common origin. Earlier reports have also suggested geographic contiguity favoring gene flow among populations [26]. Although ethnic and geographic propinquity were discernible from the allele distribution patterns across few populations in the current study, no consistent pattern across all populations of any particular group was observed. This was also evident from the analysis of molecular variance that failed to support any grouping; ethnic, linguistic, geographic or socio-cultural in contributing to the extant genetic structure of Indian populations.
The immense diversity within the ethnic and linguistic affiliations of the populations inhabiting India had always been a debatable issue, whether some of them had originated indigenously or were the results of earlier migrations [27–29]. The distinct grouping of the populations of Mizoram (Figure 3) does concord with earlier reports [4, 30] that northeastern India was peopled by migration of Tibeto-Burman speakers from East Asia. However, Tibeto-Burman speaking populations of Sikkim grouped separately and exhibited considerable gene-flow with non-Tibeto-Burman speakers. It is probable that these two regions were peopled by different waves of migration from Southeast and East Asia. Interestingly, eastern Indian populations; Saora and Gope also exhibit similarities to the populations of Mizoram indicating shared genetic ancestry. Though the Lepcha were distinct at the highest-likelihood run for K = 4 (Figure 3), in other runs with lower K, they grouped with the rest of the populations from Nepal (data not shown).
Majority of the Indian populations in general exhibited extensive admixture with each population displaying membership to multiple clusters. Populations such as Khatri, Baniya, Chenchu, Yerukula and Naikpod Gond, however, were substructured. Interestingly, populations comprising the southern Indian region exhibited substructuring with a number of populations clustering into a separate group while the rest were found similar to the general Indian population structure. This group comprising Iyenger Brahmin, Lingayat, Gowda and Muslim from Karnataka and Gounder, Vanniyar and Pallar from Tamil Nadu probably represents those populations that have resisted recent geneflow, and accumulated characteristic allele frequencies because of genetic drift leading to their differentiation from the rest of the populations. In addition, Irular of Tamil Nadu and Yerukula of Andhra Pradesh were found distinctive while Chenchu and Naikpod Gond of Andhra Pradesh grouped together. However, these populations at lower K grouped into clusters similar to those of Tanjore Kallar, Paraiyar and Chakkiliyar of Tamil Nadu and Brahmin, Raju, Komati, Kamma Chaudhury, Kappu Naidu, Kapu Reddy and Lambadi of Andhra Pradesh.
Conclusion
Our analyses failed to reveal any genetic groups that correlate to language, geography, ethnicity or socio-cultural affiliation of populations. Of course, the absence of evidence of structuring of the Indian populations based on ethnic, linguistic, geographic or socio-cultural affiliations may be related to the ascertainment bias of selection of these highly polymorphic forensic microsatellite markers. Future studies employing a large number of microsatellites/SNPs might yield higher resolution to decipher stronger associations between populations. The occurrence of few populations distinct from the general populace suggests genetic drift due to isolation of such populations have resulted in their characteristic allele frequencies. This cryptic population structure would have significant implications in forensic investigations where computations of statistical significance of a DNA match rely on ethnic identities often defined by the country of origin. The existence of substructuring in populations from northeastern and southern India also cautions against broad grouping of populations based on geographic, ethnic or linguistic affiliation that are frequently employed in population genetic studies.
Methods
A total of 3522 consenting individuals from fifty four populations belonging to three major ethnic groups and affiliated to four major language families from across the country were included in this study (Table 1) after approval of the ethical committee of the Central Forensic Science Laboratory. To ensure representations from all groups, information on geographic origin, ethnicity and linguistic affiliation were recorded for every individual sampled.
DNA was extracted either from blood or buccal swabs by standard methods [31]. Amplification was carried out using the Power Plex®16 system (Promega Corporation, Madison, USA) or AmpFl STR ®Identifiler™ PCR Amplification Kit (Applied Biosystems, Foster City, California, USA) that coamplify fifteen microsatellite loci according to manufacturers' specifications. The amplified products were separated on a denaturing 5% polyacrylamide gel using the ABI Prism™ 377 DNA Sequencer (PE Applied Biosystems, Foster City, CA, USA). The genotypes were analyzed with GeneScan® Analysis 3.1, Genotyper® 2.5 (PE Applied Biosystems, Foster City, CA, USA) and PowerTyper™ 16 Macro v2 (Promega Corporation, Madison, USA) softwares.
Analysis of molecular variance, AMOVA [32], was performed by Arlequin 2.0 software using all 15 loci to ascertain which of the attributes; ethnicity, social hierarchy, geographic or linguistic affiliation of the Indian populations contribute maximum to the extant genetic structure. Significance of the AMOVA values was estimated by use of 10,000 permutations.
We used a model-based clustering method for inferring population groups using genotype data consisting of unlinked markers as implemented in Structure 2.1 program [33]. The model assumes there are K populations (where K may be unknown), each of which is characterized by a set of allele frequencies at each locus. Individuals in the sample are assigned probabilistically to populations, or jointly to two or more populations if their genotypes indicate they are admixed. Each run used 100,000 estimation iterations for K = 2 to 8 after a 20,000 burn-in length. Each run was carried out several times to ensure consistency of the results. Posterior probabilities for each K were computed for each set of runs.
References
Singh KS: India's Communities. People of India. National Series. 1998, India: Oxford University Press, IV:
Gadgil M, Joshi NV, Shambu Prasad UV, Manoharan S, Patil S: Peopling of India. The Indian human heritage. Edited by: Balasubramanian D, Rao NA. 1997, Hyderabad, India: Universities Press, 100-129.
Cordaux R, Saha N, Bentley GR, Aunger R, Sirajuddin SM, Stoneking M: Mitochondrial DNA analysis reveals diverse histories of tribal populations from India. Eur J Hum Genet. 2003, 11: 253-264. 10.1038/sj.ejhg.5200949.
Cordaux R, Weiss G, Saha N, Stoneking M: The northeast Indian passageway: A barrier or corridor for human migrations?. Mol Biol Evol. 2004, 21: 1525-1533. 10.1093/molbev/msh151.
Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S, Majumder PP: Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003, 13: 2277-2290. 10.1101/gr.1413403.
Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP: Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet. 2001, 109: 339-350. 10.1007/s004390100577.
Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Golge M, Usanga E, Papiha SS, Cinnioglu C, King R, Cavalli-Sforza L, Underhill PA, Villems R: Thegenetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003, 72: 313-332. 10.1086/346068.
Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, Serk P, Karmin M, Behar DM, Gilbert MTP, Endicott P, Mastana S, Papiha SS, Skorecki K, Torroni A, Villems R: Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genetics. 2004, 5: 26-10.1186/1471-2156-5-26.
Kivisild T, Kaldma K, Metspalu M, Parik J, Papiha SS, Villems R: The place of the Indian mitochondrial DNA variants in the global network of maternal lineages and the peopling of the Old World. Genomic diversity. Edited by: Deka R, Papiha SS. 1999, New York: Kluwer/Academic/Plenum publishers, 135-152.
Deka R, Shriver MD, Yu LM, Heidreich EM, Jin L, Zhong Y, McGarvey ST, Agarwal SS, Bunker CH, Miki T, Hundrieser J, Yin SJ, Raskin S, Barrantes R, Ferrell RE, Chakraborty R: Genetic variation at twentythree microsatellite loci in sixteen human populations. J Genet. 1999, 78: 99-121.
Bosch E, Calafell F, Pérez-Lezaun A, Clarimón J, Comas D, Mateu E, Martínez-Arias R, Morera B, Brakez Z, Akhayat O, Sefiani A, Hariti G, Cambon-Thomsen A, Bertranpetit A: Genetic structure of north-west Africa revealed by STR analysis. Eur J Hum Genet. 2000, 8: 360-366. 10.1038/sj.ejhg.5200464.
Sun G, McGarvey ST, Bayoumi R, Mulligan CJ, Barrantes R, Raskin S, Zhong Y, Akey J, Chakraborty R, Deka R: Global genetic variation at nine short tandem repeat loci and implications on forensic genetics. Eur J Hum Genet. 2003, 11: 39-49. 10.1038/sj.ejhg.5200902.
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW: Genetic structure of human populations. Science. 2002, 298: 2381-2385. 10.1126/science.1078311.
Krithika S, Trivedi R, Kashyap VK, Bharati P, Vasulu TS: Antiquity, geographic contiguity and genetic affinity among Tibeto-Burman populations of India: A microsatellite study. Ann Hum Biol. 2006, 33: 26-42. 10.1080/03014460500424043.
Trivedi R, Sitalaximi T, Banerjee J, Singh A, Sircar PK, Kashyap VK: Molecular insights into the origins of the Shompen, a declining population of the Nicobar archipelago. J Hum Genet. 2006, 51: 217-226. 10.1007/s10038-005-0349-2.
Gaikwad S, Vasulu TS, Kashyap VK: Microsatellite diversity reveals the interplay of language and geography in shaping genetic differentiation of diverse Proto-Australoid populations of west-central India. Am J Phys Anthropol. 2006, 129: 260-267. 10.1002/ajpa.20283.
Sahoo S, Kashyap VK: Influence of language and ancestry on genetic structure of contiguous populations: a microsatellite based study on populations of Orissa. BMC Genet. 2005, 6: 4-10.1186/1471-2156-6-4.
Rajkumar R, Kashyap VK: Genetic structure of four socio-culturally diversified caste populations of southwest India and their affinity with related Indian and global groups. BMC Genet. 2004, 5: 23-10.1186/1471-2156-5-23.
Sitalaximi T, Trivedi R, Kashyap VK: Microsatellite diversity among three endogamous Tamil populations suggests their origin from a separate Dravidian genetic pool. Hum Biol. 2003, 75: 673-685.
Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MA, Hillel J, Maki-Tanila A, Tixier-Boichard M, Vignal A, Wimmers K, Weigend S: Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics. 2001, 159: 699-713.
Bamshad M, Fraley AE, Crawford MH, Cann RL, Busi BR, Naidu JM, Jorde LB: MtDNA variation in caste populations of Andhra Pradesh, India. Hum Biol. 1996, 68: 1-28.
Chakraborty R, Walter H, Mukherjee BN, Malhotra KC, Sauber P, Banerjee S, Roy M: Gene differentiation among ten endogamous groups of West Bengal, India. Am J Phys Anthropol. 1986, 71: 295-309. 10.1002/ajpa.1330710305.
Papiha SS, Mukherjee BN, Chahal MS, Malhotra KC, Roberts DF: Genetic heterogeneity and population structure in north-west India. Ann Hum Biol. 1982, 9: 235-251. 10.1080/03014468200005731.
Dutta R, Kashyap VK: Genetic variation observed at three tetrameric short tandem repeat loci – HUMTHO1, TPOX and CSF1PO in five ethnic population groups of north-eastern India. Am J Hum Biol. 2001, 13: 23-29. 10.1002/1520-6300(200101/02)13:1<23::AID-AJHB1003>3.0.CO;2-R.
Reddy BM, Sun G, Luis JR, Crawford MH, Hemam NS, Deka R: Genomic diversity at thirteen short tandem repeat loci in a sub-structured caste population, Golla, of southern Andhra Pradesh, India. Hum Biol. 2001, 73: 175-190.
Malhotra KC, Vasulu TS: Structure of human populations in India. Human population genetics: A centennial tribute to J.B. Haldane. Edited by: Majumder PP. 1993, New York: Plenum Press, 1: 207-233.
Sarkar SS: Race and race movements in India. The cultural heritage of India. Edited by: Chatterjee SK. 1958, Calcutta, India: The Ramakrishna Mission Institute of Culture, 1: 17-32.
Risley HH: The People of India. 1915, Calcutta, India: Thacker Spink
Pattanayak DP: The language heritage of India. The Indian human heritage. Edited by: Balasubramanian D, Rao NA. 1998, Hyderabad, India: University Press, 95-99.
Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L: Y chromosome haplotypes reveal prehistorical migrations to the Himalayas. Hum Genet. 2000, 107: 582-590. 10.1007/s004390000406.
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning: a laboratory manual. 1989, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, Second
Excoffier L, Smouse P, Quattro J: Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992, 131: 479-491.
Pritchard JK, Stephens M, Donnelly P: Inference of population structure using multilocus genotype data. Genetics. 2000, 155: 945-959.
Tandon M, Trivedi R, Kashyap VK: Genomic diversity at 15 fluorescent labeled short tandem repeat loci in few important populations of state of Uttar Pradesh, India. Forensic Sci Int. 2001, 128: 190-195. 10.1016/S0379-0738(02)00193-7.
Gaikwad S, Kashyap VK: Polymorphism at fifteen hypervariable microsatellite loci in four populations of Maharashtra, India. Forensic Sci Int. 2001, 126: 267-271.
Sarkar N, Kashyap VK: Genetic diversity at two pentanucleotide STR and thirteen tetranucleotide STR loci by multiplex PCR in four predominant groups of central India. Forensic Sci Int. 2002, 128: 196-201. 10.1016/S0379-0738(02)00194-9.
Ashma R, Kashyap VK: Genetic polymorphism at 15 STR loci among three important subpopulation of Bihar, India. Forensic Sci Int. 2002, 130: 58-62. 10.1016/S0379-0738(02)00346-8.
Ashma R, Kashyap VK: Genetic study of fifteen important STR loci among four major ethnic groups of Bihar, India. J Forensic Sci. 2002, 47: 1139-1142.
Guha S, Trivedi R, Kashyap VK: Concordance study on 15 STR loci in three major population of Himalayan state Sikkim. J Forensic Sci. 2002, 47: 1163-1167.
Maity B, Nunga SC, Kashyap VK: Genetic polymorphism revealed by thirteen tetrameric and two pentameric STR loci in four predominant populations of Mizoram. Forensic Sci Int. 2003, 132: 216-222. 10.1016/S0379-0738(02)00436-X.
Sahoo S, Kashyap VK: Allele frequency data for Powerplex16 loci in four major populations of Orissa, India. J Forensic Sci. 2002, 47: 912-915.
Sahoo S, Kashyap VK: Genetic variation at 15 autosomal microsatelite loci in three highly endogamous tribal populations of Orissa, India. Forensic Sci Int. 2002, 130: 189-193. 10.1016/S0379-0738(02)00349-3.
Rajkumar R, Kashyap VK: Distribution of alleles of fifteen STR loci of the Powerplex16 multiplex system in four predominant population groups of South India. Forensic Sci Int. 2002, 126: 175-179.
Sitalaximi T, Trivedi R, Kashyap VK: Autosomal microsatellite profile of three socially diverse ethnic Tamil populations of India. J Forensic Sci. 2003, 48: 211-214.
Sitalaximi T, Trivedi R, Kashyap VK: Genotype profile for thirteen tetranucleotide repeat loci and two pentanucleotide repeat loci in four endogamous Tamil population groups of India. J Forensic Sci. 2002, 47: 1168-1173.
Hima Bindu G, Trivedi R, Kashyap VK: Population genetics of seventeen microsatellite loci in three major groups of Andhra Pradesh, India. Forensic Sci Comm. 2005, 7:
Hima Bindu G, Trivedi R, Kashyap VK: Allele frequency distribution based on 17 STR markers in three major Dravidian linguistic populations of Andhra Pradesh, India. Forensic Sci Int. 2006,
Hima Bindu G, Trivedi R, Kashyap VK: Genotypic polymorphisms at fifteen tetranucleotides and two pentanucleotide repeat loci in four tribal populations of Andhra Pradesh, southern India. J Forensic Sci. 2005, 50: 978-983.
Acknowledgements
This study was supported by a grant under the X five-year plan financial assistance to CFSL, Kolkata. Saurav Guha acknowledges Directorate of Forensic Science for research fellowship. T. Sitalaximi and G. Hima Bindu were recipients of Council of Scientific and Industrial Research (CSIR) fellowships. The contributions of the researchers; Neeta Sarkar, Bhaswar Maity, Sanghamitra Sahoo, Revathi Rajkumar, Sonali Gaikwad, Richa Ashma and Manuj Tandon of CFSL, Kolkata, helped us in the preparation of this communication.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
VKK designed the course of the study and contributed significantly in manuscript preparation. SG carried out statistical analysis and participated in manuscript preparation. TS analyzed the data and drafted the manuscript. GHB performed experiments on Andhra Pradesh samples and participated in manuscript preparation. SEH provided analytical inputs for manuscript preparation. RT provided critical information for data processing and manuscript preparation.
All authors read and approved the final manuscript.
Saurav Guha, T Sitalaximi contributed equally to this work.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kashyap, V., Guha, S., Sitalaximi, T. et al. Genetic structure of Indian populations based on fifteen autosomal microsatellite loci. BMC Genet 7, 28 (2006). https://doi.org/10.1186/1471-2156-7-28
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-7-28