Abstract
Background
Section Calochroi is one of the most species-rich lineages in the genus Cortinarius (Agaricales, Basidiomycota) and is widely distributed across boreo-nemoral areas, with some extensions into meridional zones. Previous phylogenetic studies of Calochroi (incl. section Fulvi) have been geographically restricted; therefore, phylogenetic and biogeographic relationships within this lineage at a global scale have been largely unknown. In this study, we obtained DNA sequences from a nearly complete taxon sampling of known species from Europe, Central America and North America. We inferred intra- and interspecific phylogenetic relationships as well as major morphological evolutionary trends within section Calochroi based on 576 ITS sequences, 230 ITS + 5.8S + D1/D2 sequences, and a combined dataset of ITS + 5.8S + D1/D2 and RPB1 sequences of a representative subsampling of 58 species.
Results
More than 100 species were identified by integrating DNA sequences with morphological, macrochemical and ecological data. Cortinarius section Calochroi was consistently resolved with high branch support into at least seven major lineages: Calochroi, Caroviolacei, Dibaphi, Elegantiores, Napi, Pseudoglaucopodes and Splendentes; whereas Rufoolivacei and Sulfurini appeared polyphyletic. A close relationship between Dibaphi, Elegantiores, Napi and Splendentes was consistently supported. Combinations of specific morphological, pigmentation and molecular characters appear useful in circumscribing clades.
Conclusion
Our analyses demonstrate that Calochroi is an exclusively northern hemispheric lineage, where species follow their host trees throughout their natural ranges within and across continents. Results of this study contribute substantially to defining European species in this group and will help to either identify or to name new species occurring across the northern hemisphere. Major groupings are in partial agreement with earlier morphology-based and molecular phylogenetic hypotheses, but some relationships were unexpected, based on external morphology. In such cases, their true affinities appear to have been obscured by the repeated appearance of similar features among distantly related species. Therefore, further taxonomic studies are needed to evaluate the consistency of species concepts and interpretations of morphological features in a more global context. Reconstruction of ancestral states yielded two major evolutionary trends within section Calochroi: (1) the development of bright pigments evolved independently multiple times, and (2) the evolution of abruptly marginate to flattened stipe bulbs represents an autapomorphy of the Calochroi clade.
Similar content being viewed by others
Background
Calochroi is one of the most diverse lineages within the Cortinarius radiation, with estimates of between 90 and 170 species recognised so far from Europe [1], which probably represents only a portion of the taxa on a global scale. All members of this section are obligate root symbionts (ectomycorrhizal); therefore, the presence of appropriate plant hosts as well as specific edaphic factors play a crucial role in their geographical distributions. In general, the highest interspecific diversity roughly matches the distribution of coniferous (Abies, Picea) and broadleaf (Fagus, Quercus, Lithocarpus) trees across boreo-nemoral areas. Often, shared ecological specialisations makes it possible to find basidiomes of different species associated with an individual host tree on the same site. Section Calochroi, as circumscribed here, comprises species producing conspicuous basidiomes with a rounded to adruptly bulbous stipe base, a pileus with a well-developed gelatinous layer, and the presence of pigments which often change colour with the application of KOH. Most species produce basidiomes after the first autumn rains; however, in North America, certain species with a persistent veil, short stipe and subemerged basidiomes occur in spring. In oak forests of Costa Rica, a number of species occur during the rainy season in June, and sometimes later in the season as well. Traditionally, species concepts have been largely based on macromorphological traits, colour changes with KOH on the basidiomes (usually the pileus, and the stipe surface and flesh), and ecological specialisations [2–5]. However, some of these traits are heavily labile and/or affected by homoplasy, and as a result, the taxonomy of this group is extremely difficult, and much controversy exists in the delimitation and recognition of species. Because of their abundance in many forest ecosystems, and the spectacular appearance and colouration of the basidiomes, calochroid species have been the focus of numerous taxonomic studies [see, for example [2–6]], as well as the chemical structures of their pigments [7–11]. Most recently, considerable progress has been made in reconstructing phylogenetic relationships among European calochroid species through analyses of morphological features and DNA sequences. Peintner et al. [12] studied the phylogeny of sequestrate taxa related to Cortinarius based on ITS sequences and distinguished a well-supported lineage named Phlegmacium IV containing a small number of European and North American calochroid species. Garnica et al. [13] analysed the phylogenetic relationships of 54 European Phlegmacium species using ITS and D1/D2 sequences of the nuclear gene coding for the ribosomal large subunit (nucLSU rDNA) and found that members of the section Calochroi are distributed into two closely related lineages that can be assigned to Calochroi and Fulvi. Peintner et al. [14] and Garnica et al. [15] used a more extensive sampling of taxa across the genus Cortinarius and supported the monophyly of Calochroi. At least five subclades were distinguished by Peintner et al. [15]. Frøslev et al. [16] used 54 European phlegmacioid taxa and compared phylogenies resulting from RPB1, RPB2 and ITS data as single and combined data sets, and accepted three subclades named /Calochroid, /Rufoolivacei and /Fulvi. These authors showed that certain morphological features, such as a pileipellis structure (pileipellis simplex), a very broad marginate bulbous stipe base and coarse, net-like basidiospore ornamentation have evolved independently several times within the genus Cortinarius. Frøslev et al. [1] evaluated species delimitations using a total of 421 ITS sequences (including sequences from 53 type specimens) and recognised /Arcuatorum, /Calochroid, /Fulvi, /Rufoolivacei and /Sulfurinus as major subclades within section Calochroi in Europe. In several cases, these authors found that sequences obtained from collections of the same species from different geographical areas showed very little variation. Nevertheless, little is known about intra- and interspecific patterns of distribution and phylogenetic relationships within section Calochroi at a global scale. Morphology-based taxonomic studies have recorded Cortinarius species displaying calochroid and fulvoid features from several coniferous and frondose forests in North America [17, 18] and Nothofagus forests in South America [19, 20]. Furthermore, in light of similar morphological features of the basidiomes some calochroid species have been reported with European and North American disjunct distribution patterns [see [21–25]]. Therefore, to gain a more complete understanding of intra- and interspecific patterns of distribution and phylogenetic relationships at a global scale, we have attempted to use a more extensive sampling of calochroid and fulvoid Phlegmacium species, including European, North American, Tasmanian, New Zealand, and Central and South American collections.
Here, we present phylogenetic hypotheses including nearly all of the known species of Calochroi sensu M.M. Moser using nuclear DNA sequence data from the internal transcribed spacers (ITS1 and ITS2, including the 5.8S), the D1/D2 regions of the nucLSU rDNA and from the gene coding for domains A-C of RPB1 from a representative subsample. We use these phylogenies to address the following main objectives. First, we compared our own sequences with available ITS sequences from GenBank and UNITE databases to establish congruence and uniformity of species concepts for European calochroid species. Second, we estimated intra- and interspecific sequence divergences of closely related taxa by comparing the internal transcribed spacers (ITS1, ITS2) alone or including D1/D2 regions. Third, we inferred phylogenetic relationships and major morphological evolutionary trends within the section Calochroi by integrating molecular phylogenetic analyses of single and combined DNA sequence datasets with morphological, chemical and ecological features. This study provides the first insights into the geographical structure of species and populations occurring in parts of Central America, North America and Europe within a diverse and complex lineage of ectomycorrhizal fungi.
Results
Phylogenetic delimitation of the section Calochroi
The section Calochroi appeared as a well-supported monophyletic clade within the genus Cortinarius in our preliminary neighbor-joining analyses (tree not shown). These analyses also showed that C. inexpectatus from Europe, and C. rhodophyllus, C. chlorophanus, C. stephanopus and C. vaginatus from South America, which are currently classified in Calochroi and Fulvi, cluster outside of the section Calochroi as conceived here. Thus, all species assignable to section Calochroi (incl. section Fulvi) in this study are from the northern hemisphere. A total of 576 sequences including ITS and 5.8S, 230 sequences comprising ITS, 5.8S and nucLSU D1/D2 rDNA, and 56 RPB1 sequences including the conserved domains A-C were analysed in three separate datasets: dataset 1 including ITS and 5.8S sequences (Additional file 1); dataset 2 including ITS, 5.8S and D1/D2 sequences (Figures 1 and 2); dataset 3 including combined ITS, 5.8S and D1/D2, and RPB1 A-C sequences (Figure 3). The final sampling covered c. 110 species with geographical distributions across Europe, Central America and North America.
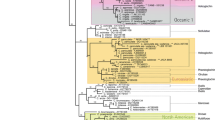
Phylogenetic relationships within Cortinarius section Calochroi as derived from ML analysis of ITS-5.8S + D1/D2 rDNA sequences. Numbers above branches are bootstrap values (values < 50% not shown) from 1000 replicates. North American Calochroi collections are printed in blue. Host tree genera are coded as follows: Betulaceae, Bet = Betula, Car = Carpinus, Cor = Corylus, Ericaceae, Arb = Arbutus; Fagaceae, Fag = Fagus, Que = Quercus, Lit = Lithocarpus; Pinaceae, Abi = Abies, Lar = Larix, Pic = Picea, Pin = Pinus, Pse = Pseudotsuga, Tsu = Tsuga; Salicaceae, Pop = Populus, Sal = Salix. Names in boldface correspond to sequences also used in the combined phylogenetic analysis (Figure 3); type collections are indicated by (T).
Phylogenetic relationships within Cortinarius section Calochroi as derived from ML analysis of ITS-5.8S + D1/D2 rDNA sequences. Numbers above branches are bootstrap values (values < 50% not shown) from 1000 replicates. North American Calochroi collections are printed in blue. Host tree genera are coded as follows: Betulaceae, Bet = Betula, Car = Carpinus, Cor = Corylus, Ericaceae, Arb = Arbutus; Fagaceae, Fag = Fagus, Que = Quercus, Lit = Lithocarpus; Pinaceae, Abi = Abies, Lar = Larix, Pic = Picea, Pin = Pinus, Pse = Pseudotsuga, Tsu = Tsuga; Salicaceae, Pop = Populus, Sal = Salix. Names in boldface correspond to sequences also used in the combined phylogenetic analysis (Figure 3); type collections are indicated by (T).
Phylogeny of the section Calochroi as derived from ML analysis of an alignment of ITS-5.8S + D1/D2 (except for C. dalecarlicus ) + RPB1 A-C sequences. Numbers above nodes are ML bootstrap values, numbers below nodes are MP bootstrap values (values < 50% not shown) from 1000 replicates, respectively. Distributions of basidiome pigments were retrieved from [7, 9, 10, 64] and are abbreviated as follows. Flavomannin pigments: ATR = atrovirin, FDM = flavomannin-6-6'dimethylether, 4-OH-FDM = 4-OH-flavomannin-6,6'-dimethylether, FLA = flavomannin, FTM = flavomannin-6,6',8-trimethylether; phlegmacin pigments: LRU = leucorufoolivacins, PHL = phlegmacin, PME B2 = phlegmacin-8'-methylether B2; triterpenoid pigments: S = sodagnitins A-F. Note that FDM is a common pigment found also in other groups in Cortinarius. A non-coding 8 bp insertion is absent in all species of the Calochroi clade and also in C. elegantissimus and C. rapaceotomentosus, but is present in the remaining species. The alignment shown includes all species of the Calochroi clade contained in the combined dataset 3 (framed box), and corresponds to positions 769–777 of intron 2 of the RPB1 sequence. Ancestral character states were reconstructed using maximum parsimony (DELTRAN strategy). Green branches indicate taxa of bright colours; taxa with abruptly marginate to flattened stipe bulbs are indicated by bold lines. Dashed branches indicate taxa where bright basidiome colours or marginate to flattened stipe bulbs are not continuously present in the populations. C. elegantior and C. humolens photos, credit: F. Röger.
Intraspecific variation and relationships between closely related taxa as inferred from ITS-5.8S and ITS-5.8S-D1/D2 sequences
ITS-5.8S sequences
Comparative intraspecific ITS-5.8S sequence analyses between our collections and those of Frøslev et al. [1, 16] revealed identical sequences in C. albertii, C. arcuatorum, C. barbaricus, C. cupreorufus, C. elegantior, C. langeorum, C. meinhardii, C. napus, C. odoratus, C. osmophorus, C. pseudoglaucopus, C. selandicus, C. subgracilis, C. sublilacinopes, C. sulfurinus and C. olivascentium; whereas, for the remaining species, an intraspecific genetic variation of up to 6 bp was observed (Additional file 1). However, it should be noted that within the same species, GenBank sequences have deviating sites across relatively conservative portions of ITS1 and ITS2 regions compared to the sequences obtained in this study. In general, the species concepts used in the present work agree with those applied by Frøslev et al. [1, 16] and Peintner et al. [12, 14, 26] (see Additional file 1). However, for nomenclatural reasons, we prefer to use the following names: C. cedretorum var. cedretorum (= C. cedretorum), C. eufulmineus (= C. quercus-ilicis), C. majusculus (= C. alcalinophilus), C. alnobetulae (= C. moseri) and C. olivascentium (= C. xanthochlorus).
ITS-5.8S-D1/D2 sequences
Many collections of the same species from different, but geographically restricted, localities (i.e. from populations in Central America, North America or Europe) shared identical sequences. Generally, sequence variation within different collections of the same species was caused by obvious polymorphic sites and/or single nucleotide indels distributed mainly across the ITS1 and ITS2 regions. There was a 0.1% sequence divergence between C. albobrunnoides var. violaceovelatus and C. napus; C. albobrunnoides var. violaceovelatus and C. subpurpureophyllus var. sulphureovelatus; and between different collections of C. aureocalceolatus, C. aureofulvus and Cortinarius sp. USA 7. Collections of C. aurantiorufus and C. haasii, C. elotoides and C. pseudoglaucopus, and C. flavaurora and C. pseudoglaucopus showed a 0.2% sequence divergence. There was a 0.2% sequence divergence within collections of C. dibaphus and C. lilacinovelatus, respectively. New World collections of C. arcuatorum clustered within three distinctive lineages designated as groups A-C in Figures 1 and 2: group A comprised the collections JFA 12037, JFA 12039 and JFA 12061 from Costa Rica with identical sequences; group B included the collection JFA 11893 from Mendocino, California, occupying an isolated position; and group C was composed of the collections JFA 11765, JFA 11766 and JFA 11803 from the Smith River drainage, Del Norte County, California, with identical DNA sequences. There was a 1.6% sequence divergence between group A and B, while group A and C differed by 1.5%, and group B and C differed by 2.1%. There were between 34–39 bp differences (approximately 2.5–2.8% sequence divergence) between New World and European collections of C. arcuatorum. New World and European Elegantiores collections were distributed into three monophyletic subgroups labeled as D-F in Figures 1 and 2: group D consists of C. elegantior var. americanus JFA 11452 from Wyoming with an isolated position; group E comprises the C. elegantior collections JFA 12438 from Wyoming and JFA 11693 from Oregon with a sequence divergence of 0.3%; and group F includes only European collections with identical sequences. Sequence divergences ranged from 0.2% to 0.5% among the groups F and D, and 0.6% to 0.7% between the groups D-E and D. Sequences from the following specimens were identical: C. albobrunnoides JFA 12426, C. napus and C. subpurpureophyllus var. sulphureovelatus; C. claroflavus and C. xanthophyllus; C. elotoides (IB 19870060 Holotypus), Cortinarius sp. JFA 11618 and JFA 11619 and C. flavaurora; C. elotus IB 1999/0192 and C. pseudoglaucopus; and C. olivellus and C. humolens. Collections of C. aurora, C. corrosus (TUB 012705, TUB 012706) and C. rufoolivaceus differed by single nucleotide indels in the ITS1 region, the collections TUB 012714 and TUB 012715 of C. haasii differed by a single nucleotide indel in the ITS2 region. Collections of Cortinarius sp. JFA 11833, JFA 11845 and C. prasinus had identical sequences and inclusively shared a polymorphism at the same position (Y = C/T).
Phylogenetic reconstruction inferred from ITS, 5.8S and D1/D2 rDNA
Our phylogenetic analyses of dataset 2 (Figures 1 and 2) supported the monophyly of the clades Calochroi, Caroviolacei, Dibaphi, Elegantiores, Splendentes and Pseudoglaucopodes. ML bootstrap support for the monophyly of Napi, Rufoolivacei and Sulfurini was below 50%; these groups appear polyphyletic in the best tree found in heuristic ML analysis (Figures 1 and 2). Cortinarius spp. USA 8, USA 13 and USA 17, C. aureocalceolatus, C. aureofulvus, C. ionodactylus, C. langeorum, C. natalis, C. osmophorus, C. rapaceotomentosus and C suaveolens could not be assigned to particular clades. More than 35% of the species analysed were nested within the Calochroi clade; the remaining taxa were distributed over various smaller lineages. The partially hypogeous taxa clustered in the clades Calochroi and Rufoolivacei, respectively.
Phylogenetic relationships inferred from ITS-5.8S-D1/D2 sequences combined with RPB1 domains A-C
Maximum likelihood and maximum parsimony analyses of dataset 3 revealed at least seven major lineages in the section Calochroi (Figure 3). The monophyly of Calochroi, Caroviolacei, Dibaphi, Elegantiores, Napi, Pseudoglaucopodes and Splendentes was supported in our maximum likelihood sequence analysis. ML and MP bootstrap support for Rufoolivacei and Sulfurini and MP bootstrap support for the monophyly of Napi was below 50%. The clades Dibaphi, Elegantiores, Napi, and Splendentes formed a well-supported monophyletic group in our maximum likelihood analysis together with C. aureocalceolatus and C. natalis, which appeared as a sister group to the remaining taxa of section Calochroi. Cortinarius citrinus, C. langeorum, C. osmophorus, C. rapaceotomentosus and C. suaveolens had isolated positions within the section Calochroi in our maximum likelihood analysis. In the maximum parsimony bootstrap analysis, C. rapaceotomentosus was supported as a sister taxon to the Calochroi clade with 70%, and C. natalis, C. splendens and C. atrovirens formed a well-supported clade (95% MP bootstrap).
Reconstruction of ancestral states
Maximum parsimony reconstruction of ancestral states on the tree with the highest likelihood found for dataset 3 suggests that the bright pigmentation of the basidiomes and an abruptly marginate to flattened stipe bulb base evolved independently several times (Figure 3). Our results indicate that an abruptly marginate to flattened stipe bulb base is an autapomorphy for the Calochroi clade. The acquisition of bright flesh pigments appears as a synapomorphy for Rufoolivacei, Pseudoglaucopodes and Sulfurini and probably evolved independently in C. citrinus and C. fulvocitrinus as well as in Splendentes and Elegantiores.
Characterisation of macro- and microscopical features
Macroscopical features, including habit, basidiome coloration and veil coloration, macrochemical reactions with 40% KOH (see Figure 4), microscopical features of the pileipellis structure and hyphal coloration in 3% KOH, and basidiospore morphology (see Figure 5), are documented and compared for the Calochroi (see Additional file 2).
Colour change reactions with 40% KOH on pileus surface and mycelia at the base of the stipe bulb in fresh and dried calochroid specimens. a. Intense pink on mycelia (fresh material) in C. insignibulbus TUB 011657 (credit: G. Smith-Stohn). b. Pink on pileus surface in C. sodagnitus TUB 011428. c. Intense pink colour on pileus surface in C. albertii TUB 011850. d. Pink on mycelia in C. sodagnitus TUB 011428. e. Vinaceous on pileus surface in C. suaveolens TUB 011876. f. Vinaceous on mycelia in C. eufulmineus TUB 011876. g. Red-brown on pileus surface in C. haasii TUB 011858. h. Black on pileus surface in C. rufoolivaceus TUB 012739. i. Black on mycelia in C. rufoolivaceus TUB 012739.
Discussion
Although there have been previous molecular phylogenetic studies of Calochroi species [1, 13, 15, 16] the present study is the first to include an extended, nearly complete sampling of the known species of this Cortinarius section across its natural range of distribution in Europe, Central America and North America (sampling does not include extensive areas of eastern North America). In general, our morphological and phylogenetic concepts for European calochroid species fit with those applied by Frøslev et al. [1, 16] and Peintner et al. [12, 14, 26] (see Additional file 1). In the majority of cases, the evaluation of ranges in species variation is based on morphological and molecular data obtained from multiple collections and analysed with ITS sequences of 37 type specimens. These findings represent considerable taxonomic progress within a taxonomically complex group, and should make it much easier to either identify specimens or to name new species occurring across Europe, Central America and North America.
Phylogenetic analyses of the three final molecular datasets resulted in congruent and complementary trees (Figures 1, 2) and 3, but backbone resolution was higher in the trees inferred from the combined dataset 3 (Figure 3). Seven major lineages were supported within the section Calochroi, which occur throughout Europe, Central America and North America, and can be mostly circumscribed by a combination of features such as colour and colouration patterns, pigment content, colour reaction with KOH and similar shared indels in the RPB1 gene. In the following discussions, we focus on the most significant features circumscribing the major clades, and their degree of interspecific variation, as well as on specific geographical patterns of distribution. For each of the lineages discussed below, we give the branch support (ML/MP bootstrap values) obtained in the phylogenetic analyses of the combined dataset 3 (see Figure 3).
Calochroiclade (99/98)
This clade is a species-rich lineage within the section Calochroi containing over thirty European and ten North American taxa (Figures 1 and 2). A combination of chemical and morphological features seems to support the monophyly of this clade, including similar colouration patterns of the context, stipe and pileus, KOH reaction, stipe shape and ecological requirements, such as edaphic and host specialisations as well as a shared 8 bp length indel in the intron 2 portion of the RPB1 gene (Figure 3). The indel is also present in two species (in C. elegantissimus and C. rapaceotomentosus) that were placed close to this lineage, but without significant support. Therefore, it is plausible that this indel represents an autapomorphy for an extended Calochroi clade. Within Calochroi as currently defined, there are some spectacular examples of similar basidiomes among distantly related species, which may be explained by similar evolutionary scenarios or by shared plesiomorphies. Thus, morphological features alone are often found to be confusing in delimiting and/or separating species, whereas DNA sequence analysis reveals the existence of a major number of still undescribed species that were overlooked in studies based solely on morphology. However, additional collections should be sequenced and thoroughly studied with respect to the degree of intraspecific variation of macroscopical, microscopical, chemical and ecological features before a formal description of any new species is completed. The fact that poor resolution within the Calochroi clade is a feature of both morphological and molecular analyses suggests that this lineage may have differentiated in a short period of time. Therefore, additional molecular markers may be needed to provide sufficient resolution within this lineage. Most species are rare, with limited distributions within boreo-nemoral areas, mainly growing on calcareous soils and associated with frondose trees. A number of species in this clade are strictly associated with frondose trees belonging to the Betulaceae (Corylus, Carpinus) and Fagaceae (Castanea, Fagus, Quercus), whereas relatively few species are known to be associated with coniferous trees of the Pinaceae (Abies, Picea, Pinus, Pseudotsuga) (see Figures 1 and 2).
Caroviolaceiclade (100/100)
This clade includes the European species C. caroviolaceus, C. saporatus and C. rapaceoides with no close relatives known to date in North America. Phylogenetic results from ITS + RPB1 + RPB2 sequences obtained by Frøslev et al. [1] placed these species in a weakly supported Rufoolivacei clade (see discussion of the Rufoolivacei clade). Moser [19] placed C. caroviolaceus and C. saporatus in separate stirpes within the subsection Multiformes. Members of this lineage are characterised by relatively large basidiomes with a distinctly rounded stipe bulb, pale context, amygdaliform basidiospores and a lack of colouration change with the application of KOH. The taxon C. caroviolaceus is exclusively associated with frondose trees belonging to the genera Carpinus, Fagus and Quercus, and C. saporatus occurs with Fagus, Quercus and Tilia in Europe.
Dibaphiclade (99/98)
This clade contains European, Central American and North American populations of C. arcuatorum and the European species C. dibaphus. Members of the Dibaphi clade share similar colour changes with KOH, pigment content and colour patterns of the basidiomes. Chromogenic triterpenoid pigments (sodagnitins) in C. arcuatorum [10] and probably in other Dibaphi may be significant chemical markers for this group. Sequences of New World populations of C. arcuatorum differ significantly from their European counterparts, and they cluster into three closely related monophyletic groups, which roughly correspond to their respective geographical ranges of distribution. Despite relatively high sequence divergences between New World and European populations of C. arcuatorum, little phenotypic variation in morphological features of the basidiomes and colouration between these collections was observed. A certain range of variability of the basidiospore size was observed between collections of C. arcuatorum from different parts of North America and Europe [23]. However, the area sampled for this study represents only part of the modern pattern of geographical distribution of this species complex. Here, it is necessary to mention that an ITS sequence (AY033120; published by Peintner et al. [14]) of C. arcuatorum from Wyoming was identical to sequences of our European collections of this taxon. Populations of C. arcuatorum are associated with tree species of the genera Fagus and Quercus in Europe, Quercus spp., in Central America, and Lithocarpus, Quercus, Pseudotsuga, and Tsuga in North America. Our study suggests that some populations of the C. arcuatorum complex have evolved very specialised habitat requirements with allopatric diversification, probably involving co-migration along a cline with their associated host trees, specifically with Douglas fir, hemlock, stone oak and oak. Future studies may show that such populations already represent separated species. Cortinarius dibaphus seems to be less host specific growing with coniferous (Abies alba) and frondose (Fagus sylvatica, Quercus spp.) trees in Europe [2].
Elegantioresclade (99/71)
This clade includes European (C. elegantior, C. eufulmineus, C. glaucescens, C. majusculus, C. murellensis, C. olearioides, C. parafulmineus, C. splendificus) and North American (C. elegantior JFA 11452 and JFA 11693, C. elegantior var.americanus JFA 12438) taxa. A phylogenetic analysis from ITS sequences published recently by Garnica et al. [25] revealed three subclades among European Elegantiores. Members of the Elegantiores clade share similar colouration of the pileus, lamellae, stipe and context, and KOH reaction. The yellow-greenish colours of the basidiomes of C. eufulmineus, C. elegantior, C. olearioides and C. splendificus correspond to flavomannin-di- and trimethylether pigments [7, 9, 11]. Populations from both sides of the distribution range of C. elegantior share remarkable morphological and colouration similarities. However, high levels of genetic diversity were found in the North American populations of C. elegantior as evidenced by various unique substitutions and indels (see Figures 1 and 2). Our phylogenetic analysis indicates that C. elegantior var.americanus represents a species separated from C. elegantior rather than a variety of it, which macroscopically differs mainly in the colour of the veil [18]. The genetic diversity found in the C. elegantior complex might suggest that populations evolved with their associated host trees. Cortinarius elegantior and C. parafulmineus are restricted to coniferous trees (Picea, Pinus, Tsuga) and the remaining taxa are associated with frondose trees of the genera Carpinus, Fagus and Quercus occurring across the boreo-nemoral areas in North America and Europe.
Napiclade (85/<50)
This clade as conceived here contains European (C. aureopulverulentus and C. napus) and Northern American (C. albobrunnoides, C. albobrunnoides var. violaceovelatus, C. aurora, and C. subpurpureophyllus var. sulphureovelatus) taxa. Members in this clade have been classified in separate sections in the subgenus Phlegmacium: C. aurora was placed in the section Aurantiovelati [18]; C. aureopulverulentus in the section Caerulescentes [4]; C. albobrunnoides, C. albobrunnoides var. violaceovelatus and C. napus in the section Phlegmacium [see, for example [4, 18, 26]], and C. subpurpureophyllus var. sulphureovelatus in the section Calochroi [17]. The groupings derived from our molecular analyses are largely in agreement with similar colouration patterns of the pileus, lamellae, context and stipe, a lack of KOH reaction (except for C. aurora and C. aureopulverulentus) and basidiospore morphology in these taxa. The close placement of C. aurora and C. aureopulverulentus in our analysis (Figure 3) fits with similarities of the veil colouration. Moser and Peintner [26] analysed the phylogenetic relationships in the Cortinarius aureopulverulentus group. These authors demonstrated the paraphyly of the section Thalliophili [5], but the precise phylogenetic placement of C. aureopulverulentus was not resolved with confidence. On the basis of differences in veil colouration, Moser and Ammirati [17, 27] distinguished the varieties C. albobrunnoides var. violaceovelatus and C. subpurpureophyllus var. sulphureovelatus. However, low sequence divergences (0.1%) among the European C. napus and the North American C. albobrunnoides, C. albobrunnoides var. violaceovelatus and C. subpurpureophyllus var. sulphureovelatus suggest that these taxa are conspecific or perhaps the ITS region is too conservative to discriminate among these species. The first alternative could suggest a surprising intraspecific lability of veil colouration. However, to clarify aspects on species delimitations, the analysis of additional fungal collections and DNA markers is necessary. Members of the Napi clade seem to have diversified together with coniferous trees of the genera Abies, Picea, Pinus, Pseudotsuga and Tsuga throughout boreo-nemoral areas in North America and Europe.
Pseudoglaucopodesclade (100/100)
This lineage comprises European (C. elotus, C. olivellus, C. pseudoglaucopus) and North American (Cortinarius sp. USA 14 and 15, C. elotoides, C. flavaurora) taxa, congruent with current classification systems [see [4, 18, 19]]. Members of this clade share similar colouration patterns of the basidiomes, basidiospore morphology and lack of KOH reaction. This study indicates the need to re-evaluate the specific status of C. elotoides and C. flavaurora (Figures 1 and 2). Members of the Pseudoglaucopodes clade seem to have co-evolved strictly with coniferous trees of the genera Picea and Pseudotsuga, and these associations are maintained through subalpine regions in the northern hemisphere (North America and Europe).
Rufoolivacei(<50/<50)
According to the present study, the monophyly of Rufoolivacei is questionable. This taxon includes the European species C. cedretorum var. suberetorum, C. cupreorufus, C. elegantissimus, C. prasinus, C. odorifer, C. olivascentium, C. rufoolivaceus, C. xanthophyllus, the North American collections Cortinarius sp. USA 7, USA 9, USA 10 and USA 12, and C. saxamontanus. Species in this group display several similarities including pigment content, KOH reaction, and colour patterns of the basidiomes. Oertel [9] and Steglich and Oertel [11] detected pigments of the phlegmacin group in the basidiomes of C. elegantissimus, C. cedretorum, and C. prasinus and of the rufoolivacin group in C. cupreorufus, C. odorifer, C. rufoolivaceus, and C. xanthophyllus, respectively. Interestingly, in the analysis of the combined dataset a cluster composed of European (C. cedretorum var. suberetorum, C. cupreorufus, C. prasinus, C. rufoolivaceus, C. xanthophyllus) and North American (C. saxamontanus) species received an ML bootstrap support of 86% (Figure 3). Rufoolivacei, as conceived here, differs substantially from the results of Frøslev et al. [1, 16]. Our present study suggests that Caroviolacei plus Pseudoglaucopodes may represent a group distinct from Rufoolivacei (Figures 1 and 2). A combination of traits (colour of the basidiomes and KOH reaction) supports this hypothesis. Most Rufoolivacei are associated with frondose trees (Fagus, Lithocarpus, Quercus) and only few species occur with coniferous trees (Picea, Pinus, Pseudotsuga) across the boreo-nemoral regions in North America and Europe.
Splendentesclade (77/88)
Splendentes includes relatively common European species (C. atrovirens f.atrovirens, C. atrovirens f.ionochlorus, C. meinhardii, C. odoratus, C. splendens) and also C. viridirubescens from North America. The phylogenetic placement of C. viridirubescens is consistent with the traditionally noted morphology, KOH reaction and colouration of the basidiomes [23]. This monophyletic group is in good agreement with similarities in pigment content, KOH reaction and colouration patterns of the basidiomes [15]. Ecologically, C. atrovirens f.ionochlorus, C. odoratus and C. splendens grow strictly associated with Fagus sylvatica; C. meinhardii with Picea abies; whereas C. atrovirens f.atrovirens and C. viridirubescens are less specific, growing with both frondose (Arbutus, Fagus, Quercus, Lithocarpus) and/or coniferous (Abies, Picea, Pinus, Pseudotsuga) trees.
Sulfurini group (<50/<50)
Concerning Sulfurini, which includes European (C. flavovirens and C. sulfurinus) and North American (C. guttatus) species, our phylogenetic hypotheses are incongruent. On the one hand, C. flavovirens and C. sulfurinus appear as sister taxa in the best tree found in the ML analysis of the combined dataset 3 (though with bootstrap support below 50%; Figure 3); on the other hand C. flavovirens was separated from the joined C. sulfurinus and C. guttatus with a bootstrap support of 92% in the analysis of dataset 2 (Figures 1 and 2). Frøslev et al. [1] recognised Sulfurini (excluding C. flavovirens); as a monophyletic group. Members of the Sulfurini group share several similarities such as KOH reaction, pigment contents and colouration patterns of the basidiomes. The yellow-greenish colours of the basidiomes correspond to phlegmacin-8'-methylether B2 (C. flavovirens and C. sulfurinus) pigments [9, 11]. Cortinarius flavovirens grows associated with frondose trees (Fagus, Quercus), and North American C. guttatus and C. sulfurinus are associated with coniferous trees (Abies, Picea, Pinus) in Europe.
Species with unresolved phylogenetic positions
The isolated positions of C. aureocalceolatus, C. aureofulvus, C. citrinus, C. langeorum, C. natalis, C. osmophorus, C. rapaceotomentosus and C. suaveolens obtained in our maximum likelihood analysis are in part consistent with previous findings by Frøslev et al. [1, 16]. Cortinarius langeorum and C. suaveolens, which were nested within the Rufoolivacei clade in the molecular analyses by these authors, were not included in Rufoolivacei, but appeared on isolated branches close to this group in our trees (Figures 1, 2 and 3). A close relationship between C. rapaceotomentosus and the Calochroi clade was supported in our MP bootstrap analysis. This hypothesis is consistent with a similar shared indel in the RPB1 sequences (Figure 3). The high MP bootstrap support for a position of C. natalis close to Splendentes is consistent with similar pigments of the basidiomes. Cortinarius aureofulvus has a wide distribution in subalpine forests across the northern hemisphere, where North American populations grow associated with Abies lasiocarpa, Picea engelmannii, P. pungens and Pseudotsuga menziesii [18], while their European counterparts occur associated with Abies alba, Picea abies and Pinus sylvestris.
Multiple origins of sequestrate species in the section Calochroi
Thiers [28] and Thiers & Smith [29] suggested that the sequestrate forms in the genus Cortinarius represent morphological adaptations to dry environmental conditions. Molecular analyses indicated that sequestrate basidiomes forms have evolved independently several times within several clades in Cortinarius [12]. The same was concluded from the geographical distribution of modern populations and glacial pluvial events in the Great Basin of North America [30]. Sequestrate species appear during spring in subalpine to alpine sites across the western USA [29, 30]. Our phylogenetic analyses suggest that sequestrate forms have evolved independently at least twice in the Calochroi clade (C. verrucisporus and Cortinarius sp. USA 16) and one time within Rufoolivacei (C. saxamontanus). Furthermore, this study emphasises the need to study the morphological and molecular data of additional specimens to clarify species concepts in these heavy veiled taxa.
Circumscribing the major lineages within section Calochroi
Our concept of section Calochroi comprises species that have been classified in the sections Calochroi and Fulvi [2, 31]; in the sections Coerulescentes, Fulgentes, Glaucopodes, Laeticolores, Multiformes and Thalliophili [5, 32]; and in the sections Calochroi, Coerulescentes, Fulvi and Laeticolores [4], or Calochroi, Coerulescentes, Fulvi and Scauri [19]. Our phylogenetic results differ from those of Frøslev et al. [1, 16] mainly regarding the number and the inner topology of the major clades recognised within the section Calochroi: (i) Frøslev et al. [1, 16] recognised three (/Calochroid, /Fulvi and /Rufoolivacei) and five (/Arcuatorum, /Calochroid, /Fulvi, /Rufoolivacei and /Sulphurinus) major subclades within Calochroi, respectively; (ii) the members of the clades Dibaphi, Elegantiores and Splendentes in the present study were nested within the subclades /Fulvi and /Rufoolivacei of Frøslev et al. [16] and /Arcuatorum, /Fulvi and /Rufoolivacei of Frøslev et al. [1]; (iii) Caroviolacei and Pseudoglaucopodes were placed within a relatively weakly supported subclade assigned as /Rufoolivacei by Frøslev et al. [1, 16]; (iv) the members of Splendentes were placed in isolated positions in the analyses of Frøslev et al. [16]. Some relationships among the Dibaphi, Elegantiores, Napi and Splendentes clades are clearly resolved for the first time in our combined analysis (Figure 3). Including additional molecular markers in future phylogenetic analyses may be the key to resolving more basal nodes in the Calochroi phylogeny.
Many species in the section Calochroi are characterised by bright colours on the one hand and by abruptly marginate to flattened bulbous stipe bases (see Figure 3). Most of the species in this clade are associated with broadleaf trees and generally grow under a layer of fallen leaves. Thus they are shielded from sunlight, whereas the presence of a broader stipe base allows more stability of larger basidiomes, especially in sites with a relatively thin layer of organic material. In our phylogenetic hypothesis obtained from the combined dataset (Figure 3) the species with brightly coloured pigments distributed throughout the whole basidiome and usually with rounded to marginate bulbous stipe base were placed in the basal parts of the tree, while members of the Calochroi clade and closely related species are characterised by their relatively pale coloured basidiomes (predominantly whitish to cream flesh and sometimes with greyish to violet tinges restricted above the lamellae, stipe apex and/or cortex) and abruptly marginate to flattened bulbous stipe base. Interestingly, there is striking homoplasy and phenotypic plasticity of basidiome colouration with age in species of the clades Calochroi, Napi and Pseudoglaucopodes. In contrast, the basidiomes of species of Rufoolivacei and the Sulfurini and Splendentes clades exhibit relatively little colour change with age. A possible explanation for this may be that the more highly methylated pigments of the latter group are more stable in relation to environmental factors (e.g. sunlight).
Our analyses show that colour reaction with KOH and colour of the basidiome context are the most phylogenetically informative characters to circumscribe major clades within the section Calochroi with only a few exceptions, whereas characters related to size and shape of the basidiomes, stipe shape, basidiospore size and shape, and pileipellis structure show high levels of homoplasy and/or phenotypic plasticity. Furthermore, characteristic colour reactions with KOH related to the presence/absence of sodagnitins either on the pileus and/or mycelium covering the stipe bulb offer a framework for discriminating among species with similar macroscopical appearance and ecological specialisation. This study demonstrates that dried and adequately preserved herbarium specimens are also useful for performing macrochemical tests with KOH. Similar observations were made in Dermocybe species on herbarium specimens stored for several years [33].
Patterns of species diversification and biogeographic distributions
ITS DNA sequence divergence among several collections of species restricted to Europe, North America or Central America was low (0–0.1%). In general, our morphological and phylogenetic concepts for European calochroid species fit with those applied by Frøslev et al. [1, 16] and Peintner et al. [12, 14, 27] (Additional file 1). Similar sequence variation was observed between collections from disjunct populations in the clade Napi (C. napus/C. albobrunnoides/C. albobrunnoides var. violaceovelatus/C. subpurpureophyllus var. sulphureovelatus) and C. aureofulvus, where sequence divergence was poorly related to geographical distances. For these examples, the ITS sequence divergence between North American and European populations is relatively low compared with the range of variation that has been detected in some biological species of Agaricales [34–37]. A possible explanation for the very low genetic variation between the populations mentioned above is that they were established through recent founder events and have not had time to evolve a higher ITS sequence divergence. Although long-distance dispersal explains well a conspicuous percentage of today's trans-Atlantic disjunct distributions in angiosperms (see [38] for a recent review), it seems unlikely that long-distance dispersal through basidiospores between continents is the reason for the low sequence divergence between North American and European populations of the taxa mentioned above, since a combination of several factors such as the presence of a compatible haploid mycelium, appropriated edaphic and climatic conditions, as well as the availability of adequate host tree(s) is necessary for successful colonisation and the development of basidiomes in the new habitat. Higher levels of sequence divergence, on the other hand, may indicate that populations have evolved faster or/and had a more ancient separation. Substantial sequence divergences have occurred for example within the lineages Calochroi, Dibaphi and Elegantiores, resulting in well-resolved phylogenetic relationships within these clades. Within the lineages Dibaphi and Elegantiores, we found that intracontinental relationships of European and North American populations were closer than the relationships between populations from distinct continents. Similar levels of genetic divergence were found in widely distributed species within the genus Amanita [39]. However, species/populations within the C. arcuatorum and C. elegantior complexes have undergone very little morphological changes since their divergence, making it impossible to separate them on the basis of morphological characters alone. Selection in these taxa may have predominantly occurred at the physiological level rather than at the morphological level. It is evident from these results that future studies including more representative population samplings are needed to clarify questions concerning distributional patterns and ranges of genetic divergence.
Host tree distributions seem to constitute one of the driving forces in the evolution of the genus Cortinarius. Main geographical distributions of species/populations in the section Calochroi include regional radiations (Elegantiores clade), as well as clinal (Dibaphi), boreo-nemoral (Calochroi, Pseudoglaucopodes) or intercontinental (Napi, C. aureofulvus) patterns throughout Europe, Central and North America. However, to understand modern geographical distributions in the section Calochroi better, it is necessary to consider the biogeographical history of their host trees [40–42] as well as geological events [see, for example, [43, 44]]. Although the history of the flora is still somewhat controversial, generally it is accepted that migration either via the Bering or North Atlantic bridges during the late Cretaceous and the Tertiary periods has played a crucial role in modern distributions of plants [45]. Therefore, it is reasonable to assume that closely related Cortinarius species occurring on separate continents originated in the migration of a common ancestor together with its associated tree host.
The high diversification found in the Calochroi clade roughly matches the distribution of several broadleaf trees throughout boreo-nemoral areas [40] in Europe, Central and North America. An explanation for this high diversification may be that the edaphic factors (e.g., pH, soil aeration, organic and inorganic composition) of broadleaf forests are more favourable for many calochroid species than the coniferous forests, or simply the fact that there is a higher species diversity in the angiosperm trees compared to the conifers. The diversification of angiosperms and gymnosperms is thought to have driven the diversification of other ectomycorrhizal fungal groups [see e.g., [46–48]] and the Calochroi diversification might follow similar patterns. Despite their crucial role for the ecology of a wide spectrum of trees in the northern hemisphere (ectomycorrhiza), we are only beginning to understand the factors modelling the evolutionary history of the Calochroi. Future studies with a focus on molecular dating and phylogeographic analyses including a broad sampling of taxa occurring disjunctly in the northern hemisphere are required to estimate the age and geographical origins of the calochroid lineages as well as to further elucidate the historical dispersal and diversification.
Conclusion
This study demonstrates the great potential of comparative DNA sequence analyses of multiple populations from different geographical origins for understanding patterns of genetic and morphological variation at the species/population level within a biogeographical and taxonomic context. Our phylogenetic analyses support the division of the section Calochroi into seven major clades (Calochroi, Caroviolacei, Dibaphi, Elegantiores, Napi, Pseudoglaucopodes, and Splendentes), whereas Rufoolivacei and Sulfurini were not supported as monophyletic entities. Major phylogenetic groups agree partially with morphology-based and earlier molecular phylogenetic studies. Some phylogenetic relationships within these clades are unexpected based on phenotypic features, which may be explained by convergent evolution of similar morphologies within phylogenetically divergent lineages in Calochroi. This study shows that combinations of features such as colour and colouration patterns, colour reactions with KOH, pigment contents, veil, and pileipellis colouration of basidiomes are useful for circumscribing clades, whereas characters related to size and shape of the basidiomes, stipe shape, basidiospore size and shape, and pileipellis structure display high levels of homoplasy and/or phenotypic plasticity. Bright pigments evolved independently multiple times in the section Calochroi, and abruptly marginate to flattened stipe bulbs represents an autapomorphy for the clade Calochroi.
Furthermore, our study suggests the need for future phylogenetic analyses using multiple molecular markers of those lineages with poorly understood phylogenies as well as the critical re-evaluation of species concepts especially within the clades Calochroi, Dibaphi, Elegantiores, Napi and Pseudoglaucopodes. Main geographical distributional patterns for Calochroi species include clinal, regional and disjunct intercontinental distributions across Europe, Central and North America, where species/populations seem to follow the natural distribution of their host trees. Species in the section Calochroi show different degrees of host fidelity: some are associated exclusively either with coniferous or frondose trees; while others show relatively high host fidelity at least at the host genus level, suggesting potential co-evolutionary/co-migratory patterns.
Methods
Taxon sampling
For preliminary molecular analyses we used a taxonomically representative global sampling of collections of Cortinarius, subgenus Phlegmacium, as a phylogenetic framework to filter out calochroid species which do not belong to our focal group, the monophyletic section Calochroi sensu Frøslev et al. [1, 16] and Garnica et al. [15]. In a second step, thorough analyses were performed to estimate phylogenetic relationships within our focal group. Our final taxon sampling included collections of all known species of Calochroi and Fulvi sensu M.M. Moser [4, 19] and Brandrud [2] with the exception of C. chlorophanus, C. inexpectatus, C. rhodophyllus, C. stephanopus and C. vaginatus. In addition, to establish congruence and uniformity of species concepts, our own sequences were compared with ITS sequences of European calochroid species retrieved from GenBank http://www.ncbi.nlm.nih.gov and UNITE http://unite.ut.ee/ databases. In many cases, multiple collections per species from different geographical origins were used to analyse intraspecific morphological and genetic variation. In addition to the specimens collected and identified in fresh condition by the authors and collaborators, selected herbarium collections from Austria (IB), France (PC), Norway (O), Spain (Arangu-Cort) and USA (MICH) were examined. Host tree genera were assigned to the collections based on field observations. A specimen list including geographical origin, host tree(s), voucher number and GenBank http://www.ncbi.nlm.nih.gov accession numbers of the Calochroi species included in this study is available in the Supplementary Table (see Additional file 3).
Extraction of genomic DNA, PCR amplification, and sequencing
The total genomic DNA was extracted from dried lamella fragments using the DNAeasy Plant Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The ITS region (including the gene coding for the 5.8 S ribosomal subunit) and the D1/D2 regions of the nucLSU were amplified with the primer combination ITS1F (5'-CTTGGTCATTTAGAGGAAGTAA-3', [49])/NL4 (5'-GGTCCGTGTTTCAAGACGG-3', [50]). Alternatively, DNA from older or poorly preserved specimens was amplified with the primer combinations ITS1F/ITS4 (5'-TCCTCCGCTTATTGATATGC-3', [51]) and 5.8SR (5'-TCGATGAAGAACGCAGCG-3')/LR3 (5'-CCGTGTTTCAAGACGGG-3') [52]. PCR amplifications of RPB1 domains A-C were made with the primer combination RPB1-A (5'-GARTGYCCDGGDCAYTTYGG-3') and RPB1-C (5'-CCNGCDATNTCRTTRTCCATRTA-3') [53]. In a few cases, weak amplifications of the RPB1 gene were used as a template for a second PCR with the same primer combination. Amplified PCR products were purified using the QIAquick protocol (Qiagen). Cycle sequencing was accomplished using the ABI PRISM Dye-Terminator Cycle Sequencing Kit version 3.1 (Applied Biosystems, Foster City, CA, USA). For sequencing of the RPB1 A-C gene, the forward primer RPB1-A sg (5'-YTSAARGCYGGTGAGT-3') and the reverse primer RPB1-B sg (5'-TCCGCRCCYTCYTTGG-3') were used [54]. Sequencing was performed with an ABI 3100 automated sequencer (Applied Biosystems). Forward and reverse sequences were assembled and edited using Sequencher version 4.1 (Gene Codes Corporation, Ann Arbor, MI, USA).
Alignments
Initially, contiguous sequences including ITS1 and ITS2, 5.8S and approximately 600 bases of the 5' terminal domain (D1/D2 regions) of the nucLSU of a broad sampling of Phlegmacium species by Garnica et al. [[13, 15], unpublished data] were automatically aligned in MAFFT v5.7 [55] and analysed by neighbor joining [56] in PAUP* 4.0b10 [57] using Kimura 2-parameter distances [58]. A final sequence sampling was obtained by successive pruning towards the focal fungal group of this study. Those sequences lacking either ITS1 or ITS2 regions were excluded from the final matrix. We also aligned the nucleotide sequences of ITS region from our final alignment with those downloaded from GenBank http://www.ncbi.nlm.nih.gov and UNITE http://unite.ut.ee/ databases following the methodology indicated above (dataset 1). In addition, to estimate genetic variation between collections of the same species and also between closely related species more accurately, we aligned and compared our own sequences with their respective chromatograms and also those sequences retrieved from the GenBank and UNITE databases using Sequencher. Intraspecific genetic differences were counted from pairwise alignments. Furthermore, percentages of internal transcribed spacer region (ITS1, ITS1, and 5.8S subunit) and D1/D2 region of the rDNA sequence divergence within and among closely related species were estimated. We then realigned a final set of 230 ITS, 5.8S and D1/D2 sequences using the FFT-NS-i strategy of MAFFT (dataset 2). Sixty-one RPB1 A-C sequences were aligned using the E-INS-i strategy of MAFFT v5.7 [54], with minor manual adjustments made in SeAl v2.11 [59]. This alignment was concatenated with aligned ITS-5.8S-D1/D2 sequences from the same specimens to obtain dataset 3. Leading or trailing gaps were coded as missing data. Gblocks Version 0.91b [60] was used for eliminating gapped and too divergent portions of the datasets 2 and 3 with a minimum block length of 4 positions and gaps allowed in any alignment position for no more than half of the sequences. The full alignments may be obtained from TreeBase http://www.treebase.org/treebase/index.html. After exclusion of the sites indicated above, the alignments had a length of 1144 bp (dataset 2 with 234 sequences) and 2381 bp (dataset 3 with 61 sequences).
Molecular phylogenetic analyses
To establish congruence and uniformity of species concepts for European calochroid species (dataset 1) and to estimate phylogenetic relationships (datasets 2 and 3), maximum-likelihood (ML) analysis was performed with RAxML version 7.0.0 [61]. Doing so, we followed a new approach implemented in the software, which integrates bootstrap analysis [62] and heuristic searches for an optimal tree. First, 1000 bootstrap replicates were run, starting from maximum parsimony trees and using the GTRCAT approximation. Then, every 5th bootstrap tree was used as a starting point for thorough ML analysis, using the GTRCAT model for heuristic searches, and optimising branch lengths and likelihood values of the final trees using the GTR + Γ model of DNA substitution. Tree graphics were generated in PAUP* [57] (Macintosh Classic version). For dataset 3 we additionally performed a maximum parsimony (MP) bootstrap analysis in PAUP*, involving 1000 replicates and treating gaps as missing data. In each replicate, ten rounds of heuristic search were run, with starting trees obtained by successive addition of sequences in random order and using the TBR branch swapping algorithm (storing multiple trees, steepest descent option in effect). The outgroup taxa (Cortinarius caerulescens UL 98/88, C. glaucopus TUB 011414, C. variicolor TUB 011416 and C. violaceus TUB 011561) were chosen on the basis of previous phylogenetic studies within genus Cortinarius [14, 15].
Ancestral state reconstruction
Ancestral states were reconstructed for two characters related to pigmentation and morphology; flesh colour: pale (whitish to cream, sometimes with greyish to violet tinges are present above the lamellae, stipe cortex and/or in stipe apex)/bright (yellow, olive, yellow green); shape of the stipe base: rounded to marginate/abruptly marginate to flattened. For this we used unweighted Wagner parsimony as implemented in PAUP* according to the ACCTRAN and the DELTRAN strategies [63] on the tree with the highest likelihood found. Since the used outgroup species differ from members of section Calochroi in their pigments and have a different stipe morphology, these taxa were excluded for the reconstruction of ancestral states (grey branches). For Cortinarius sp. USA 16 no data about these characters are available. Since the outgroup species differ from members of section Calochroi in their pigments and have a different stipe morphology, these taxa were excluded for the reconstruction of ancestral states.
Macrochemical tests and morphological analyses
Fresh basidiomes were documented using comprehensive macroscopical descriptions accompanied with colour photographs. Macrochemical reactions with 40% KOH were performed by placing a drop directly on the tissue (surface and flesh of the pileus and stipe, and the mycelium covering the base of stipe bulb) of fresh and dried specimens, observing immediate and longer (5–10 minutes) colour changes. Changes in colour were documented from dried specimens with a NIKON COOLPIX 5400 digital camera. Microscopical characters were observed from both fresh and dried specimens mounted in H2O or 3% KOH solution. Pileipellis portions of dried specimens were soaked in 3% KOH for a few minutes before sectioning. The pileipellis structure from expanded pilei at a position halfway to the centre was studied from radial (longitudinal) free-hand sections using a razor blade and observed with ×40 and ×100 (oil immersion) lenses. Hyphal colour was recorded in 3% KOH. Scanning electron microscopy (SEM) of the basidiospores of selected species were made from approx. 5 mm2 lamellae fragments fixed on double adhesive tape and sputter-coated with gold-palladium. Samples were examined with a Cambridge Stereoscan 250 Mk2 scanning electron microscope.
References
Frøslev TG, Jeppesen TS, Laessøe T, Kjøller R: Molecular phylogenetics and delimitation of species in Cortinarius section Calochroi (Basidiomycota, Agaricales) in Europe. Molecular Phylogenetics and Evolution. 2007, 44: 217-227. 10.1016/j.ympev.2006.11.013.
Brandrud TE, Lindström H, Marklund H, Melot J, Muskos S: Cortinarius, Flora Photographica. (German Version). Cortinarius HB, Matfors 1990-Vols 1 (1990), 2 (1992), 3 (1995) & 4 (1998). 1998
Henry R: Suite et complément à l'étude des Phlegmacia. Bull Soc Myco Fr. 1939, 55: 61-94.
Moser M: Die Gattung Phlegmacium (Schleimköpfe). 1960, Julius Klinkhardt, Bad Heilbrunn, Germany
Moënne-Loccoz P, Reumaux P, Bidaud A: Atlas des Cortinaires. 1990, Editions Féderation Mycologique Dauphiné-Savoie, Annecy, 1–14:
Oertel B, Laber D: Die Laugenreaktion an der Unterseite der Stielknolle bei Fruchtkörpern der Gattung Cortinarius, Untergattung Phlegmacium (Agaricales). Zeitschrift für Mykologie. 1986, 52: 139-154.
Brandrud TE: Cortinarius subgen. Phlegmacium sect. Fulvi – chemotaxonomy versus morphological taxonomy. Journées Eurpéennes du Cortinaire. 1998, 0: 4-9.
Mueller M, Lamottke K, Steglich W, Busemann S, Reichert M, Bringmann G, Spiteller P: Biosynthesis and stereochemistry of phlegmacin-type fungal pigments. European Journal of Organic Chemistry. 2004, 23: 4850-4855. 10.1002/ejoc.200400518.
Oertel B: Untersuchungen zur Konstitution von Dihydroanthracenonen und Angaben zu ihrer Verbreitung in Pilzen. PhD Dissertation. 1984, Mathematisch-Naturwissenschaftliche Fakultät, Rheinische Friedrich-Wilhelms-Universität, Bonn
Sontag B, Fröde R, Bross M, Steglich W: Chromogenic triterpenoids from Cortinarius fulvoincarnatus, C. sodagnitus, and related toadstools (Agaricales). European Journal of Organic Chemistry. 1999, 1: 255-260. 10.1002/(SICI)1099-0690(199901)1999:1<255::AID-EJOC255>3.0.CO;2-6.
Steglich W, Oertel B: Untersuchungen zur Konstitution und Verbreitung der Farbstoffe von Cortinarius, Untergattung Phlegmacium (Agaricales). Sydowia. 1985, 37: 284-295.
Peintner U, Bougher NL, Castellano MA, Moncalvo J-M, Moser MM, Trappe JM, Vilgalys R: Multiple origins of sequestrate fungi related to Cortinarius (Cortinariaceae). American Journal of Botany. 2001, 88: 2168-2719. 10.2307/3558378.
Garnica S, Weiß M, Oertel B, Oberwinkler F: Phylogenetic relationships of European Phlegmacium species (Cortinarius, Agaricales). Mycologia. 2003, 95: 1155-1170. 10.2307/3761917.
Peintner U, Moncalvo J-M, Vilgalys R: Toward a better understanding of the infrageneric relationships in Cortinarius (Agaricales, Basidiomycota). Mycologia. 2004, 96: 1042-1058. 10.2307/3762088.
Garnica S, Weiß M, Oertel B, Oberwinkler F: A framework for a phylogenetic classification in the genus Cortinarius (Basidiomycota, Agaricales) derived from morphological and molecular data. Canadian Journal of Botany. 2005, 83: 1457-1477. 10.1139/b05-107.
Frøslev TG, Matheny PB, Hibbett DS: Lower level relationships in the mushroom genus Cortinarius (Basidiomycota, Agaricales): A comparison of RPB1, RPB2, and ITS phylogenies. Molecular Phylogenetics and Evolution. 2005, 37: 602-618. 10.1016/j.ympev.2005.06.016.
Moser M, Ammirati JF: Studies in North American Cortinarii VI. New and interesting taxa in subgenus Phlegmacium from the pacific states of North America. Mycotaxon. 2000, 74: 1-36.
Moser M, McKnight KH, Ammirati JF: Studies on North American Cortinarii I. New and interesting taxa from the Greater Yellowstone area. Mycotaxon. 1995, 55: 301-346.
Moser M: Cortinarius and Dermocybe. The Agaricales in modern taxonomy. Edited by: Singer R. 1986, Koenigstein, Germany: Koeltz Scientific Books, 618-656. 4
Moser M, Horak E: Cortinarius Fr. und nahe verwandte Gattungen in Südamerika. Beihefte zur Nova Hedwigia. 1975, 52: 1-628.
Kauffman CH: Cortinarius Fries. North American Flora. 1932, 10: 282-348.
Smith AH: Studies in the genus Cortinarius I. Contributions from the University of Michigan Herbarium. 1939, 2: 1-42.
Moser M, Ammirati JF: Studies on North American Cortinarii IV. New and interesting Cortinarius species (subgenus Phlegmacium) from oak forests in Northern California. Sydowia. 1997, 49: 25-48.
Moser M, McKnight KH, Sigl M: Plant and their Environments. The genus Cortinarius (Agaricales) in the Greater Yellowstone area. Mycorrhizal host associations and taxonomic considerations. Proceedings of the First Biennial Scientific Conference on the Greater Yellowstone Ecosystem. Edited by: Despain DG. 1994, 239-246.
Garnica S, Münzmay T, Saar G: Cortinarius Subsektion Elegantiores in Europa. Journées Européennes du Cortinaire. 2006, 8: 79-101.
Moser M, Peintner U: Die phylogenetischen Beziehungen der Cortinarius aureopulverulentus Gruppe. Journées Européennes du Cortinaire. 2002, 4: 28-38.
Moser M, Ammirati JF: Studies in North American Cortinarii II. Interesting and new species collected in the North Cascade Mountains, Washington. Mycotaxon. 1996, 58: 387-412.
Thiers HD: The secotioid syndrome. Mycologia. 1984, 76: 1-8. 10.2307/3792830.
Thiers HD, Smith AH: Hypogeous Cortinarii. Mycologia. 1969, 61: 526-536. 10.2307/3757242.
Fogel R: Materials for a hypogeous mycoflora of the Great Basin and adjacent cordilleras of the Western United States II. Two subemergent species, Cortinarius saxamontanus, sp. nov., and C. magnivelatus, plus comments on their evolution. Mycologia. 1994, 86: 795-801. 10.2307/3760594.
Melot J: Une classification du genre Cortinarius (Pers.) S.F. Gray. Documents Mycologiques. 1990, 20: 43-59.
Bidaud A, Moënne-Loccoz P, Reumaux P: Atlas des Cortinaires, clé génerale des sous-genres, sections, sous-sections et séries. 1994, Editions Féderation mycologique Dauphiné-Savoie, Annecy
Ammirati JF: The section Dermocybe of Cortinarius in North America. PhD thesis. 1972, University of Michigan
Hughes KW, McGhee LL, Metheven AS, Johnson JE, Petersen RH: Patterns of geographic speciation in the genus Flammulina based on sequences of the ribosomal ITS1-5.8S-ITS2 area. Mycologia. 1999, 91: 978-986. 10.2307/3761628.
Jin J, Hughes KW, Petersen R: Biogeographical patterns in Panellus stypticus. Mycologia. 2001, 93: 308-315. 10.2307/3761652.
Vilgalys R, Sun BL: Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proceedings of the National Academy of Sciences of the USA. 1994, 91: 4599-4603. 10.1073/pnas.91.10.4599.
Zervakis I, Moncalvo J-M, Vilgalys R: Molecular phylogeny, biogeography and speciation of the mushroom species Pleurotus cystidiosus and allied taxa. Microbiology. 2004, 150: 715-726. 10.1099/mic.0.26673-0.
Renner SS: Plant dispersal across the tropical Atlantic by wind and sea currents. International Journal of Plant Sciences. 2004, 165 (4, Suppl): S23-S33. 10.1086/383334.
Oda T, Tanaka C, Tsuda M: Molecular phylogeny and biogeography of the widely distributed Amanita species, A. muscaria and A. pantherina. Mycological Research. 2004, 108: 885-896. 10.1017/S0953756204000620.
Manos PS, Stanford AM: The historical biogeography of Fagaceae: tracking the tertiary history of temperate and subtropical forests of the Northern Hemisphere. International Journal of Plant Sciences. 2001, 162: S77-S93. 10.1086/323280.
Mueller GM, Wu QX, Huang YQ, Guo SY, Aldana-Gomez R, Vilgalys R: Assessing biogeographic relationships between North American and Chinese macrofungi. Journal of Biogeography. 2001, 28: 271-281. 10.1046/j.1365-2699.2001.00540.x.
Wang X-Q, Tank DC, Sang T: Phylogeny and divergence times in Pinaceae: Evidence from three genomes. Mol Biol Evol. 2000, 17 (5): 773-781.
Hewitt GM: Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996, 58: 247-276.
Hewitt GM: Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society. 1999, 68: 87-112. 10.1111/j.1095-8312.1999.tb01160.x.
Milne RI: Northern hemisphere plant disjunctions: a window on tertiary land bridges and climate change?. Annals of Botany. 2006, 98: 465-472. 10.1093/aob/mcl148.
Wu QX, Mueller GM, Lutzoni FM, Huang YQ, Guo SY: Phylogenetic and biogeographic relationships of Eastern Asian and Eastern North American disjunct Suillus species (Fungi) as inferred from nuclear ribosomal RNA ITS sequences. Molecular Phylogenetics and Evolution. 2000, 17: 37-27. 10.1006/mpev.2000.0812.
Chapela IH, Garbelotto M: Phylogeography and evolution in matsutake and close allies inferred by analyses of ITS sequences and AFLPs. Mycologia. 2004, 96: 730-741. 10.2307/3762107.
Halling RE: Ectomycorrhizae: Co-Evolution, significance, and biogeography. Annals of the Missouri Botanical Garden. 2001, 88: 5-13. 10.2307/2666128.
Gardes M, Bruns D: ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993, 2: 113-118. 10.1111/j.1365-294X.1993.tb00005.x.
O'Donnell K: Fusarium and its near relatives. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Edited by: Reynolds DR, Taylor JW. 1993, Wallingford, UK: CAB International, 225-233.
White TJ, Bruns T, Lee S, Taylor J: Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A guide to methods and amplifications. Edited by: Innis MA, Gelfand H, Sninsky JS, White TJ. 1990, Academic Press, New York, 315-322.
Vilgalys R, Hester M: Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990, 172: 4238-4246.
Matheny PB, Liu YJ, Ammirati JF, Hall BD: Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). American Journal of Botany. 2002, 89: 688-698. 10.3732/ajb.89.4.688.
Garnica S, Weiß M, Walther G, Oberwinkler F: Reconstructing the evolution of agarics from nuclear gene sequences and basidiospore ultrastructure. Mycological Research. 2007, 111: 1019-1029. 10.1016/j.mycres.2007.03.019.
Katoh K, Kuma K, Toh H, Miyata T: MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research. 2005, 33: 511-518. 10.1093/nar/gki198.
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4 (4): 406-425.
Swofford DL: PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10. 2002, Sunderland, Massachusetts: Sinauer Associates
Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980, 16: 111-120. 10.1007/BF01731581.
Rambaut A: Se-Al. Sequence Alignment Editor. 2002, Department of Zoology, University of Oxford, Oxford, UK
Castresana J: Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000, 17 (4): 540-552.
Stamatakis A: RAxML-VI-HPC: maximum likelihood-based phylogentic analyses with thousands of taxa and mixed models. Bioinformatics. 2006, 22: 2688-2690. 10.1093/bioinformatics/btl446.
Felsenstein J: Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985, 39: 783-791. 10.2307/2408678.
Swofford DL, Maddison WP: Reconstructing ancestral characters states under Wagner Parsimony. Mathematical Biosciences. 1987, 87: 199-229. 10.1016/0025-5564(87)90074-5.
Gill M, Steglich W: Pigments of Fungi (Macromycetes). Progress in the Chemistry of Organic Natural Products. 1987, 51: 1-317.
Høiland K, Holst-Jensen A: Cortinarius phylogeny and possible taxonomic implications of ITS rDNA sequences. Mycologia. 2000, 92: 694-710. 10.2307/3761427.
Acknowledgements
The authors thank José Antonio Cadiñanos, Andreas Gminder, Reiner Hintzen, Manfred Huth, Doris Laber, Antonio Ortega, Rafael Mahiques, Frank Röger, Günter Saar, Michelle Seidl, Karl Soop, Geert Schmidt-Stohn, James Trappe, Jordi Vila and Ben Wood for providing accurately documented specimens. Additional herbarium specimens were provided by Arangu-Cort (Sociedad Micológica Aranguren Herbarium, Spain), IB (Innsbruck Herbarium, Austria), MICH (The University of Michigan Herbarium, USA), O (Oslo Herbarium, Norway) and PC (Herbier de Mycologie, Paris, France). We also thank Miguel Angel Jara, Marianne Valdebenito and Oscar Valdebenito for help in the collection of Cortinarius specimens from Nothofagus forests in Southern America (Argentina and Chile), and the editor and two anonymous referees for their helpful comments. This research was financed by the Deutsche Forschungsgemeinschaft (OB 24/27-1, 29) and supported by the Universidad de Costa Rica and Instituto Nacional de Biodiversidad (INBio) Costa Rica, the New York Botanical Garden (Roy Halling), and the D. E. Stuntz Memorial Foundation, Seattle, Washington.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
SG designed the study, generated the DNA sequences, carried out morphological analyses and wrote the manuscript; MW and SG performed the phylogenetic analyses; MW, JA, BO and FO revised several versions of this manuscript. All authors approved the final manuscript.
Electronic supplementary material
12862_2008_915_MOESM1_ESM.jpeg
Additional file 1: RAxML ML phylogram from an alignment of 576 calochroid sequences of ITS-5.8S. Support values are derived from 1000 bootstrap replicates. GenBank accession numbers: DQ08XXXX are from [16]; DQ32XXXX, DQ3508242 and DQ66XXXX are from [1]; AY17XXXX are from [13], AY66XXXX from [15], EFO1XXXX from [25]; AF50XXXX are from [26]; AF3XXXXX and AY03XXXX are from [12] and [14], respectively; AJ236067 is from [65]; EUXXXXXX are from this study. UNITE accession numbers are indicated as UDBXXXXXX. Note: short sequences lacking either ITS1 or ITS2 were excluded from the final analysis. (JPEG 6 MB)
12862_2008_915_MOESM2_ESM.doc
Additional file 2: Distribution of macro- and microscopical features among calochroid species. Macro- and microscopical features referring to habit, basidiome and veil coloration, macrochemical reaction with 40% KOH, pileipellis structure, hyphal coloration in 3% KOH and spore morphology were documented. (DOC 41 KB)
12862_2008_915_MOESM3_ESM.doc
Additional file 3: Specimens used in this study and their respective collection sites, host tree(s), herbarium numbers and GenBank accession numbers. Herbarium abbreviations: Arangu-Cort = Herbarium Sociedad Micológica Aranguren, Spain; IB = Herbarium Innsbruck, Austria; JFA = J. F. Ammirati, MES = Matthew Smith; MTS = Michelle Seidl, ST = Steve Trudell, BW = Ben Wood, University of Washington Herbarium (WTU), USA; KS-CO = private herbarium of Karl Soop; O = Herbarium Oslo, Norway; TUB = Herbarium Tubingense, University of Tübingen, Germany; OSC = Oregon State University; S = Herbarium Stockholm, Sweden; SCR = Sierra Research Center (Matthew Smith), University of California Berkeley (UCB), USA. aSequence from [13], bSequence from [15], cSequence from [25]. (DOC 373 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Garnica, S., Weiß, M., Oertel, B. et al. Phylogenetic relationships in Cortinarius, section Calochroi, inferred from nuclear DNA sequences. BMC Evol Biol 9, 1 (2009). https://doi.org/10.1186/1471-2148-9-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-9-1