Abstract
Background
Extant placental mammals are divided into four major clades (Laurasiatheria, Supraprimates, Xenarthra and Afrotheria). Given that Afrotheria is generally thought to root the eutherian tree in phylogenetic analysis of large nuclear gene data sets, the study of the organization of the genomes of afrotherian species provides new insights into the dynamics of mammalian chromosomal evolution. Here we test if there are chromosomal bands with a high tendency to break and reorganize in Afrotheria, and by analyzing the expression of aphidicolin-induced common fragile sites in three afrotherian species, whether these are coincidental with recognized evolutionary breakpoints.
Results
We described 29 fragile sites in the aardvark (OAF) genome, 27 in the golden mole (CAS), and 35 in the elephant-shrew (EED) genome. We show that fragile sites are conserved among afrotherian species and these are correlated with evolutionary breakpoints when compared to the human (HSA) genome. Inddition, by computationally scanning the newly released opossum (Monodelphis domestica) and chicken sequence assemblies for use as outgroups to Placentalia, we validate the HSA 3/21/5 chromosomal synteny as a rare genomic change that defines the monophyly of this ancient African clade of mammals. On the other hand, support for HSA 1/19p, which is also thought to underpin Afrotheria, is currently ambiguous.
Conclusion
We provide evidence that (i) the evolutionary breakpoints that characterise human syntenies detected in the basal Afrotheria correspond at the chromosomal band level with fragile sites, (ii) that HSA 3p/21 was in the amniote ancestor (i.e., common to turtles, lepidosaurs, crocodilians, birds and mammals) and was subsequently disrupted in the lineage leading to marsupials. Its expansion to include HSA 5 in Afrotheria is unique and (iii) that its fragmentation to HSA 3p/21 + HSA 5/21 in elephant and manatee was due to a fission within HSA 21 that is probably shared by all Paenungulata.
Similar content being viewed by others
Background
Analyzing how mammalian genomes are organized and how chromosomal rearrangements are involved in speciation and macroevolution are fundamental to understanding the dynamics of mammalian chromosomal evolution. Phylogenetic analysis of both nuclear and mitochondrial DNA [1–6], among others, as well as the insertion sites of multiple long interspersed elements (LINE, [7]) and long terminal repeats (LTRs, [8]), all support the division of extant placental mammals into four major clades. These are Laurasiatheria and Supraprimates that together form Boreoeutheria (with a northern hemisphere origin), and Xenarthra and Afrotheria which have a Gondwanan (southern hemisphere) genesis, although this biogeographic hypothesis is not without detractors [9]. Although Afrotheria is usually depicted as basal in sequence based phylogenies, the first divergence in the placental tree has been a matter of concern for some time. For example, some argue [8] for a basal Xenarthra (the so-called Epitheria hypothesis, [10]) on insertion sites of retroposed elements (but see [11] for a contrary view), while data from 218 genes encompassing 205 kb of sequence resulted in a highly supported phylogeny that places the root between Afrotheria and other Placentalia [6], consistent with the basal Afrotheria or Exafroplacentalia hypothesis [4]. Most recently, however, Xenarthra and Afrotheria have been placed on a common basal branch (the Atlantogenata or Xenafrotheria hypothesis), to the exclusion of Boreoeutheria [12].
Afrotheria, the focus of our study, includes six mammalian orders all with an Afro-Arabian origin that exhibit extreme morphogical diversity and niche preference thought to result from the long period of isolation when Africa was an island continent 105-25 mya [13]. The six orders are Proboscidea (elephant), Sirenia (manatees and dugongs), Hyracoidea (hyrax), Tubulidentata (aardvark), Macroscelidea (elephant shrews) and Afrosoricida (golden moles and tenrecs). In cases where the analysis of primary sequences generates ambiguous phylogenetic results, "rare genomic changes" (RGCs, [14]) such as indels, LINEs, SINEs and chromosomal rearrangements have been widely viewed as markers that could, given their low levels of homoplasy, provide additional resolution to seemingly intractable phylogenetic problems (see [15] for application of SINEs in vertebrate phylogenetics). So far, afrotherian monophyly is supported by a suite of sequence-based characters that include a 9 bp deletion in exon 11 of the BRCA1 gene [16], 5' and 3' deletions in exon 26 of apolipoprotein B [5], the presence of a unique family of SINEs (AfroSINEs, [17–19]), long interspersed elements (LINE 1, [7]) and long terminal repeat (LTR) elements [8]. Consistent with the view that chromosomal rearrangements are similarly rarely homoplasious, and therefore robust indicators of evolutionary change (a default rate of 1–2 changes per 10 million years of mammalian evolution has been suggested, [20–22]), it comes as no surprise that in addition to providing evidence in support of the uniqueness of Afrotheria, chromosomal syntenies have also proved useful for clarifying the phylogenetic relationships within the group [23, 24].
One of the most important features of mammalian chromosomal evolution is the suggestion of a non-random distribution of regions implicated in evolutionary chromosomal reorganization, the so-called "fragile-breakage" hypothesis [25]. Related to this, recent experimental data have demonstrated a correlation between the location of fragile sites and evolutionary breakpoints [26–28] suggesting that these unstable regions could be one of many factors implicated in the evolutionary process. At the cytogenetic level, fragile sites are expressed as non-stained gaps and breaks when cells are cultured under specific conditions [29]. In general, fragile sites can be expressed by agents such as aphidicolin, BrdU and 5-azacytidine among others, which delay or inhibit DNA replication or repair [30]. According to their frequency in the human population, as well as their mechanisms of expression, fragile sites have been conventionally classified into two groups: common and rare [31]. Common fragile sites in particular have been studied in different mammalian species confirming the initial hypothesis that they are structural characteristics of mammalian chromosomes [31]. Common fragile sites have been expressed in rodents [32–37], pig and cow [38–41], horse [42], cat [43–45], dog [46, 47] and different primate species [26, 48–51]. However, in all instances the species studied group within Laurasiatheria and Supraprimates, the most distant relatives of Xenarthra and Afrotheria that are thought to have diverged ~93 mya [52]. There is, at this point, no comparable data for the deeper divergences such as the Afrotheria whose separation from Boreoeutheria and Xenarthra is estimated at ~105 mya [52].
In an attempt to test if there are loci with a high tendency to break and reorganize in the Afrotheria, and whether these are coincidental with evolutionary breakpoints, we have analyzed the expression of aphidicolin-induced common fragile sites in fibroblast cultures from different specimens of three afrotherian species. These are the aardvark (Orycteropus afer, OAF, Tubulidentata), golden mole (Chrysochloris asiatica, CAS, Afrosoricida) and elephant-shrew (Elephantulus edwardii, EED, Macroscelidea). Given the position of Afrotheria near the root of Placentalia (species on the so-called "eutherian" side of the "metatherian-eutherian" dichotomy), the analysis of chromosomal instability in these species provides a unique opportunity to further our understanding of the mechanisms underpinning mammalian chromosomal evolution.
Results
Fragile site expression
(i) Orycteropus afer
A total of 652 metaphase spreads were analysed in two specimens, 374 from cultures treated with 0.2 μM APC, and 278 from control cultures (table 1). As expected, cells treated with aphidicolin presented the highest number of chromosomal aberrations (51.34% of the total metaphases), a 13-fold increase with respect to the control cultures. A total of 287 chromosomal abnormalities were detected of which the most common aberrations were chromatid breaks (61–94% of all aberrations detected, see table 2).
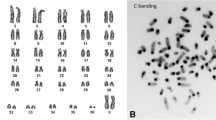
For each data set, the FSM program used in our analysis of fragile sites provides a critical value (Cα) that indicates the minimum number of breaks needed for a chromosomal band to be considered fragile. This value was ≥3 for aardvark cultures treated with 0.2 μM aphidicolin. The number of fragile sites detected ranged from 23 in OAF specimen 1 to 12 in OAF specimen 2. Given the intraspecific variability of fragile site expression in mammalian species, we combined all expressed sites into a single species-specific analysis (this was also done for the golden mole and elephant shrew, see below). On this basis, a total of 29 sites were considered fragile in the aardvark (figure 1a and table 3) showing that there are regions in this species' genome that are prone to breakage under specific culture conditions; of these, six were expressed in both aardvark specimens (1p11, 1q18, 1q44, 2q13, 2q15 and 3p11, table 3).
Ideograms representative of O. afer (A), C. asiatica (B) and E. edwardii (C) chromosomes. The regions of homology with human chromosomes [see 24, 53, 54] are depicted in red and indicated by the numbers to the left of each chromosomal schematic. In aardvark, homology with the African elephant chromosomes is shown in blue. The location of aphidicolin-induced fragile sites (fs) found in all specimens studied is indicated to the right of each chromosomal schematic in the three afrotherian species. Asterisks mark fragile sites conserved in at least two of the three species studied. Heterochromatic regions are marked by diagonal lines.
(ii) Chrysochloris asiatica
A total of 1047 metaphase spreads were analysed from the three specimens included in our investigation: 482 cells from cultures treated with 0.2 μM APC, and 565 from control cultures (table 1). Cells treated with aphidicolin presented the highest number of chromosomal aberrations (68.67% of the total metaphases scored) reflecting a 10-fold increase with respect to the control cultures. A total of 501 chromosomal abnormalities were detected of which chromatid breaks were the most frequent class of aberration encountered in this species (71–94%, table 2).
Analysis of the aphidicolin induced aberrations using the FSM program indicate that chromosomal bands characterised by ≥3 or ≥4 abnormalities per band (depending on the specimen analysed) could be considered fragile. Using these values, a total of 27 fragile sites were detected in the golden mole genome (figure 1b and table 4), with the number of fragile sites ranging from 12 in CAS specimen 1, to 21 in CAS specimen 3 (table 4). Eight of the fragile sites (1p13, 1q13, 1q32, 2p13, 2q22, 2q31, 6q23 and 8q17) were found to be expressed in all three specimens examined (table 4).
(iii) Elephantulus edwardii
A total of 1239 metaphase spreads were analysed in the four specimens of this species of which 620 were from cultures treated with 0.2 μM APC, and 619 from control cultures (table 1). We detected 824 chromosomal abnormalities of which chromatid breaks were the most frequent class of aberration scored (67–92%, table 2).
Mirroring the results in the golden mole, the critical value generated by the FSM program for the elephant shrew was ≥3 or, depending on the specimen studied, ≥4 chromosomal abnormalities per band. The number of fragile sites detected ranged from 14 in EED specimen 4 to 20 in EED specimen 3 (table 5); in total 35 sites were considered fragile in the elephant-shrew genome (figure 1c and table 5). Only three of these (2p16, 3q26 and 5q25) are expressed in all four specimens studied (table 5).
Distribution of evolutionary breakpoints and conservation of fragile sites
We plotted all human chromosomal homologies previously described in [53] and [24] onto the ideogram of each of the afrotherian species studied so as to identify bands that delimit human syntenic blocks. Using this approach we identified a set of evolutionary chromosomal bands that correspond to junctions defining human chromosomal syntenies in the three afrotherian species studied herein. These are: (a) Aardvark – 1p11, 1q18, 1q28, 1q35, 2q31, 2q34, 3q29, 4q15, 4q24, 5q21, 6q22 and 6q23, (b) Golden mole – 1q18, 1q21, 2q21, 2q22, 3q28, 4p14, 4q12, 4q22, 6q12, 9q14, 11p13 and 11p12 and (c) Cape rock elephant shrew – 1p26, 1q15, 1q21, 2q17, 2q19, 4q14, 4q24, 5q12, 5q22, 7q12, 10q12, 11p12 and 11q12 (see figure 1). Additionally, we were able to plot the African elephant/aardvark chromosomal syntenies described in [54] that are based on reciprocal painting of these two species thereby providing insights into the association between evolutionary breakpoints and fragile sites among these two species compared to the older, phylogenetically more distant human/Afrotheria evolutionary comparison. Nineteen evolutionary chromosomal bands were detected when comparing the aardvark and African elephant genomes: 1p11, 1q15, 1q18, 1q24, 1q26, 1q28, 1q35, 1q38, 2q13, 2q15, 2q21, 2q25, 2q27, 2q31, 3q12, 3q15, 4p11, 5q14 and 6q23 (figure 1a). On combining these data (human and elephant chromosomal syntenies) 25 distinct evolutionary breakpoints could be defined in the aardvark; of these, six are common to both data sets (1p11, 1q18, 1q28, 1q35, 2q31 and 6q23).
Given these findings, we proceeded to determine if there is a correlation between the position of evolutionary breakpoints and the location of fragile sites in each of the afrotherian species studied using contingency analysis. Of the 12 evolutionary breakpoints identified in the aardvark by reciprocal painting with human painting probes [53], seven are coincidental with regions of fragility as defined by fragile site location (1.8 bands expected if the distribution was random, p = 0.0004) (figure 1a). Of the 19 evolutionary breakpoints identified in the aardvark by reciprocal painting with the African elephant painting probes [54], eight are coincidental with regions of fragility (three bands are expected if the distribution was random, p = 0.0032) (figure 1a). It is noteworthy that of the six bands (1p11, 1q18, 1q28, 1q35, 2q31 and 6q23) that delimit human and elephant chromosomal syntenic blocks in the aardvark genome (see above), all but one (1q35) express fragile sites. Additionally, 27 fragile sites were expressed in the golden mole of which four (1.6 expected, p = 0.07) show correspondence with the 12 evolutionary breakpoints detected by chromosome painting (figure 1b). Of the 35 aphidicolin induced fragile sites in the elephant shrew, six (2.5 expected, p = 0.02) (figure 1c) were coincidental with the 13 evolutionary breakpoints previously identified in this species using human chromosome painting probes [24].
We conducted a more refined analysis of the afrotherian fragile sites by comparing those that are (i) expressed in a single species (i.e., species-specific fragile sites), and (ii) those fragile sites conserved between two or more species (i.e., conserved fragile sites). As above, we assessed each category of fragile site for correspondence with evolutionary breakpoints. Our aim was to test if conserved fragile sites, which are more likely to be ancient fragile sites, might show an enrichment of evolutionary breakpoints. This was borne out by the data which show that of the 12 evolutionary breakpoints identified in the aardvark by reciprocal painting with human painting probes [53], three are coincidental with aardvark-specific fragile sites (1.15 bands are expected if the distribution was random, p = 0.001) and four are coincidental with conserved fragile sites (0.7 bands are expected if the distribution was random, p = 0.001). Similarly, a significant association was found in the elephant shrew (p = 0.03). However, in the case of the golden mole, the tendency was not significant. These data suggest, therefore, that evolutionary breakpoints tend to concentrate more frequently in conserved fragile sites than in those that are species-specific, although only significantly so in two of the three species analysed. Finally, three conserved fragile sites were shared between all three afrotherian species (located in bands 1q28, 1q44 and 3q24 in the aardvark, bands 1q28, 1q32 and 3q24 in the golden mole, and bands 1p22, 1q15 and 1q32 in the elephant shrew). One of these (corresponding to band 1q28 in the aardvark, 1q18 in the golden mole and 1q15 in the elephant shrew) was coincidental with the site of an evolutionary breakpoint in all three species – that corresponding to HSA 2/8, the only chromosomal synteny which strongly supports the Afroinsectiphillia (golden moles, elephant shrews and aardvark) to the exclusion of the elephant [24]).
Discussion
Fragile sites and chromosomal evolution
This investigation confirms and extends earlier observations that fragile sites form part of the chromosomal structure in mammals, and that the characteristics underlying their susceptibility to breakage have been conserved during evolution [26, 36, 37, 51, 55, 56]. Using data from fragile site expression, G-banding analysis, and cross-species chromosome painting, we have identified fragile sites in aardvark, golden mole and elephant-shrew (Afroinsectiphillia) that are located in homologous chromosomal positions in these species. We detected 11 conserved fragile sites in aardvark genome, eight in golden mole, and 10 fragile sites in the elephant-shrew (figure 1). Fragile sites detected in more than one species were regarded as "conserved fragile sites" in order to distinguish them from those that were species-specific.
Although fragile sites have been considered "hot spots" for evolutionary reorganization in a variety of mammalian species (i.e. are regions where chromosomal rearrangements such as fusion/fissions and inversions can repeatedly occur), the data are limited to a single clade, Boreoeutheria [26–28]. This begs the question whether this fragility is a more general phenomenon in mammals, and whether the evolutionary breakpoints that characterise human syntenies detected in the basal Afrotheria similarly correspond at the chromosomal band level with fragile sites detected in other species when using a chemical challenge. Twenty nine fragile sites were detected in aardvark, 27 in golden mole and 35 in the elephant-shrew (Figure 1). A contingency analysis shows that there is a significant association for bands that contain evolutionary breakpoints to accumulate fragile sites in the aardvark (p = 0.0004), as well as in the Cape rock elephant shrew (p = 0.02) genomes. The association was not statistically significant in the golden mole, but there is, nonetheless, a tendency for bands that contain evolutionary breakpoints to accumulate fragile sites in this species (p = 0.07).
The inclusion of the elephant/aardvark chromosome painting data into the analysis offered an opportunity to compare old (aardvark vs. human) and phylogenetically younger evolutionary breakpoints (aardvark vs. elephant) and their correlation with chromosomal fragility. We reasoned that younger breakpoints may show a greater correlation with afrotherian fragile sites than their more ancient counterparts identified in the human vs. afrotherian comparisons. The analysis, however, did not reveal marked differences between the two types of evolutionary breakpoints. When the human chromosomal homologies are plotted against the aardvark genome, seven of the 12 evolutionary breakpoints co-localize with fragile sites (1.8 expected, p = 0.0004). Plotting the elephant chromosomal homologies to the aardvark genome revealed that eight of 19 evolutionary breakpoints co-localize with fragile sites (3 expected, p = 0.0032) indicating that the mechanism causing the fragility is conserved. Given these findings, we then proceeded to determine whether the data inform previous conclusions on chromosomal syntenies thought to underpin the recognition of Afrotheria as one of the four major supraordinal clades of placental mammals (Placentalia).
Chromosomal signatures in Afrotheria
The recognition of a monophyletic afrotherian clade was initially based on DNA sequence comparisons [13, 57] and subsequently supported by the analysis of large concatenations of nuclear and mitochondrial genes [1, 2, 4, 5, 16, 58, 59], unique insertion and deletion events (indels) [8, 18, 19], comparative cytogenetic studies [23, 24, 60], morphology [61, 62], placentation [63] and, most recently, by whole genome assemblies [11]. Given that Afrotheria is thought to be near the root of the eutherian tree, the organization of their genomes could provide unique insights into the dynamics of mammalian chromosomal evolution.
Two human syntenies have been proposed by chromosome painting studies to link afrotherians to the exclusion of other placental mammals (1/19p [23, 53] and 3/21/5 [24]). In terms of the former, it is noteworthy that HSA 1/19 has been reported in a xenarthran species (Tamandua tetradactyla, [64, 65]), the insectivore shrew-hedgehog (Neotetracus sinensis, [66]), the pig [67], as well as in the prosimians Galago moholi, Otolemur crassicaudatus and Nycticebus coucang [68, 69], the New World monkeys Saimiri sciureus [70] and Callicebus lugens [71], and the Old World monkeys Presbytis cristata [72], Pygathrix nemaeus [73], Nasalis larvatus [74], Trachypithecus francoisi [74] and T. phayrei [74]. However, only three of these studies relied on reciprocal chromosomal painting [23, 53, 69] with a fourth [67] based on unidirectional painting, but complemented by comparative gene mapping (figure 2). These data are a prerequisite for allowing unequivocal identification of the chromosomal arms (either 19p or 19q) involved in the 1/19 syntenies. Of these, G. moholi and N. coucang [69] show an HSA 1q/19q association and the pig HSA 1p/19q [67]. In contrast, the African elephant and the aardvark share HSA 1/19p [23, 53] begging more detailed analysis of whether 1/19p is truly an afrotherian specific chromosomal signature (figure 2).
Phylogenetic tree showing the HSA 1/19 chromosomal syntenies in different mammalian species. In this scheme we place Afrotheria at the root. There are other competing hypotheses for the basal resolution of Placentalia (see text for details). The ancestral chromosomal forms corresponding to HSA 1, HSA 19p and HSA 19q/16q are represented as single conserved entities in both the therian (Marsupialia + Placentalia) and boreoeutherian ancestors. Data based on chromosomal painting in the African elephant (Afrotheria, [23]), the anteater (Xenarthra, [64, 65]), loris (Primates, [69]), the shrew-hedgehog (Eulipotyphla, [66]) and the pig (Cetartiodactyla, [67]) are represented. The human/opossum homologies are determined from ENSEMBL genome sequence alignments [82]. Question marks indicate instances of ambiguity where reciprocal chromosomal painting has not been performed and therefore unequivocal identification of chromosomal arms is not possible. Inset shows G-banding comparisons between elephant chromosome 2 (LAF 2) and human chromosomes 1 and 19 (HSA 1 and HSA 19); syntenic boundaries are derived from reciprocal chromosome painting data [23]. Note that HSA 1 is inverted to facilitate comparisons with LAF 2. Centromeres are marked by asterisks.
By computationally scanning the genomic assemblies of human and opossum (a marsupial outgroup to Afrotheria and other Placentalia) we sought to validate the HSA 1/19p synteny as a lineage specific, rare genomic change underpinning the monophyly of Afrotheria. In attempting to address this it is important to point out that the defining character in a conserved segmental association is the presence of the breakpoint (i.e. the junctions 1/19p and 3/21/5) since, as has been noted elsewhere [28, 60], the size of segments may be altered by subsequent translocations to other regions in the genome, and FISH provides no insight to gene order within the syntenic block which may be altered by intrachromosomal rearrangement. Furthermore, in deciding the most parsimonious pathway to derive a specific chromosomal rearrangement we follow [22] in viewing the independent disruption of a syntenic group to be more likely than the same adjacent synteny being independently reassembled in different lineages. While the opossum genome shows a HSA 1p/19q/1p/19q/1p/19q/1p association on its chromosome 4 (figure 3), this adjacent synteny is different to the HSA 1q/19q found in the prosimian species G. moholi and N. coucang ([69] and figure 2) further reinforcing the finding that HSA 1, reportedly the largest physical unit in the eutherian ancestral genome, has suffered multiple independent fissions [75].
Importantly, however, and of substantial phylogenetic significance, the human chromosomal segment involved in the HSA 1/19p afrotherian synteny is currently ambiguous since the painting data do not allow inference on whether the junction is between HSA 1q or HSA 1p in those species for which there are reciprocal painting data (i.e., elephant and aardvark [23, 53]). G-banding homology on the other hand favours HSA 1p/19p (see insert figure 2). If correct, this association would support the monophyly of Afrotheria [23, 53] on current information. However, should further analysis reveal its presence in Xenarthra, this would give credence to the recognition of Atlantogenata [76], a clade containing Afrotheria and Xenarthra to the exclusion of Boreoeutheria [11]. Both outcomes underscore the importance of resolving this critical chromosomal synteny for clarifying deep divergences in the eutherian tree although recent strong support for a sister group relationship for Afrotheria and Xenarthra (based on a ~2.2 mega-base data set of protein coding sequences), clearly tips the odds in favour of HSA 1(p?)/19p being a shared synteny for Atlantogenata [12].
Moreover, it is of interest that the human syntenies HSA 1, HSA 16q/19q and HSA 19p have all previously been proposed for the boreoeutherian ancestor [22, 28, 77] which, if present in the eutherian ancestor, would require a fusion (possibly promoted by an ancient fragile site retained in aardvark OAF 3q29 – figure 1a) to derive the HSA 1/19p synteny that possibly unites Afrotheria to the exclusion of other Placentalia (figure 2). If the same syntenies were present at the therian root all that would be required is a breakpoint in HSA19q with a fusion to HSA1 to give the opossum HSA 1p/19q and HSA1 6q/19q combinations (figure 2).
Using a similar approach we examined the second synteny thought to underpin Afrotheria monophyly, HSA 3/21/5. It was previously argued [24] that the ancestral association HSA 3/21 [53] should be expanded to include segments homologous to human chromosome 5 forming an HSA 3/21/5 segmental combination defining Afrotheria. The authors posit that the most parsimonious explanation for the observed patterns is that HSA 21 appears to have fissioned within Paenungulata; in this regard it is noteworthy that 2q31 is expressed as a fragile site in aardvark (figure 1a), perhaps indicating an ancestral locus. Their reasoning was that the HSA 3/21/5 configuration is present in aardvark, golden mole and elephant-shrew with all three chromosomes retained as intact, conserved entities in the two former species. In the case of the elephant, the fissioning of HSA 21 gave rise to HSA 5/21 (LAF 3) and HSA 1/3/21/3 on LAF 21 (see [23] and inset in figure 4). This rearrangement (the modification of 3/21/5 to HSA 3/21 + HSA 5/21) was recently confirmed [54] in manatee and elephant (data on the hyrax are incomplete) by reciprocal painting with paenungulate species-specific painting probes, and through inferences made from human and aardvark [53]. New information on the Florida manatee (Trichechus manatus latirostris, TMA, [78]) resulting from unidirectional painting experiments with human probes similarly show HSA 5/21 on TMA 1, and HSA 2/3/21 on TMA 15.
Phylogenetic tree showing the HSA 3/21 chromosomal syntenies in different mammalian species. In this scheme we place Afrotheria at the root. There are other competing hypotheses for the basal resolution of Placentalia (see text for details). The ancestral chromosomal forms corresponding to HSA 3/21 are represented as single conserved entities in the amniote, therian, eutherian and boreoeutherian ancestors. Source references for the species shown in the tree are: African elephant [23], domestic pig [84], rabbit [85], tree shrew [86], grey squirrel [87], domestic cat [88], aardvark [53], the xenarthran taxa the amadillo and lesser anteater [64], and the ENSEMBL genome database [82] for the rat, mouse, cattle, dog, opossum and chicken sequence alignments. The asterisks indicate the unequivocal identification of the HSA 3p/21 synteny based on reciprocal chromosome painting or data from the ENSEMBL genome database [82]. The estimated divergence dates follow [11]. The opossum and the chicken are included as representative outgroups species. Inset shows G-banding comparisons between elephant chromosomes 3 (LAF 3) and 21 (LAF 21) and aardvark chromosome 2 (OAF 2) showing the regions of homology to human chromosomes 3, 5 and 21 (HSA 3, HSA 5 and HSA 21); syntenic boundaries are derived from reciprocal chromosome painting data [23].
The widely accepted boreutherian ancestral syntenies (HSA 4/8/4, 7a/16p, 10/12a/22a, 12b/22b, 14/15 and 16q/19q, [22, 77]) are all in the opossum genome suggesting their presence in a therian ancestor, and retention for >180 mya (divergence based on stem branches between crown placentals and crown marsupials, [79–81]). One further ancestral synteny (HSA 3/21) deserves special comment, especially with respect to its importance for Afrotheria. Froenicke [22] provided evidence that neighbouring segments homologous to HSA 3 and HSA 21 have been found in all eutherian orders for which there is information, and that the combined analysis of reciprocal chromosome painting data in conjunction with draft genome sequence information for mouse and human showed that the breakpoint is located in HSA 3p, the region closest to the centromere of this chromosome. While there is no evidence of this synteny in opossum, electronic screening of the chicken genome assembly indicates its retention in this species (the HSA 3p segment extends from 76Mb-90 Mb, [82], see figure 3). Opossum has a 21/Xp/3q/Xp/3q synteny in chromosome 4 and, importantly, 3p/3q/Xp in chromosome 7 indicating different breakpoints in Marsupialia (figure 4). In summary therefore, it would seem that HSA 3p/21 was present in the common ancestor of Amniota (i.e. common to turtles, lepidosaurs, crocodilians, birds and mammals) supporting its identification as an ancestral synteny that was present >310 MYA [79], but which was disrupted in the lineage leading to the marsupials. The expansion to include HSA 5 in the afrotherian ancestor is unique [24] and defines the monophyly of this ancient African clade of mammals.
Conclusion
Using data from fragile site expression, G-banding analysis, and cross-species chromosome painting, we have described a suite of afrotherian common fragile sites that are correlated with evolutionary breakpoints when compared to the human genome. By computationally scanning the newly released opossum and chicken genomes as outgroups to Placentalia, we have shown that the primitive HSA 3p/21 synteny was present in the amniote ancestor, and its expansion to include HSA 5 validates the HSA 3/21/5 synteny as a robust cytogenetic signature that defines the monophyly of Afrotheria. Its fission into two segments (HSA 3p/21 + HSA 5/21) is probably shared by all Paenungulata and may have been facilitated by an ancient fragile site that is still expressed in aardvark. Further, if the human syntenies HSA 1, HSA 16q/19q and HSA 19p (all previously proposed for the boreoeutherian ancestor) were present at the eutherian root, a single fusion (the breakpoint junction being coincidental with a fragile site retained in aardvark at OAF 3q29) would be required to derive the HSA 1/19p synteny that may, with further refined analysis, be found to unite Afrotheria to the exclusion of other Placentalia.
Methods
Cell culture and fragile site expression
Fibroblast cell cultures were established from two aardvark (one male and one female), three golden mole (two males and one female) and four elephant-shrew specimens (two females and two males). Twenty four hours before harvesting 50 μl of aphidicolin (APC, 2 mM) dissolved in DMSO was added to cell cultures at a final concentration of 0.2 μM. Concurrent control cultures were established for each experiment. Cells were harvested and chromosomal preparations obtained using standard protocols.
Ideograms were constructed for each of the species according to the standardised karyotype for O. afer (2n = 20, OAF [53]), C. asiatica (2n = 30, CAS [24]) and E. edwardii (2n = 28, EED [24]). (Note that the karyotype presented in [24] was originally incorrectly identified as E. rupestris; DNA from the same specimen was subsequently extracted and sequenced, verifying its status as E. edwardii; unpublished data). Our banding data show that 177 chromosomal bands define the O. afer ideogram, whereas the C. asiatica and E. edwardii ideograms have 200 and 182 bands, respectively.
All metaphases were sequentially solid-stained and then G-banded to establish the location of the breakpoints. Digital images were captured with a BX60 Olympus microscope utilizing the GENUS imaging System (version 2.75) (Applied Imaging Corporation).
Analysis of fragile sites
In order to identify which chromosomal bands could be considered fragile regions, a statistical analysis of the distribution of chromosomal abnormalities detected in each specimen analysed was undertaken using the programme FSM (version 995, [83]). In this program, the standardized χ2 and G2 test statistics are used for all chromosomal bands that express non-random breaks or gaps. The hypothesis tested by the programme is that the probability of breakage is equal for all chromosomal bands in a given karyotype using a 0.05 level of significance (α). The FSM analysis gives a critical value for each data set analysed, abbreviated to Cα, which indicates the lowest frequency of breakage per band that exceeds the level of significance [35, 83]. Any chromosomal band with a number of breaks greater than the critical value is considered a fragile site. This value ranged from 3 to 4 depending on the number of chromosomal bands determined in each karyotype (177 bands for the aardvark, 200 bands for the golden mole and 182 bands for the elephant shrew), and the number of breaks detected in each species' data set. In our analyses, chromatid and chromosomal breaks and gaps were treated equally as representing single chromosomal events, and these were mapped to the respective ideogram of each afrotherian species.
Computational analysis of afrotherian and boreoeutherian lineage-specific syntenies
Data from different studies [24, 53, 54, 78] were used as sources for determining homologies between the human genome and those of the aardvark, golden mole and Cape rock elephant shrew. Evolutionary breakpoints were defined as the limit between each adjacent human homologous region. Evolutionary bands are chromosomal bands that contain evolutionary breakpoints (see [27] for further clarification of evolutionary breakpoints and evolutionary bands). We omitted the centromeres from the analysis and plotted all fragile sites and evolutionary breakpoints onto the ideograms of each of the afrotherian species analysed. A contingency analysis was used (JMP package version 5.1.2; SAS Institute Inc.) to evaluate if evolutionary breakpoints concentrate significantly (p ≤ 0.05) in chromosomal bands containing fragile sites. This is based on the assumption that the chromosomal bands of the genomes of each afrotherian species analysed have the same probability to be affected, independently of the size of the band involved.
The Ensembl genome browser of the Sanger Center and EMBL data base [82] were used for determining homologies between the human genome and those of the opossum and chicken. We used the completed human/chicken (WASHUC 1) and human/opossum (MonDom 4.0) whole-genome sequence assemblies that are available on the Ensembl genome browser [82] to determine syntenic regions between the human genome (NCBI Build 36) and those of opossum and chicken
References
Murphy WJ, Eizirik E, Jonson WE, Zhang YP, Ryder OA, O'Brien SJ: Molecular phylogenetics and the origins of placental mammals. Nature. 2001, 409: 614-618. 10.1038/35054550.
Murphy WJ, Eizirik E, O'Brien SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS: Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001, 294: 2348-2351. 10.1126/science.1067179.
Waddell PJ, Kishino H, Ota R: A phylogenetic foundation for comparative mammalian genomics. Genome Inform. 2001, 12: 141-154.
Waddell PJ, Shelley S: Evaluating placental inter-ordinal phylogenies with novel sequences including RAG1, gamma-fibrinogen, ND6, and mt-tRNA, plus MCMC-driven nucleotide, amino acid, and codon models. Mol Phylogenet Evol. 2003, 28: 197-224. 10.1016/S1055-7903(03)00115-5.
Amrine-Madsen H, Koepfli K-P, Wayne RK, Springer MS: A new phylogenetic marker, apolipoprotein B, provides compelling evidence for eutherian relationships. Mol Phylogenet Evol. 2003, 28: 225-240. 10.1016/S1055-7903(03)00118-0.
Nikolaev S, Montoya-Burgos JI, Margulies EH, NISC Comparative Sequencing Program, Rougemont J, Nyffeler B, Antonarakis SE: Early History of Mammals Is Elucidated with the ENCODE Multiple Species Sequencing Data. PLoS Genet. 2007, 3: e2-10.1371/journal.pgen.0030002.
Waters PD, Dobigny G, Waddell PJ, Robinson TJ: Evolutionary History of Line-1 in the Major Clades of Placental Mammals. PLoS One. 2007, 2: e158-10.1371/journal.pone.0000158.
Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J: Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006, 4: e91-10.1371/journal.pbio.0040091.
Hunter JP, Janis CM: Spiny Norman in the Garden of Eden? Dispersal and early biogeography of Placentalia. J Mammal Evol. 2006, 13: 89-123. 10.1007/s10914-006-9006-6.
McKenna MC: Toward a phylogenetic classification of the Mammalia. Phylogeny of the Primates. Edited by: Luckett WP, Szalay FS. 1975, New York: Plenum Press, 21-46.
Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W: Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007, 17: 413-421. 10.1101/gr.5918807.
Hallström B, Kullberg M, Nilsson M, Janke A: Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sistergroups. Mol Biol Evol. 24 (9): 2059-2068. 10.1093/molbev/msm136.
Springer MS, Cleven GC, Madsen O, de Jong WW, Waddell VG, Amrine HM, Stanhope MJ: Endemic African mammals shake the phylogenetic tree. Nature. 1997, 388: 61-64. 10.1038/40386.
Rokas A, Holland PW: Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000, 15: 454-459. 10.1016/S0169-5347(00)01967-4.
Shedlock AM, Okada N: SINE insertions: a powerful tools for molecular systematics. BioEssays. 2000, 22: 148-160. 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z.
Madsen O, Scally M, Douady CJ, Kao DJ, DeBry RW, Adkins R, Amrine HM, Stanhope MJ, de Jong WW, Springer MS: Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001, 409: 610-614. 10.1038/35054544.
Nikaido M, Nishihara H, Hukumoto Y, Okada N: Ancient SINEs from African endemic mammals. Mol Biol Evol. 2003, 20: 522-527. 10.1093/molbev/msg052.
Nishihara H, Satta Y, Nikaido M, Thewissen JGM, Stanhope MJ, Okada N: A Retroposon Analysis of Afrotherian Phylogeny. Mol Biol Evol. 2005, 22: 1823-1833. 10.1093/molbev/msi179.
Nishihara H, Hasegawa M, Okada N: Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci USA. 2006, 103: 9929-9934. 10.1073/pnas.0603797103.
O'Brien SJ, Stanyon R: Ancestral primate viewed. Nature. 1999, 402: 365-366. 10.1038/46450.
Wienberg J: The evolution of eutherian chromosomes. Curr Opin Genet Dev. 2004, 14: 657-666. 10.1016/j.gde.2004.10.001.
Froenicke L: Origins of primate chromosomes – as delineated by Zoo-FISH and alignments of human and mouse draft genome sequences. Cytogenet Genome Res. 2005, 108: 122-138. 10.1159/000080810.
Froenicke L, Wienberg J, Stone G, Adams L, Stanyon R: Towards the delineation of the ancestral eutherian genome organization: comparative genome maps of human and the African elephant (Loxodonta africana) generated by chromosome painting. Proc R Soc Lond B Biol Sci. 2003, 270: 1331-1340. 10.1098/rspb.2003.2383.
Robinson TJ, Fu B, Ferguson-Smith MA, Yang F: Cross-species chromosome painting in the golden mole and elephant-shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc R Soc Lond B Biol Sci. 2004, 271: 1477-1484. 10.1098/rspb.2004.2754.
Pevzner P, Tesler G: Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci USA. 2003, 100: 7672-7677. 10.1073/pnas.1330369100.
Ruiz-Herrera A, Ponsà M, García F, Egozcue J, Garcia M: Fragile sites in human and Macaca fascicularis are breakpoints in chromosome evolution. Chromosome Res. 2002, 10: 33-44. 10.1023/A:1014261909613.
Ruiz-Herrera A, Castresana J, Robinson TJ: Is mammalian evolution driven by regions of genome fragility?. Genome Biol. 2006, 7: R115-10.1186/gb-2006-7-12-r115.
Robinson TJ, Ruiz-Herrera A, Froenicke L: Dissecting the mammalian genome – new insights into chromosomal evolution. Trends Genet. 2006, 22: 297-301. 10.1016/j.tig.2006.04.002.
Sutherland GR: Fragile sites on human chromosomes: demonstration of their dependence on the type of tissue culture medium. Science. 1977, 197: 265-266. 10.1126/science.877551.
Sutherland GR, Baker E: The clinical significance of fragile sites on human chromosomes. Clin Genet. 2000, 58: 157-161. 10.1034/j.1399-0004.2000.580301.x.
Sutherland GR, Richards RI: Fragile sites-cytogenetic similarity with molecular diversity. Am J Hum Genet. 1999, 64: 354-359. 10.1086/302267.
Robinson TJ, Elder FF: Multiple common fragile sites are expressed in the genome of the laboratory rat. Chromosoma. 1987, 96: 45-49. 10.1007/BF00285882.
Tewari R, Juyal RC, Thelma BK, Das BC, Rao SRV: Folate-sensitive fragile sites on the X-chromosomes heterochromatin of the Indian mole rat Nesokia indica. Cytogenet Cell Genet. 1987, 44: 11-17.
Elder FFB, Robinson TJ: Rodent common fragile sites, are they conserved? Evidence from mouse and rat. Chromosoma. 1989, 97: 459-464. 10.1007/BF00295030.
McAllister B, Greenbaum IF: How common are common fragile sites: variation of aphidicolin-induced chromosomal fragile sites in a population of the deer mouse (Peromyscus maniculatus). Hum Genet. 1997, 100: 182-188. 10.1007/s004390050487.
Rozier L, El-Achkar E, Apiou F, Debatisse M: Characterization of a conserved aphidicolin-sensitive common fragile site at human 4q22 and mouse 6C1: possible association with an inherited disease and cancer. Oncogene. 2004, 23: 6872-6880. 10.1038/sj.onc.1207809.
Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T: Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006, 16: 1222-1230. 10.1101/gr.5335506.
Riggs PK, Kuczek T, Chrisman CL, Bidwell CA: Analysis of aphidicolin-induced chromosome fragility in the domestic pig (Sus scrofa). Cytogenet Cell Genet. 1993, 62: 110-116.
Yang MY, Long SE: Folate sensitive common fragile sites in chromosomes of the domestic pig (Sus scrofa). Res Vet Sci. 1993, 55: 231-235.
Ronne M: Localization of fragile sites in the karyotype of Sus scrofa domestica: present status. Hereditas. 1995, 122: 153-162. 10.1111/j.1601-5223.1995.00153.x.
Rodriguez V, Llambi S, Postiglioni A, Guevara K, Rincon G: Localisation of aphidicolin-induced break points in Holstein-Friesian cattle (Bos taurus) using RBG-banding. Genet Sel Evol. 2002, 34: 649-656. 10.1051/gse:2002029.
Ronne M: Putative fragile sites in the horse karyotype. Hereditas. 1992, 117: 127-136.
Stone DM, Stephens KE, Doles J: Folate-sensitive and aphidicolin-inducible fragile sites are expressed in the genome of the domestic cat. Cancer Genet Cytogenet. 1993, 65: 130-134. 10.1016/0165-4608(93)90221-7.
Ronne M: Localization of fragile sites in the karyotypeof Felis catus. Hereditas. 1995, 122: 279-283. 10.1111/j.1601-5223.1995.00279.x.
Kubo K, Matsuyama S, Sato K, Shiomi A, Ono K, Ito Y, Ohashi F, Takamori Y: Novel putative fragile sites observed in feline fibroblasts treated with aphidicolin and fluorodeoxyuridine. J Vet Med Sci. 1998, 60: 809-813. 10.1292/jvms.60.809.
Stone DM, Jacky PB, Hancock DD, Prieur DJ: Chromosomal fragile site expression in dogs: I. Breed specific differences. Am J Med Genet. 1991, 40: 214-22. 10.1002/ajmg.1320400219.
Stone DM, Stephens KE: Bromodeoxyuridine induces chromosomal fragile sites in the canine genome. Am J Med Genet. 1993, 46: 198-202. 10.1002/ajmg.1320460220.
Schmid M, Ott G, Haaf T, Scheres JMJC: Evolutionary conservation of fragile sites induced by 5-azacytidine and 5-azadeoxycytidine in man, gorilla and chimpanzee. Hum Genet. 1985, 71: 342-350. 10.1007/BF00388461.
Smeets DF, Van de Klundert FA: Common fragile sites in man and three closely related primate species. Cytogenet Cell Genet. 1990, 53: 8-14.
Fundia A, Gorostiaga M, Mudry M: Expression of common fragile sites in two Ceboidea species: Saimiri boliviensis and Alouatta caraya (Primates: Platyrrhini). Genet Sel Evol. 2000, 32: 87-97. 10.1051/gse:2000108.
Ruiz-Herrera A, Garcia F, Giulotto E, Attolini C, Egozcue J, Ponsà M, Garcia M: Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet GenomeRes. 2005, 108: 234-247. 10.1159/000080822.
Springer MS, Murphy WJ, Eizirik E, O'Brien SJ: Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003, 100: 1056-1061. 10.1073/pnas.0334222100.
Yang F, Alkalaeva EZ, Perelman PL, Pardini AT, Harrison WR, O'Brien PC, Fu B, Graphodatsky AS, Ferguson-Smith MA, Robinson TJ: Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral karyotype. Proc Natl Acad Sci USA. 2003, 100: 1062-1066. 10.1073/pnas.0335540100.
Pardini AT, O'Brien PC, Fu B, Bonde RK, Elder FFB, Ferguson-Smith MA, Yang F, Robinson TJ: Chromosome painting among Proboscidea, Hyracoidea and Sirenia: support for Paenungulata (Afrotheria, Mammalia) but not Tethytheria. Proc R Soc Lond B Biol Sci. 2007, 274: 1333-1340. 10.1098/rspb.2007.0088.
Glover TW, Hoge AW, Miller DE, Ascara-Wilke JE, Adam AN, Dagenais SL, Wilke CM, Dierick HA, Beer DG: The murine Fhit gene is highly similar to its human orthologue and maps to a common fragile site region. Cancer Res. 1998, 58: 3409-3414.
Uboldi C, Guidi E, Roperto S, Russo V, Roperto F, Di Meo GP, Iannuzzi L, Floriot S, Boussaha M, Eggen A, Ferretti L: Comparative genomic mapping of the bovine Fragile Histidine Triad (FHIT) tumour suppressor gene: characterization of a 2 Mb BAC contig covering the locus, complete annotation of the gene, analysis of cDNA and of physiological expression profiles. BMC Genomics. 2006, 7: 123-10.1186/1471-2164-7-123.
Stanhope MJ, Waddell VG, Madsen O, de Jong W, Hedges SB, Cleven GC, Kao D, Springer MS: Molecular evidence for multiple origins of Insectivora and for a new order of endemic African insectivore mammals. Proc Natl Acad Sci USA. 1998, 95: 9967-9972. 10.1073/pnas.95.17.9967.
Malia MJ, Adkins RM, Allard MW: Molecular support for Afrotheria and the polyphyly of Lipotyphla based on analyses of the growth hormone receptor gene. Mol Phylogenet Evol. 2002, 24: 91-101. 10.1016/S1055-7903(02)00219-1.
Murata Y, Nikaido M, Sasaki T, Cao Y, Fukumoto Y, Hasegawa M, Okada N: Afrotherian phylogeny as inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2003, 28: 253-260. 10.1016/S1055-7903(03)00035-6.
Robinson TJ, Seiffert E: Afrotherian origins and interrelationships: new views and future prospects. Curr Topics Dev Biol. 2004, 63: 37-60.
Sanchez-Villagra MR, Narita Y, Kuratani S: Thoracolumbar vertebral number: The first skeletal synapomorphy for afrotherian mammals. Syst Biodiversity. 2007, 5: 1-7. 10.1017/S1477200006002258.
Tabuce R, Marivaux L, Adaci M, Bensalah M, Hartenberger J-L, Mahboubi M, Mebrouk F, Tafforeau P, Jaeger JJ: Early Tertiary mammals from North Africa reinforce the molecular Afrotheria clade. Proc R Soc Lond B Biol Sci. 2007, 274: 1159-1166. 10.1098/rspb.2006.0229.
Carter AM, Blankenship TN, Enders AC, Vogel P: The fetal membranes of the otter shrews and a synapomorphy for Afrotheria. Placenta. 2006, 27: e268-
Svartman M, Stone G, Stanyon R: The ancestral Eutherian karyotype is present in Xenarthra. PLoS Genet. 2006, 2: e109-10.1371/journal.pgen.0020109.
Yang F, Graphodatsky AS, Li T, Fu B, Dobigny G, Wang J, Perelman PL, Serdukova NA, Su W, O'Brien PC, Wang Y, Ferguson-Smith MA, Volobouev V, Nie W: Comparative genome maps of the pangolin, hedgehog, sloth, anteater and human revealed by cross-species chromosome painting: further insight into the ancestral karyotype and genome evolution of eutherian mammals. Chromosome Res. 2006, 14: 283-296. 10.1007/s10577-006-1045-6.
Ye J, Biltueva L, Huang L, Nie W, Wang J, Jing M, Su W, Vorobieva NV, Jiang X, Graphodatsky AS, Yang F: Cross-species chromosome painting unveils cytogenetic signatures for the Eulipotyphla and evidence for the polyphyly of Insectivora. Chromosome Res. 2006, 14: 151-159. 10.1007/s10577-006-1032-y.
Biltueva LS, Yang F, Vorobieva NV, Graphodatsky AS: Comparative maps between the domestic dog and pig. Mamm Genome. 2004, 15: 809-818. 10.1007/s00335-004-2391-3.
Stanyon R, Koehler U, Consigliere S: Chromosome painting reveals that galagos have highly derived karyotypes. Am J Phys Anthropol. 2002, 117: 319-326. 10.1002/ajpa.10047.
Nie W, O'Brien PCM, Fu B, Wang J, Su W, Ferguson-Smith MA: Chromosome painting between human and lorisform prosimians: evidence for the HSA 7/16 synteny in the primate ancestral karyotype. Am J Phys Anthropol. 2006, 129: 250-259. 10.1002/ajpa.20299.
Stanyon R, Consigliere S, Müller S, Morescalchi A, Neusser M, Wienberg J: Fluorescent in situ hybridization (FISH) maps chromosomal homologies between the dusty titi and squirrel monkey. Am J Primatol. 2000, 50: 95-107. 10.1002/(SICI)1098-2345(200002)50:2<95::AID-AJP1>3.0.CO;2-8.
Stanyon R, Bonvicino CR, Svartman M, Seuanez HN: Chromosome painting in Callicebus lugens, the species with the lowest diploid number (2n = 16) known in primates. Chromosoma. 2004, 112: 201-206. 10.1007/s00412-003-0261-5.
Bigoni F, Stanyon R, Koehler U, Morescalchi AM, Wienberg J: Mapping homology between human and black and white colobine monkey chromosomes by fluorescent in situ hybridization. Am J Primatol. 1997, 42: 289-298. 10.1002/(SICI)1098-2345(1997)42:4<289::AID-AJP4>3.0.CO;2-T.
Bigoni F, Houck ML, Ryder OA, Wienberg J, Stanyon R: Chromosome painting shows that Pygathrix nemaeus has the most basalkaryotype among Asia Colobinae. Int J Primatol. 2004, 25: 679-688. 10.1023/B:IJOP.0000023580.46185.9c.
Bigoni F, Stanyon R, Wimmer R, Schempp W: Chromosome painting shows that the proboscis monkey (Nasalis larvatus) has a derived karyotype and is phylogenetically nested within Asian Colobines. Am J Primatol. 2003, 60: 85-93. 10.1002/ajp.10095.
Murphy WJ, Froenicke L, O'Brien SJ, Stanyon R: The origin of human chromosome 1 and its homologs in placental mammals. Genome Res. 2003, 13: 1880-1888.
Waddell PJ, Cao Y, Hasegawa M, Mindell DP: Assessing the Cretaceous superordinal divergence times within birds and placental mammals by using whole mitochondrial protein sequences and an extended statistical framework. Syst Biol. 1999, 48: 119-137. 10.1080/106351599260481.
Froenicke L, Caldes MG, Graphodatsky A, Müller S, Lyons LA, Robinson TJ, Volleth M, Yang F, Wienberg J: Are molecular cytogenetics and bioinformatics suggesting contradictory models of ancestral mammalian genomes?. Genome Res. 2006, 16: 306-310. 10.1101/gr.3955206.
Kellogg ME, Burkett S, Dennis TR, Stone G, Gray BA, McGuire PM, Zori RT, Stanyon R: Chromosome painting in the manatee supports Afrotheria and Paenungulata. BMC Evol Biol. 2007, 7: 6-10.1186/1471-2148-7-6.
Kumar S, Hedges SB: A molecular timescale for vertebrate evolution. Nature. 1998, 392: 917-920. 10.1038/31927.
Woodburne MO, Rich TH, Springer MS: The evolution of tribospheny and the antiquity of mammalian clades. Mol Phylogenet Evol. 2003, 28: 360-385. 10.1016/S1055-7903(03)00113-1.
van Rheede T, Bastiaans T, Boone DN, Hedges SB, de Jong WW, Madsen O: The platypus is in its place: nuclear genes and indels confirm the sister group relation of monotremes and therians. Mol Biol Evol. 2006, 23: 587-597. 10.1093/molbev/msj064.
The Ensembl genome browser of Sanger Center and EMBL database. [http://www.ensembl.org]
Böhm U, Dahm PF, McAllister BF, Greenbaum IF: Identifying chromosomal fragile sites from individuals: a multinomial statistical model. Hum Genet. 1995, 95: 249-256. 10.1007/BF00225189.
Goureau A, Yerle M, Schmitz A, Riquet J, Milan D, Pinton P, Frelat G, Gellin J: Human and porcine correspondence of chromosome segments using bidirectional chromosome painting. Genomics. 1996, 36: 252-62. 10.1006/geno.1996.0460.
Korstanje R, O'Brien PCM, Yang F, Rens W, Bosma AA, van Lith AH, van Zutphen LFM, Ferguson-Smith MA: Complete homology maps of the rabbit (Oryctolagus cuniculus) and human by reciprocal chromosome painting. Cytogenet Cell Genet. 1999, 86: 317-322. 10.1159/000015325.
Müller S, Stanyon R, O'Brien PCM, Ferguson-Smith MA, Plesker R, Wienberg J: Defining the ancestral karyotype of all primates by multidirectional chromosome painting between tree shrews, lemurs and humans. Chromosoma. 1999, 108 (6): 393-400. 10.1007/s004120050391.
Stanyon R, Stone G, Garcia M, Froenicke L: Reciprocal chromosome painting shows that squirrels, unlike murid rodents, have a highly conserved genome organization. Genomics. 2003, 82: 245-249. 10.1016/S0888-7543(03)00109-5.
Wienberg J, Stanyon R, Nash WG, O'Brien PC, Yang F, O'Brien SJ, Ferguson-Smith MA: Conservation of human vs. feline genome organization revealed by reciprocal chromosome painting. Cytogenet Cell Genet. 1997, 77: 211-217.
Acknowledgements
Financial support to TJR from the National Research Foundation, South Africa and a postdoctoral fellowship from the Spanish Ministry of Education and Science (MEC) to ARH are gratefully acknowledged. The authors thank Lutz Froenicke for providing insightful comment on an earlier draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declares that there are no competing interests.
Authors' contributions
ARH and TR conceived and designed the experiments. ARH performed the experiments and analysed the fragile site data. Both authors scrutinised evolutionary breakpoint for use in a phylogenetic context, and contributed equally to writing the manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ruiz-Herrera, A., Robinson, T.J. Chromosomal instability in Afrotheria: fragile sites, evolutionary breakpoints and phylogenetic inference from genome sequence assemblies. BMC Evol Biol 7, 199 (2007). https://doi.org/10.1186/1471-2148-7-199
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-7-199