Abstract
Background:
Duplicated genes are common in vertebrate genomes. Their persistence is assumed to be either a consequence of gain of novel function (neofunctionalisation) or partitioning of the function of the ancestral molecule (sub-functionalisation). Surprisingly few studies have evaluated the extent of such modifications despite the numerous duplicated receptor and ligand genes identified in vertebrate genomes to date. In order to study the importance of function in the maintenance of duplicated genes, sea bream (Sparus auratus) PAC1 receptors, sequence homologues of the mammalian receptor specific for PACAP (Pituitary Adenylate Cyclase-Activating Polypeptide), were studied. These receptors belong to family 2 GPCRs and most of their members are duplicated in teleosts although the reason why both persist in the genome is unknown.
Results:
Duplicate sea bream PACAP receptor genes (sbPAC1A and sbPAC1B), members of family 2 GPCRs, were isolated and share 77% amino acid sequence identity. RT-PCR with specific primers for each gene revealed that they have a differential tissue distribution which overlaps with the distribution of the single mammalian receptor. Furthermore, in common with mammals, the teleost genes undergo alternative splicing and a PAC1Ahop1 isoform has been characterised. Duplicated orthologous receptors have also been identified in other teleost genomes and their distribution profile suggests that function may be species specific. Functional analysis of the paralogue sbPAC1s in Cos7 cells revealed that they are strongly stimulated in the presence of mammalian PACAP27 and PACAP38 and far less with VIP (Vasoactive Intestinal Peptide). The sbPAC1 receptors are equally stimulated (LOGEC50 values for maximal cAMP production) in the presence of PACAP27 (-8.74 ± 0.29 M and -9.15 ± 0.21 M, respectively for sbPAC1A and sbPAC1B, P > 0.05) and PACAP38 (-8.54 ± 0.18 M and -8.92 ± 0.24 M, respectively for sbPAC1A and sbPAC1B, P > 0.05). Human VIP was found to stimulate sbPAC1A (-7.23 ± 0.20 M) more strongly than sbPAC1B (-6.57 ± 0.14 M, P < 0.05) and human secretin (SCT), which has not so far been identified in fish genomes, caused negligible stimulation of both receptors.
Conclusion:
The existence of functionally divergent duplicate sbPAC1 receptors is in line with previously proposed theories about the origin and maintenance of duplicated genes. Sea bream PAC1 duplicate receptors resemble the typical mammalian PAC1, and PACAP peptides were found to be more effective than VIP in stimulating cAMP production, although sbPAC1A was more responsive for VIP than sbPAC1B. These results together with the highly divergent pattern of tissue distribution suggest that a process involving neofunctionalisation occurred after receptor duplication within the fish lineage and probably accounts for their persistence in the genome. The characterisation of further duplicated receptors and their ligands should provide insights into the evolution and function of novel protein-protein interactions associated with the vertebrate radiation.
Similar content being viewed by others
Background
Increased gene number and complexity are generally assumed to have contributed to the success of vertebrates. The evolutionary driving forces behind this are still under debate however gene and/or genome duplications and exon shuffling events are proposed to have been of fundamental importance [1–5]. The increased complexity of metazoan genomes have been attributed to rounds of gene or whole genome duplication [1, 6–9]. Analysis of metazoa genomes reveals that a remarkable percentage of duplicated genes exist [10–13] and whilst some genes decay to non-functionality and are subsequently eliminated from the genome, others are maintained either through the acquisition of novel functions (neofunctionalisation) or by partitioning the function of the ancestral molecule between the duplicated isoforms (subfunctionalisation).
The secretin family of G-protein coupled receptors (GPCRs) (a.k.a. family 2 GPCRs) is a large hormone and neuropeptide receptor gene family present in metazoan genomes. Members of this family have been identified in both protostomes and deuterostomes [14–16] and their conserved sequence and gene organisation has led to the proposal that they evolved from a common ancestral gene as a consequence of total, or partial genome duplication [14]. Vasoactive Intestinal Peptide (VIP) and Pituitary Adenylate Cyclase Activating Polypeptide (PACAP) receptors (VPAC and PAC1, respectively) are closely related members of family 2 GPCRs. They are important pharmaceutical targets as their ligands, the brain-gut peptides VIP and PACAP, control a number of important physiological functions in mammals [17, 18].
In humans three receptors exist, PAC1, VPAC1 and VPAC2 and binding studies reveal that VPACs are able to bind the ligands, VIP and PACAP with similar affinities, while PAC1 preferentially binds PACAP [18, 19]. In vertebrates, activation of PAC1/VPAC receptors occurs via three intracellular transduction mechanisms. These involve either cyclic AMP (cAMP) production, IP turnover via PLC or/and calcium mobilization as a consequence of the activation of a G-protein complex [18, 20–22]. In mammals, five PAC1 isoforms, which result from the insertion of one or two, 28 (hip or hop1 variant) or 27 (hop2 variant) amino acid cassettes in intracellular loop 3 (IL3) have been identified [23, 24]. In addition, two VPAC1 and VPAC2 isoforms have been recently described which lack TM6 and TM7 [25]. In other vertebrates PAC1 splice isoforms have been isolated, however no alternative splice forms of VPAC have yet been identified and in vertebrates these latter receptors do not activate the IP3 signalling pathway [26–28].
Teleosts, which diverged from the tetrapod lineage approximately 450 million years ago (MYA), represent one of the most successful vertebrate groups with over 25,000 species. The existence of a variety of teleost genome sizes and ploidy levels has made them very useful for studies of gene evolution and function [29]. Recently duplicated PAC1 and VPAC receptor genes have been identified in teleosts using in silico approaches [16] and in Takifugu they have a differential tissue distribution [15]. In the present study duplicate PAC1 cDNAs were isolated from the marine teleost, sea bream (Sparus auratus) and their tissue expression and functional profile characterised using the peptides VIP, PACAP27, PACAP38 and SCT (secretin). The persistence of duplicate sbPAC1 receptors in teleost genomes is discussed in relation to the current proposed theories for gene evolution in vertebrates.
Results
Sea bream duplicate PAC1 receptors
Two complete putative sbPAC1 cDNAs, namely sbPK713 (2368 bp) and sbPP1C (2791 bp), were isolated from a sea bream kidney and pituitary cDNA libraries, respectively (Figures 1 and 2). The isolated cDNAs encode putative proteins of 444 and 448 amino acids respectively, and share an overall amino acid identity of 77% due to the highly conserved TM domains and C-terminal regions; the N-terminal domain is only 48% identical. The sbPK713 and sbPP1C cDNAs share 91% and 86% amino acid sequence identity respectively, with the previously identified Takifugu PAC1A and PAC1B and for this reason were designated sbPAC1A (AJ514930) and sbPAC1B (AJ514931). Analysis of sea bream genomic DNA by RT-PCR coupled with Southern blot (data not shown) confirmed that both sea bream PAC1 cDNAs are encoded by single genes and are not the result of alternative splicing. This observation is confirmed by mapping of sbPAC1A and sbPAC1B genes in the recently released sea bream gene-based radiation hybrid (RH) map, to group 6 (RH6) and 2 (RH2), respectively [30].
Nucleotide and predicted protein sequence of sbPAC 1 A receptor. Seven TM domains are highlighted with black lines and the conserved N-terminal cysteine residues and putative N-glycosylation sites are annotated respectively by a circle and by "+". The microsatellite identified in the 5'UTR is underlined and the stop codon is indicated by "*".
Nucleotide and predicted protein sequence of sbPAC 1 B receptor. The seven TM domains are highlighted with black lines and the conserved N-terminal cysteine residues and putative N-glycosylation sites are annotated respectively by a circle and by "+". The microsatellite identified in the 5'UTR is underlined and stop codon is indicated by "*".
A microsatellite sequence is present in the 5'UTR region of both receptors and sbPAC1A contains an imperfect (AC)32 dinucleotide repeat and sbPAC1B an imperfect (GA)28 repeat upstream of the initiation codon (Figures 1 and 2). Genotyping of the microsatellites using genomic DNA from sea bream caught at different geographic locations and a family panel, revealed both microsatellites are polymorphic and the locus sbPAC1B scores more alleles (8 alleles) than sbPAC1A (4 alleles) (Additional file 1).
Sequence comparisons and phylogenetic analysis
Sequence comparison of sbPAC1A and sbPAC1B with vertebrate homologue receptors revealed six conserved cysteine residues and two putative N-glycosylation sites at the N-terminal domain which are implicated in ligand-binding [20](Figure 3). Additional conserved motifs characteristic of PAC1 are also present. Within intracellular loop three (IL3) the amino acid motifs, P-D-M and R-L-A-R (Basic-L/A-L/A/V/S-Basic), important for functional coupling with the Gsα protein, are conserved in the teleost and tetrapod receptors. Amino acid residues K322 and E394, which are involved in activation of the cAMP pathway in humans [21] are also conserved suggesting that a similar receptor signalling pathway may exists in sea bream. A putative signal peptide sequence is also present along with the consensus signature motif of TM7 in mammalian PACAP and VIP receptors: FQGBBVXXBYCFXNXEVXQ (where X is any amino acid and B is a basic amino acid [15, 31]. Duplicate PAC1 genes also exist in other teleosts genomes, such as stickleback (ENSGACP00000022696 and ENSGACP00000007143), Tetraodon (GSTENP00011829001 and GSTENP00034495001), Medaka (ENSORLP00000022250 and ENSORLP00000016315) and zebrafish (NM_001013444 and XM_701685) (Table 1). In contrast, a single copy of a PAC1 gene was identified in the Atlantic salmon (Salmon salar, CK885244) and rainbow trout (Oncorhynchus mykiss, AY706216). It seems likely that further PAC1 receptors exist in salmonids and it is expected that these will be identified as sequence coverage of their genomes is enriched.
Multiple sequence alignment of vertebrate PAC 1 receptors. The seven TM domains are boxed, the P-D-I/M motif indicated by "○"and the R-L-A-R motif by "*". The alternative receptor hop splice isoform is indicated by a dotted box and the putative signal peptide sequence with a double ended arrow. Conserved cysteines and putative N-glycosylation sites in the N-terminal domain are indicated by "●" and "+", respectively. Accession numbers of sequences used in the multiple amino acid sequence alignment were; human (Homo sapiens, P41586), rat (Rattus norvegicus, P32215), bovine (Bos taurus, Q29627), Rana (Rana ridibunda, Q90Y07), Xenopus (Xenopus laevis, Q9PTK1), Goldfish (Carassius auratus, O7376, Takifugu (Takifugu rubripes, AJ494861 for PAC1A and AJ490804 for PAC1B) and sea bream sbPAC1A (AJ514930) and sbPAC1B (AJ514931).
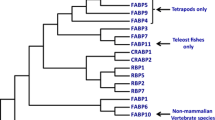
Phylogenetic analysis of sbPAC1A and sbPAC1B receptors with the vertebrate PAC1, VPAC1, VPAC2, PRP, GHRH and SCT receptors confirmed their identity as duplicated members of the vertebrate PAC1 family (Figure 4, Additional file 2). The tree topology indicates that vertebrate PAC1, VPAC (1 and 2), PRP and GHRH receptors evolved from a common ancestral gene and that a teleost specific duplication has occurred which is in line with the proposed partial or whole genome duplication event within their lineage [9, 12, 15, 16, 32, 33].
Phylogenetic analysis of the vertebrate PAC 1 members. This figure represents the upper quartile of the consensus tree, for full image and details please refer to Additional file 2. Sea bream PAC1 receptors are in bold and the novel teleost PAC1 receptors identified are refereed by their Ensembl nomenclature. PAC1 accession numbers are: Human, P41586; Mouse, P70205; Rana, Q90Y07; Takifugu 1A, AJ494861, Takifugu 1B, AJ490804; zebrafish1A, NM_001013444 and zebrafish1B, XM_701685.
Linkage analysis
Gene environment comparisons of the PAC1 homologue genomic regions revealed it is highly conserved across vertebrates (Figure 5). In human, PAC1 is localized on chromosome 7 and in chicken it maps to chromosome 2 and is in close proximity with the chicken GHRH and VPAC1 receptors. In teleosts the duplicate genes map to different genome regions and are localised in the Takifugu scaffolds N000080 and N00239 and in zebrafish chromosomes 10 and 2. Three genes (RT-like protein, PDE1C and NeuroD6-B genes) were found to be shared between the Takifugu, chicken and human genomic regions analysed. The PAC1 gene is linked with the PRPR gene in Takifugu and chicken suggesting these genes arose by tandem duplication, although it has been lost in the human. In Xenopus, in which the genome assembly is incomplete, PAC1 maps to the final region of scaffold_333 and NeuroD6-B and PDE1C to scaffold_588 suggesting that these two regions are contiguous in the genome although so far no homologue of PRPR was identified.
Short-range linkage analysis of the PAC 1 receptors. The gene environment of Takifugu, chicken and human PAC1 regions is represented. Homologue genes present in all three study organisms are represented by open boxes and black boxes represent flanking genes that had no corresponding homologues. Genes are named according to the HUGO annotation and PAC1 genes are in bold. Takifugu scaffolds (assembly 4) are represented using the NIX annotation [40]. The gene environment of PAC1 in Xenopus is not represented since it was very incomplete. The Takifugu gene environment is represented and the scale corresponds approximately to 10 Kb and the relative position of linked genes in the chromosomes of the chicken and human is given.
Tissue expression of sbPAC1 receptors
PAC1 tissue distribution was carried out in several sea bream tissues by RT-PCR using specific primers for each receptor (Figure 6). These primers were designed in order to amplify all potential receptor transcripts. The sbPAC1A was found to be expressed in the pituitary, kidney, duodenum and skin and weakly in heart and gonads whilst sbPAC1B was restricted to brain and pituitary. A larger sbPAC1A PCR product was also amplified which had an overlapping tissue distribution with sbPAC1A with the exception of heart and gonads. Sequence analysis indicates that the larger sbPAC1A PCR product corresponds to the homologue of the mammalian PAC1hop1 isoform and contains an insertion of 28 amino acids within IL3 region. No other PAC1A isoforms were amplified and no alternative splice forms for sbPAC1B were detected.
Tissue expression of the duplicate sbPAC 1 A and sbPAC 1 B receptors. Expression was carried out by RT-PCR using specific primers to amplify potential receptor transcripts. PCR products of approximately 1200 bp, 1000 bp and 1280 bp were obtained and corresponded to sbPAC1A, sbPAC1B and sbPAC1Ahop1 isoform, respectively.
Functional studies of duplicate sbPAC1
The sbPAC1A and sbPAC1B were successfully expressed in Cos7 and Hek293 cell lines as confirmed by immunofluorescence and Western blot analysis. Cos7 cell extracts subject to Western blot contained a specific immunoreactive fusion protein of approximately 52 kDa in transfected cells, which corresponds to the estimated molecular weight in silico of the fusion proteins, (T7PAC1A is 52.75 kDa and T7PAC1B is 52.32 kDa). Cos7 and Hek293 cell lines expressing the recombinant vector were activated by Forskolin and gave maximal cAMP production. Negative control experiments in which Cos7 and Hek293 cells were transfected with pcDNA3 without insert and incubated with the maximum concentration used of test peptide (10-6 M), revealed that Hek293 cells are responsive to PACAP and VIP peptides. This suggests the existence of endogenous receptors in Hek293 which is confirmed by available proteome data [34]. For this reason only the results obtained with transfected Cos7 cells are presented.
The cAMP potency profile of Cos7 transfected cells expressing the recombinant sbPAC1 receptors in the presence of different concentrations of VIP and PACAP (PACAP38 and PACAP27) peptides is depicted in Figure 7. All mammalian peptides were able to stimulate cAMP production in a dose dependent manner. The peptide activation profile for the sbPAC1A gene was as follows PACAP27 ≈ PACAP38 > VIP as assessed by the LOGEC50 values for cAMP production for each peptide (-8.74 ± 0.29 M, -8.54 ± 0.18 M and -7.23 ± 0.20 M, respectively). The stimulation of cAMP production by VIP was significantly lower (P < 0.05) than for PACAP peptides. The peptide activation profile of sbPAC1B was also tested and found to be similar to sbPAC1A and is as follows PACAP27 ≈ PACAP38 > VIP and the LOGEC50 values for maximal cAMP production for each peptide are -9.15 ± 0.21 M, -8.92 ± 0.24 M and -6.57 ± 0.14 M, respectively. The stimulation of cAMP by VIP was significantly lower than PACAP27 and PACAP38 (P < 0.001). Human SCT causes a negligible increase above the basal concentration of cAMP production in non-stimulated Cos7 cells transformed with either sbPAC1A or sbPAC1B (data not shown). Comparison of the potency profile of each peptide (PACAP27, PACAP38 and VIP) for the duplicate sea bream receptors revealed that PACAP stimulates both receptors equally. In contrast, VIP is significantly more potent for sbPAC1A (-7.23 ± 0.20 M) compared to sbPAC1B (-6.57 ± 0.14 M, P < 0.05).
Production of maximal cAMP by the recombinant Cos7 cell expressing sbPAC 1 A (A) and sbPAC 1 B (B). Transfected cells expressing the receptors were incubated with different concentrations of PACAP27 (□) PACAP38 (△) and VIP (○). Data was normalized and receptor potency profiles is given as percentage of intracellular cAMP produced per well and error bars indicate the ± SEM of a minimum of three independent experiments performed in duplicate. Only the lower error bars are represented.
Discussion
In the present study, duplicate sea bream PAC1 genes (sbPAC1A and sbPAC1B) have been isolated and functionally characterised. Structural motif identification and expression assays reveal they are functional family 2 GPCR members. The sbPAC1B is predominantly found in brain and pituitary while sbPAC1A has a widespread distribution but is absent from brain. The overall tissue distribution of the sbPAC1 receptors is similar to the single mammalian PAC1 receptor and suggests they may have similar functions in the endocrine, nervous, gastrointestinal and reproductive systems [17, 18]. In common with the mammalian PAC1 receptor both sbPAC1 receptors are highly stimulated by PACAP but poorly by VIP and SCT. The tissue distribution of PAC1 receptors in sea bream is different from that in Takifugu in which PAC1B has a widespread tissue distribution and PAC1A distribution is restricted to brain, gill and gonads [15]. Therefore, despite the high sequence conservation between teleost PAC1A or PAC1B genes (91% and 86%, respectively), the paralogue genes seems to have evolved in a species specific manner in relation to tissue distribution and possibly function.
Duplicate PAC1 genes have been identified in other teleosts and also for other family 2 GPCR members [14, 15] as expected in light of the proposed partial or full genome duplication event suggested to have occurred within the teleost lineage [32, 33, 35–37].
Analysis of teleost genomes indicates that duplicate PAC1 genes are linked with the recently reclassified GHRH-like receptor which based upon ligand binding characteristics has been reassigned as a teleost PRP receptor (PRPR) [38, 39]. In Takifugu, PAC1A and PRPR genes are localised on scaffold N000080 and PAC1B/PRPR in scaffold N002399 and in the zebrafish genome on chromosome 10 and 2, respectively [40, 41]. In terrestrial vertebrates, with the exception of mammals, co-localization of both receptors is also observed suggesting that PAC1 and PRPR genes arose by tandem gene duplication prior to the teleost divergence and that PRPR gene was subsequently lost in the mammalian lineage (Figure 6).
Stimulation of cAMP production and not peptide affinity or activation of alternative signalling pathways has been investigated and revealed that sbPAC1 receptors are highly stimulated and have identical potency profiles for the mammalian PACAP27 and PACAP38 peptides compared to VIP. The latter peptide was found to be more potent for sbPAC1A in comparison to sbPAC1B. Although, the identification in teleosts of duplicate genes for the ligands and the existence of two potentially active PACAP peptides raises further issues in relation to PAC1 receptor activation and function. Two copies of PACAP are present in Takifugu (DQ659331 and DQ659332), Tetraodon (Q4RN19 and Q4RH43) and zebrafish (NW_652622 and NW_634478), but functional studies with the duplicate receptors and their ligands are scarce. The ligand binding characteristics of the duplicated zebrafish zfPACAP27 peptides for the zebrafish PAC1 receptor, the sequence homologue of sbPAC1A, was tested and both peptides strongly stimulate in a similar way the cAMP and phospholipase pathways [28, 42] suggesting conservation of function. A further measure of complexity is also introduced by the identification of duplicate VIP genes in teleost genomes [38, 43] and it will be of importance in future to establish the affinity of the duplicate peptides for the duplicate receptors, as well as compare their tissue distribution in teleosts.
Moreover, a microsatellite is identified for the first time in the 5'UTR of PAC1 receptors. Genotyping analysis of the microsatellite reveals it is polymorphic in sea bream and raises intriguing questions about its potential influence on gene expression. Analysis of homologue regions in other teleosts [41] also reveals the presence of a microsatellite in the 5'UTR. For example, in the stickleback PAC1A and PAC1B respectively, two perfect microsatellite repeats (TG)21 and (CA)34 and an imperfect (CA)21 dinucleotide repeat are present. The PAC1A gene in medaka contains a (TG)8 repeat, although no microsatellite is present in the paralogue gene (Additional file 3). So far no microsatellites have been described or identified in the mammalian, chicken, frog and goldfish homologue receptors [28, 44–46]. The importance of PACAP in growth and development [17, 18] and the presence of a microsatellite in its receptor suggests it may be a useful tool for genomic and functional analysis. A previous study of early growth variation in the Artic charr (Salvelinus alpinus, [47]) revealed a strong marker-trait association with a single nucleotide polymorphism (G/A) of the 18th base pair of the intronic region between the exons that code for PACAP-related peptide and PACAP in the PRP/PACAP gene precursor.
The human SCT peptide failed to significantly stimulate cAMP production by the sbPAC1 receptor which may indicate either a failure to activate the cAMP signalling pathway or a failure to bind the receptor. The preceding results are at odds with the physiological role attributed to secretin in fish, pancreatic stimulation via cholecystokinin and oxyntomodulin [48]. However, SCT has only been identified by immunohistochemistry in the gastrointestinal tract of the flower fish (Pseudophoxinus antalyae) [49]. The failure to identify in teleosts a gene for the SCT receptor or its ligand suggests they probably evolved subsequent to teleost divergence or were lost during the teleost radiation and the significant sequence differences between SCT and VIP or PACAP probably explains the absence of activity of SCT in the present study [16].
Gene duplicates and generation of alternative splice isoforms are major contributors to functional diversity of the vertebrate proteome. In teleosts, the existence of gene duplicates is proposed to reduce the incidence of gene isoforms since they are assumed to generate functional redundancy of single-copy gene splice isoform [50], and this may explain reduced number of PAC receptor splice isoforms detected in the sea bream. In sea bream, a PAC1Ahop receptor isoform was identified with an overlapping tissue distribution with the shorter PAC1A receptor transcript, but it was different from the tissue distribution reported for alternative splice isoforms in zebrafish and goldfish [26, 28]. Recently in zebrafish, two novel IL3 splice isoforms were characterised, a hop2 isoform (insertion of 87 bp) present in ovaries and a novel skip isoform (resulting in a truncated protein) in the gills [28]. Characterisation of a recently isolated PAC1 and hop1 receptor isoform in the goldfish reveals that they have similar activation profiles raising question about their functional role [26]. The mounting evidence for the presence of hop1 receptor isoforms in all vertebrates suggests that a homologue transcript was probably also expressed by the PAC1 receptor gene in ancestral metazoa [50]. In order to understand the functional relevance of duplicate receptors and isoforms it will be important to establish if they are co-localised in vivo in order to establish possible interactions.
Expression studies of the sbPAC1 genes demonstrated they are functional family 2 GPCRs and PACAP and VIP peptides were able to stimulate cAMP production in a dose dependent manner. Such assays also indicate that both sea bream receptors are specific PACAP receptors and have different activation profiles for VIP although, as only cAMP production was measured, it remains to be established if a similar activation profile occurs for the alternative IP3 signalling pathway. One hypothesis for the apparent functional divergence and differential expression of sea bream paralogue receptors may be related to functional specialisation related to the existence in teleosts of duplicate ligands which might also explain the significant amino acid changes (48% sequence identity) observed in the N-terminal region. In mammals, important amino acid motifs involved in receptor binding have been identified and mutation studies reveal the importance of the N-terminus [20]. Amino acid motifs involved in ligand binding are conserved between tetrapod and teleost homologue receptors and include the motifs W-D, G-W-S and the following amino acids W, P and P (Figure 3). However, the amino acid motifs that account for potential ligand selectivity of the teleost duplicated receptors are still unknown, future receptor mutation studies should help to clarify this issue.
An intriguing aspect about the sbPAC1 genes is their divergent tissue distribution and this may be the basis of their specific physiological functions and persistence in the genome. Understanding the biological function of receptors is complex as it is not only the availability and concentration of receptors but also of ligands and accessory factors at a given site which will determine receptor preference/activity and ultimately biological function. Physiological studies of the ligands, PACAP and VIP, in teleosts are not very numerous and certainly do not encompass all the actions assigned in mammals making it difficult currently to assign possible biological roles to the duplicate receptors. However, one major function attributed to PACAP is its role in GH-release [46, 51, 52]. In common with mammals, PACAP38 is the predominant isoform in teleost brain and this peptide is found to have a more potent stimulatory effect on fish GH secretion by pituitary cells when compared to GHRH and GnRH (potent mammalian GH releasing factors). In contrast, VIP has little or no effect on GH release [46, 51, 52] and this has been taken to suggest that teleost GH-release is mediated via PAC1 pituitary receptors and the sea bream PAC1B may play a central role in this process. Relatively few studies have been carried out to characterise the biological activity of fish VIP and as in mammals it is proposed to have an important role in the gastrointestinal system [51, 53, 54]. In cod, Gadus morhua, VIP stimulates gastric and pancreatic secretion [55, 56] and in tilapia it is involved in ion and water absorption by the intestine [57]. In sea bream, tissue distribution of PACAP and VIP transcripts indicate that PACAP is mainly restricted to nervous tissue whilst VIP is abundant in the gastrointestinal system but is also present in a wide range of other tissue (unpublished data). The widespread distribution in non nervous tissue of sbPAC1A indicates that this receptor may have a broader physiological role. Unquestionably much more work is required to elucidate the physiological relevance of duplicate sbPAC1 and alternative approaches such as ligand mutational studies; gene knock-down strategies and physiological experiments will be needed.
Conclusion
Duplicate PAC1 receptors genes are present in the majority of teleost genomes, although the reason for their persistence is not yet clearly established. The present study with duplicate sbPAC1 receptors suggests that their maintenance may be due to a process of neofuncionalisation as a consequence of the accumulation of mutations in the ligand binding domain of the receptors after duplication. Such a proposal is supported by the poor sequence conservation (48%) in the N-terminal ligand binding domain of the receptor. The divergent tissue distribution of the receptors, with one form predominantly found in nervous tissue and the other with a more widespread distribution is highly suggestive of functional divergence. The isolation and characterisation of ligands for the receptors in teleosts will be an important step in establishing receptor function, as will improved characterisation of the tissue distribution of both receptors and ligands and the factors regulating their expression.
Methods
Sea bream cDNA library screening
A homologous PAC1 receptor probe (620 bp) was obtained by RT-PCR using sea bream brain cDNA and degenerate primers flanking transmembrane regions 2 and 7 (Table 2). PCR reactions were performed in a 25 μl reaction volume with 1 × PCR buffer (Biocat, Italy), 1,5 mM MgCl2 (Biocat), 0,2 mM dNTPs (GE healthcare, UK), 1 mM of each primer, EuroTaq DNA Polymerase 5 U/μl (Biocat) and DNase Free water (Sigma-Aldrich). The PCR product was used to screen sea bream pituitary and kidney cDNA libraries constructed using the λ-Zap vector II kit (Stratagene, USA) [58]. The phage libraries were titred to obtain approximately 500,000 pfu per plate and blotting procedures were performed in duplicate using Nylon membranes (Hybond-N, GE healthcare, UK). Probes were radioactively labelled using α-P32dCTP (GE healthcare, UK) and the Rediprime kit (GE healthcare, UK) following manufacturer's instructions. Membrane filters were hybridised overnight at 58°C with Church-Gilbert hybridisation solution (1 mM EDTA pH8; 0.5 M NaPO4 pH7.2, 7% (w/v) SDS) and washed under high stringency conditions at 58°C and exposed to X-OMAT film (Kodak, USA) at -80°C for 24 hours. Positive clones were selected based on the intensity of signals and their presence on the duplicate plate. Single clones were excised into phagemid vectors using the manufacturer's protocol and DNA was extracted and sequenced to confirm its identity.
RT-PCR analysis of receptor tissue distribution
Tissue distribution and analysis of PAC1 splice variants was carried out by RT-PCR. Total RNA was extracted from sea bream pituitary, brain, kidney, gills, gut, heart, gonads, liver and skin using TRI reagent (Sigma-Aldrich, Spain). Mature sea bream of 11–13 months old weighing approximately 350–500 g were sacrificed by decapitation and their tissues collected and immediately frozen at -80°C. cDNA was synthesised using 1 μg of sea bream total RNA and a reverse transcription system kit (Promega, Spain) and hexameric oligonucleotides following the manufacture's instructions. The quality of cDNA obtained was verified by PCR with sea bream EF1-α (a housekeeping gene) using the following thermocycle; 94°C for 2 minutes; 25 cycles (94°C for 1 minute, 58°C for 1 minute and 72°C for 1 minute) and a final extension step at 72°C for 5 minutes. Specific primers for each sbPAC1 gene were designed (Table 2) and PCR was performed as previously described using the following cycle: 94°C for 2 minutes; 34 cycles (94°C for 1 minute, 62°C for 1 minute and 72°C for 1 minute) and a final step at 72°C for 5 minutes. The products obtained were sequenced to confirm their identity.
Genotype analysis
The variability of a microsatellite identified in the 5' UTR of sbPAC1A and sbPAC1B was assessed using specific primers (Table 2) and sea bream genomic DNA from fish of diverse geographic origins (Morocco, Portugal, France and Adriatic sea) and from a genomic panel composed of parents and 50 first generation progeny. PCR reactions were performed using fluorescently labelled forward primers (0.3 μM; 6-FAM and TET; Metabion International AG) and unlabelled reverse primers (0.3 μM) using the following thermocycle: 95°C for 2 minutes; 35 cycles (95°C for 30 seconds, 57°C for 30 seconds and 72°C for 30 seconds) and a final step at 72°C for 5 minutes. PCR products were separated using high resolution 6% Long Ranger acrylamide gels (Cambrex, USA) on a automated ABI 377 sequencer (Applied Biosystems, USA) and data was analysed with the GenScan software (Applied Biosystems, USA).
Database searches, sequence alignments and phylogenetic analysis
The conserved amino acid sequence of sbPAC1 transmembrane (TM) domains was used to search for homologous receptors in teleost genomes. Briefly, the amino acid sequences of the seven TM domains were extracted, concatenated and used in BLAST sequence similarity searches [59] against the Tetraodon nigroviridis [41], medaka (Oryzias latipes), stickleback(Gasterosteus aculeatus), zebrafish (Danio renio) [41] and atlantic salmon (Salmon salar) [60] genome databases and NCBI EST database [61]. The amino acid sequence of the TM domains of a total of 52 receptors were concatenated and a multiple sequence alignment was produced using the ClustalX vs1.83 (Blosum matrix and Gap opening penalty 10 and Gap extension 0.2) [62] and percentage of identity/similarity calculated using Genedoc [63]. The alignment produced (length 170, with 162 informative sites) did not require the insertion of gaps and was used to construct phylogenetic trees using both maximum parsimony and neighbour joining methods [64] with 1000 bootstrap replicates, complete gap deletion and Poisson correction in MEGA 3.1 phylogenetic programme [65].
Short-range linkage analysis
The gene environment of the Takifugu, Xenopus, chicken and human PAC1 homologue regions were compared using a sequence similarity approach. The human and chicken gene environments were accessed using the NCBI Mapview interfaces [61] and Xenopus using the Ensembl database [41]. The gene environment of the Takifugu scaffolds (release17/05) was accessed using NIX annotation [40] and the neighbouring genes were used to search for orthologues in human, chicken and Xenopus genomes using the tblastn algorithm [61].
Construction of the recombinant expression vector
Specific primers for each sbPAC1 gene were designed to amplify the mature receptor sequence (Table 2) and were cloned into pcDNA3 vector (Invitrogen, UK) containing a signal peptide of CD33 and a T7-epitope tag (CD33-T7-pcDNA3, [66]). In order to facilitate cloning, restriction digestion sites for BglII and EcoRI enzymes were incorporated in the forward and reverse primers respectively. Template amplification was carried out using the thermocyle; 95°C for 2 minutes; 35 cycles of (95°C for 1 minute, 68°C for 1 minute and 70°C 2 minutes); 72°C for 10 minutes with a proof-reading Pfu DNA polymerase (Promega, Spain) in a 25 μl reaction volume containing 1 × PCR buffer (Promega, Spain), 1,5 mM MgCl2 (Promega, Spain), 0,2 mM dNTPs (Amersham), 1 mM of each primer and 1,5 U Pfu DNA polymerase (3 U/μl)) and DNase Free water (Sigma-Aldrich, Spain). The amplified PCR products were cloned into pGEMT-easy vector (Promega, Spain), sequenced and subcloned into CD33-T7-pcDNA3 vector in frame with the T7-epitope tag. The recombinant constructs produced (CD33-T7-pcDNA3+sbPAC1A and CD33-T7-pcDNA3+sbPAC1B) were sequenced and used to transfect mammalian Cos7 and Hek293 cells lines. The CD86 (NM_175862) membrane protein of the immunoglobulin superfamily cloned in CD33-T7-pcDNA3 expression vector was used as positive control for cell transfection (de Vet et al., 2001).
Mammalian cell transfections
Mammalian Cos7 and Hek293 cell lines were transfected using the Effectene transfection kit (Qiagen, Germany) and the success of transfection was assessed by western blot and immunofluorescence assays using antisera raised against the T7-epitope tag protein [66]. One day prior to transfection, approximately 60,000 to 70,000 Cos7 and Hek293 cells were seeded and transfections carried out using approximately 0.4 μg of recombinant construct (pcDNA3+sbPAC1A or B) and cells were grown for 2 days at 37°C. The viability of transfected cells was determined by dye exclusion using Trypan blue (0,4% solution, Sigma) and the success of transfection calculated by fluorescent confocal microscopy using DAPI (4',6-Diamidino-2-phenylindole) and FITC (Fluorescein) staining for Cos7 cells and the FACS (Fluorescence Activated Cell Sorting) method for Hek293 cells. The percentage of cell transfection was estimated to be approximately 30% in both cell lines.
Immunofluorescence and western blot assays
An anti-T7 epitope monoclonal antibody (Novagen, UK) was used in immunofluorescence and western blots assays to assess the success of transfection and production of recombinant fusion protein (T7+sbPAC1) as described in de Vet et al., 2001 [66]. Briefly, for the immunofluorescence assay, cells were grown in 6 well plates (Greiner, Germany) on sterile cover slips. Approximately 120,000 cells were used per well and cell transfections were carried out using 0.4 μg of DNA as described above. Immunofluorescence localization studies were performed [67] and DAPI and FITC staining examined using a microscope linked to a confocal imaging system (Bio-Rad, UK). Western blots were carried out by lysing transfected cells (30 μl) in 1× Laemmli SDS-PAGE loading buffer [68] and proteins were fractionated on a 10% SDS-PAGE gel with a constant current of 35 mA. The fractionated proteins were transferred to a nitrocellulose membrane (GE healthcare, UK) and blocked (2% milk powder in 1 × PBS/0.1%Tween) for 1 hour at room temperature. Incubations were carried out with anti-T7 tag monoclonal antibody (Novagen, UK) and a secondary antibody coupled to Horse-Radish peroxidase as described in de Vet., 2001 [66]. Immunoreactive proteins were detected using the ECL system (PerkinElmer Life Sciences, UK). The size of the immunoreactive protein was comparable to the estimated sizes of the recombinant receptor protein determined in silico using the Swiss Prot database interface [69].
Ligand-binding studies
Ligand binding studies were performed two days after transfections, in three independent experiments. Transfected cells were incubated in duplicate with different concentrations (10-6 to 10-11M) of the human VIP, PACAP38 and secretin (SCT) peptides and ovine PACAP27 peptide for 30 minutes (Sigma-Aldrich, Spain). Homologous sea bream peptides are unavailable but their predicted amino acid sequences are 82%, 100%and 92% identical to human VIP, PACAP27 and PACAP38 respectively and important amino acids at N-terminus potential involved in receptor binding are totally conserved (unpublished data). Prior to ligand-binding assays, Cos7 and Hek293 cells were washed 3 times with DMEM medium without fetal calf serum and incubated in a CO2 incubator with 1 mM of IBMX (3-Isobutyl-1-Methylxantine) for 30 minutes. Ligand-binding assays were performed by incubating the cells in fresh medium containing the peptides in the presence of 1 mM IBMX for 30 minutes at 37°C in the CO2 incubator. After incubation, cells were lysed and stored at -80°C until required. Mammalian Cos7 and Hek293 cells transfected with wild type pcDNA3 were used as negative controls which were incubated with the highest tested peptide concentration (10-6mM). Prior to assays the responsiveness of transfected Cos7 and Hek293 cells was tested by stimulating cAMP production using Forskolin (10 mM, 1 mM and 0.1 mM; Sigma-Aldrich).
Radioimmunoassay (RIA) and statistical analyses
The quantification of cAMP produced was determined by radioimmunoassay using the TRK432 kit (GE Healthcare, UK) following the manufacturer's instructions. Cos7 and Hek293 cells were lysed by sonication, centrifuged and the supernatant heat denatured for 10 minutes at 100°C. The concentration of cAMP produced (pmol/well) was determined in duplicate for each sample and calculations performed based on a linear regression curve constructed using standard concentrations of labelled (3H) cAMP. The cAMP data was normalized as a percentage of stimulation above basal levels and plotted as a percentage of cAMP production per well (%cAMP/well). The results are presented as the mean ± SEM of three independent experiments in duplicate and analysis was performed using the SigmaPlot9.01 programme. Data was analyzed by comparing the potency profile of each sbPAC1 receptor in the presence of different concentrations of test peptides and by comparing the potency profile of both receptors in the presence of identical peptide concentrations. The presence of significant differences in cAMP production was assessed with the SigmaStat3.11 programme using two way Anova and the Holm-Sidak method for pairwise multiple comparisons (P < 0.05 and P < 0.001 were considered statistically significant).
References
Ohno S: Evolution by gene duplication. 1970, Berlin, New York, Springer-Verlag
Iwabe N, Kuma K, Miyata T: Evolution of gene families and relationship with organismal evolution: rapid divergence of tissue-specific genes in the early evolution of chordates. Mol Biol Evol. 1996, 13 (3): 483-493.
Patthy L: Genome evolution and the evolution of exon-shuffling--a review. Gene. 1999, 238 (1): 103-114. 10.1016/S0378-1119(99)00228-0.
Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV: Selection in the evolution of gene duplications. Genome Biol. 2002, 3 (2): RESEARCH0008-10.1186/gb-2002-3-2-research0008.
Seoighe C: Turning the clock back on ancient genome duplication. Curr Opin Genet Dev. 2003, 13 (6): 636-643. 10.1016/j.gde.2003.10.005.
Ferris SD, Whitt GS: Loss of duplicate gene expression after polyploidisation. Nature. 1977, 265 (5591): 258-260. 10.1038/265258a0.
Holland PW, Garcia-Fernandez J, Williams NA, Sidow A: Gene duplications and the origins of vertebrate development. Dev Suppl. 1994, 125-133.
Holland PW: Gene duplication: past, present and future. Semin Cell Dev Biol. 1999, 10 (5): 541-547. 10.1006/scdb.1999.0335.
Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H: Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004, 431 (7011): 946-957. 10.1038/nature03025.
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J: Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999, 151 (4): 1531-1545.
Betran E, Long M: Expansion of genome coding regions by acquisition of new genes. Genetica. 2002, 115 (1): 65-80. 10.1023/A:1016024131097.
Amores A, Suzuki T, Yan YL, Pomeroy J, Singer A, Amemiya C, Postlethwait JH: Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 2004, 14 (1): 1-10. 10.1101/gr.1717804.
Hughes AL: Adaptive evolution after gene duplication. Trends Genet. 2002, 18 (9): 433-434. 10.1016/S0168-9525(02)02755-5.
Cardoso JC, Pinto VC, Vieira FA, Clark MS, Power DM: Evolution of secretin family GPCR members in the metazoa. BMC Evol Biol. 2006, 6: 108-10.1186/1471-2148-6-108.
Cardoso JC, Power DM, Elgar G, Clark MS: Duplicated receptors for VIP and PACAP (VPAC1R and PAC1R) in a teleost fish, Fugu rubripes. J Mol Endocrinol. 2004, 33 (2): 411-428. 10.1677/jme.1.01575.
Cardoso JC, Clark MS, Viera FA, Bridge PD, Gilles A, Power DM: The secretin G-protein-coupled receptor family: teleost receptors. J Mol Endocrinol. 2005, 34 (3): 753-765. 10.1677/jme.1.01730.
Sherwood NM, Krueckl SL, McRory JE: The origin and function of the pituitary adenylate cyclase-activating polypeptide (PACAP)/glucagon superfamily. Endocr Rev. 2000, 21 (6): 619-670. 10.1210/er.21.6.619.
Vaudry D, Gonzalez BJ, Basille M, Yon L, Fournier A, Vaudry H: Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol Rev. 2000, 52 (2): 269-324.
Schafer H, Zheng J, Morys-Wortmann C, Folsch UR, Schmidt WE: Structural motifs of pituitary adenylate cyclase-activating polypeptide (PACAP) defining PAC1-receptor selectivity. Regul Pept. 1999, 79 (2-3): 83-92. 10.1016/S0167-0115(98)00147-5.
Laburthe M, Couvineau A, Marie JC: VPAC receptors for VIP and PACAP. Receptors Channels. 2002, 8 (3-4): 137-153. 10.1080/10606820213680.
Couvineau A, Lacapere JJ, Tan YV, Rouyer-Fessard C, Nicole P, Laburthe M: Identification of cytoplasmic domains of hVPAC1 receptor required for activation of adenylyl cyclase. Crucial role of two charged amino acids strictly conserved in class II G protein-coupled receptors. J Biol Chem. 2003, 278 (27): 24759-24766. 10.1074/jbc.M301916200.
Dickson L, Aramori I, McCulloch J, Sharkey J, Finlayson K: A systematic comparison of intracellular cyclic AMP and calcium signalling highlights complexities in human VPAC/PAC receptor pharmacology. Neuropharmacology. 2006, 51 (6): 1086-1098. 10.1016/j.neuropharm.2006.07.017.
Spengler D, Waeber C, Pantaloni C, Holsboer F, Bockaert J, Seeburg PH, Journot L: Differential signal transduction by five splice variants of the PACAP receptor. Nature. 1993, 365 (6442): 170-175. 10.1038/365170a0.
Journot L, Spengler D, Pantaloni C, Dumuis A, Sebben M, Bockaert J: The PACAP receptor: generation by alternative splicing of functional diversity among G protein-coupled receptors in nerve cells. Semin Cell Biol. 1994, 5 (4): 263-272. 10.1006/scel.1994.1032.
Bokaei PB, Ma XZ, Byczynski B, Keller J, Sakac D, Fahim S, Branch DR: Identification and characterization of five-transmembrane isoforms of human vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide receptors. Genomics. 2006, 88 (6): 791-800. 10.1016/j.ygeno.2006.07.008.
Kwok YY, Chu JY, Vaudry H, Yon L, Anouar Y, Chow BK: Cloning and characterization of a PAC1 receptor hop-1 splice variant in goldfish (Carassius auratus). Gen Comp Endocrinol. 2006, 145 (2): 188-196. 10.1016/j.ygcen.2005.08.011.
Alexandre D, Vaudry H, Grumolato L, Turquier V, Fournier A, Jegou S, Anouar Y: Novel splice variants of type I pituitary adenylate cyclase-activating polypeptide receptor in frog exhibit altered adenylate cyclase stimulation and differential relative abundance. Endocrinology. 2002, 143 (7): 2680-2692. 10.1210/en.143.7.2680.
Fradinger EA, Tello JA, Rivier JE, Sherwood NM: Characterization of four receptor cDNAs: PAC1, VPAC1, a novel PAC1 and a partial GHRH in zebrafish. Mol Cell Endocrinol. 2005, 231 (1-2): 49-63. 10.1016/j.mce.2004.12.002.
Venkatesh B: Evolution and diversity of fish genomes. Curr Opin Genet Dev. 2003, 13 (6): 588-592. 10.1016/j.gde.2003.09.001.
Sarropoulou E, Franch R, Louro B, Power DM, Bargelloni L, Magoulas A, Senger F, Tsalavouta M, Patarnello T, Galibert F, Kotoulas G, Geisler R: A gene-based radiation hybrid map of the gilthead sea bream Sparus aurata refines and exploits conserved synteny with Tetraodon nigroviridis. BMC Genomics. 2007, 8: 44-10.1186/1471-2164-8-44.
Chow BK, Yuen TT, Chan KW: Molecular evolution of vertebrate VIP receptors and functional characterization of a VIP receptor from goldfish Carassius auratus. Gen Comp Endocrinol. 1997, 105 (2): 176-185. 10.1006/gcen.1996.6818.
Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B: Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004, 21 (6): 1146-1151. 10.1093/molbev/msh114.
Taylor JS, Braasch I, Frickey T, Meyer A, Van De Peer Y: Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003, 13: 382-390. 10.1101/gr.640303.
HEK293 Cell Database. [http://www.mbi.ufl.edu/~shaw/293.html]
Taylor JS, Van de Peer Y, Braasch I, Meyer A: Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci. 2001, 356: 1661-1679. 10.1098/rstb.2001.0975.
Wittbrodt J, Meyer A, Schartl M: More genes in fish? . BioEssays. 1998, 20: 511-515. 10.1002/(SICI)1521-1878(199806)20:6<511::AID-BIES10>3.0.CO;2-3.
Meyer A, Van de Peer Y: From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. 2005, 27 (9): 937-945. 10.1002/bies.20293.
Lee LT, Siu FK, Tam JK, Lau IT, Wong AO, Lin MC, Vaudry H, Chow BK: Discovery of growth hormone-releasing hormones and receptors in nonmammalian vertebrates. Proc Natl Acad Sci U S A. 2007, 104 (7): 2133-2138. 10.1073/pnas.0611008104.
Wang Y, Li J, Ying Wang CY, Ho Yan Kwok AJ, Leung FC: Identification of the endogenous ligands for chicken growth hormone-releasing hormone (GHRH) receptor: evidence for a separate gene encoding GHRH in submammalian vertebrates. Endocrinology. 2007, 148 (5): 2405-2416. 10.1210/en.2006-1013.
Fugu Genome database. [http://fugu.biology.qmul.ac.uk]
Ensembl database. [http://www.ensembl.org]
Wang Y, Wong AO, Ge W: Cloning, regulation of messenger ribonucleic acid expression, and function of a new isoform of pituitary adenylate cyclase-activating polypeptide in the zebrafish ovary. Endocrinology. 2003, 144 (11): 4799-4810. 10.1210/en.2003-0501.
Cardoso J, Vieira FA, Gomes A, Power D: PACAP, VIP and their receptors in the metazoa: insights about the origin and evolution of the ligand-receptor pair. Peptides. 2007
Aino H, Hashimoto H, Ogawa N, Nishino A, Yamamoto K, Nogi H, Nagata S, Baba A: Structure of the gene encoding the mouse pituitary adenylate cyclase-activating polypeptide receptor. Gene. 1995, 164 (2): 301-304. 10.1016/0378-1119(95)00391-I.
Couvineau A, Maoret JJ, Rouyer-Fessard C, Carrero I, Laburthe M: Cloning and functional characterization of the human VIP1/PACAP receptor promoter. Ann N Y Acad Sci. 1998, 865: 59-63. 10.1111/j.1749-6632.1998.tb11163.x.
Wong AO, Leung MY, Shea WL, Tse LY, Chang JP, Chow BK: Hypophysiotropic action of pituitary adenylate cyclase-activating polypeptide (PACAP) in the goldfish: immunohistochemical demonstration of PACAP in the pituitary, PACAP stimulation of growth hormone release from pituitary cells, and molecular cloning of pituitary type I PACAP receptor. Endocrinology. 1998, 139 (8): 3465-3479. 10.1210/en.139.8.3465.
Tao WJ, Boulding EG: Associations between single nucleotide polymorphisms in candidate genes and growth rate in Arctic charr (Salvelinus alpinus L.). Heredity. 2003, 91 (1): 60-69. 10.1038/sj.hdy.6800281.
Nelson LE, Sheridan MA: Gastroenteropancreatic hormones and metabolism in fish. Gen Comp Endocrinol. 2006, 148 (2): 116-124. 10.1016/j.ygcen.2006.01.011.
Cinar K, Senol N, Ozen MR: Immunohistochemical study on distribution of endocrine cells in gastrointestinal tract of flower fish (Pseudophoxinus antalyae). World J Gastroenterol. 2006, 12 (42): 6874-6878.
Su Z, Wang J, Yu J, Huang X, Gu X: Evolution of alternative splicing after gene duplication. Genome Res. 2006, 16 (2): 182-189. 10.1101/gr.4197006.
Wong AO, Li WS, Lee EK, Leung MY, Tse LY, Chow BK, Lin HR, Chang JP: Pituitary adenylate cyclase activating polypeptide as a novel hypophysiotropic factor in fish. Biochem Cell Biol. 2000, 78 (3): 329-343. 10.1139/bcb-78-3-329.
Montero M, Yon L, Kikuyama S, Dufour S, Vaudry H: Molecular evolution of the growth hormone-releasing hormone/pituitary adenylate cyclase-activating polypeptide gene family. Functional implication in the regulation of growth hormone secretion. J Mol Endocrinol. 2000, 25 (2): 157-168. 10.1677/jme.0.0250157.
Holmgren S, Vaillant C, Dimaline R: VIP-, substance P-, gastrin/CCK-, bombesin-, somatostatin- and glucagon-like immunoreactivities in the gut of the rainbow trout, Salmo gairdneri. Cell Tissue Res. 1982, 223 (1): 141-153. 10.1007/BF00221505.
Fouchereau-Peron M, Laburthe M, Besson J, Rosselin G, Le Gal Y: Characterization of the vasoactive intestinal polypeptide (VIP) in the gut of fishes. Comp Biochem Physiol. 1980, 65A: 489-492. 10.1016/0300-9629(80)90064-X.
Thwaites DT, Young J, Thorndyke MC, Dimaline R: The isolation and chemical characterization of a novel vasoactive intestinal peptide-related peptide from a teleost fish, the cod, Gadus morhua. Biochim Biophys Acta. 1989, 999 (2): 217-220.
Holstein B, Humphrey CS: Stimulation of gastric acid secretion and suppression of VIP-like immunoreactivity by bombesin in the Atlantic codfish, Gadus morhua. Acta Physiol Scand. 1980, 109 (2): 217-223.
Mainoya JR, Bern HA: Influence of vasoactive intestinal peptide and urotensin II on the absorption of water and NaCl by the anterior intestine of the tilapia, Sarotherodon mossambicus. Zool Sci. 1984, 1 (100-105):
Santos CR, Brinca L, Ingleton PM, Power DM: Cloning, expression, and tissue localisation of prolactin in adult sea bream (Sparus aurata). Gen Comp Endocrinol. 1999, 114 (1): 57-66. 10.1006/gcen.1998.7228.
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ: Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25 (17): 3389-3402. 10.1093/nar/25.17.3389.
Salmon Genome Project. [http://www.salmongenome.no]
NCBI database. [http://www.ncbi.nih.gov]
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25 (24): 4876-4882. 10.1093/nar/25.24.4876.
Genedoc software. [http://www.psc.edu/biomed/genedoc]
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987, 4 (4): 406-425.
Kumar S, Tamura K, Nei M: MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004, 5 (2): 150-163. 10.1093/bib/5.2.150.
de Vet EC, Aguado B, Campbell RD: G6b, a novel immunoglobulin superfamily member encoded in the human major histocompatibility complex, interacts with SHP-1 and SHP-2. J Biol Chem. 2001, 276 (45): 42070-42076. 10.1074/jbc.M103214200.
Aguado B, Campbell RD: Characterization of a human lysophosphatidic acid acyltransferase that is encoded by a gene located in the class III region of the human major histocompatibility complex. J Biol Chem. 1998, 273 (7): 4096-4105. 10.1074/jbc.273.7.4096.
Laemmli UK: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970, 227 (5259): 680-685. 10.1038/227680a0.
ExPASy Proteomics Server. [http://www.expasy.ch]
Acknowledgements
This work was supported by a FCT/CCMAR pluriannual grant. JCRC was funded by FCT grant BPD/14449/03. The authors would like to acknowledge the technical assistance of Elisabete Caldas for performing part of the teleost sequence database searches and to Elsa Couto for helping with the cAMP radioimmunoassays.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
The majority of the work described was carried by JCRC and the cell expression studies were carried out in collaboration with ECJMV and GE. BL carried out the genotype analysis. MSC and DMP planned the study, discussed the results and critically revised the manuscript for important intellectual content and data analysis. All authors read and approved the final manuscript.
Electronic supplementary material
12862_2007_512_MOESM1_ESM.pdf
Additional file 1: Genotyping analysis of the sbPAC 1 microsatellites in a geographic (A) and family (B) panels. Score alleles for sbPAC1A are represented from A-D and for sbPAC1B from A'-F'. In A, individuals were sampled in four distinct geographic regions. Three in the Atlantic Ocean (Atl) in the Moroccan (Mr), Portuguese (Pt) and French (Fr) coast and in the Adriatic Sea (Adr) in Italian coast. In B, M represent males and F females and 50 progeny individuals were analysed and the percentage of allele scores is indicated. (PDF 11 KB)
12862_2007_512_MOESM2_ESM.pdf
Additional file 2: Phylogenetic analysis of PAC 1 /VPAC/PRPR/GHRHR family members. Sea bream PAC1 genes are indicated in bold and human secretin receptor (SCTR, Q8IV17) is underlined. The receptors identified in Tetraodon (GSTENP00011829001, GSTENP00034495001 and GSTENP00011830001), Stickleback (ENSGACP00000002397), Medaka (ENSORLP00000007394), zebrafish (ENSDARP00000054330 and ENSDARP00000070262), salmon (CK885244 and CB511922) and trout (CU069615) were not used in the analysis since they were found to have incomplete TM domains. Accession numbers for Takifugu and zebrafish receptors were obtained from Table 1 and the novel genes identified in ENSEMBL are indicated. Receptors accession numbers used are: Human PAC1, P41586; Mouse PAC1, P70205; Rana PAC1, Q90Y07; Human VPAC1, P32241; Rat VPAC1, P30083; Rana VPAC1, Q9YHC6; Human VPAC2, P41587; Rat VPAC2, P35000; Chicken PRPR, XM_425958; Goldfish PRPR, O73768; Human GHRHR, Q02643; Mouse GHRHR, Q8AXV2. Bootstrap values under 50 were removed. (PDF 22 KB)
12862_2007_512_MOESM3_ESM.pdf
Additional file 3: Sequence of the putative 5'UTR regions of the Stickleback and Medaka duplicate PAC 1 s genes. Sequences were obtained from ENSEMBL database [41] and 3000 bases localised upstream the initial methionine are represented. Microsatellite repeats identified are underlined and in italics and in bold upper cases the sequence of the first predicted exon is represented. (PDF 25 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Cardoso, J.C., de Vet, E.C., Louro, B. et al. Persistence of duplicated PAC1 receptors in the teleost, Sparus auratus. BMC Evol Biol 7, 221 (2007). https://doi.org/10.1186/1471-2148-7-221
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-7-221