Abstract
Background
A diversity of hypotheses have been proposed based on both morphological and molecular data to reveal phylogenetic relationships within the order Cetacea (dolphins, porpoises, and whales), and great progress has been made in the past two decades. However, there is still some controversy concerning relationships among certain cetacean taxa such as river dolphins and delphinoid species, which needs to be further addressed with more markers in an effort to address unresolved portions of the phylogeny.
Results
An analysis of additional SINE insertions and SINE-flanking sequences supported the monophyly of the order Cetacea as well as Odontocete, Delphinoidea (Delphinidae + Phocoenidae + Mondontidae), and Delphinidae. A sister relationship between Delphinidae and Phocoenidae + Mondontidae was supported, and members of classical river dolphins and the genera Tursiops and Stenella were found to be paraphyletic. Estimates of divergence times revealed rapid divergences of basal Odontocete lineages in the Oligocene and Early Miocene, and a recent rapid diversification of Delphinidae in the Middle-Late Miocene and Pliocene within a narrow time frame.
Conclusions
Several novel SINEs were found to differentiate Delphinidae from the other two families (Monodontidae and Phocoenidae), whereas the sister grouping of the latter two families with exclusion of Delphinidae was further revealed using the SINE-flanking sequences. Interestingly, some anomalous PCR amplification patterns of SINE insertions were detected, which can be explained as the result of potential ancestral SINE polymorphisms and incomplete lineage sorting. Although a few loci were potentially anomalous, this study demonstrated that the SINE-based approach is a powerful tool in phylogenetic studies. Identifying additional SINE elements that resolve the relationships in the superfamily Delphinoidea and family Delphinidae will be important steps forward in completely resolving cetacean phylogenetic relationships in the future.
Similar content being viewed by others
Background
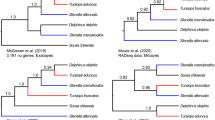
Extant cetaceans (whales, dolphins and porpoises), which consist of approximately 89 species in 14 families, are ecologically diverse, ranging from coastal to oceanic and from tropical to polar waters [1]. The order Cetacea has traditionally been divided into two highly distinct suborders: Mysticeti (the filter-feeding baleen whales) and Odontoceti (the echolocating toothed whales). Cetaceans differ dramatically from other mammals in terms of morphology, behavior and ecology, representing one of the most fascinating evolutionary transitions within vertebrates. The phylogeny of Cetacea has long attracted interest of evolutionary biologists and has been investigated using both morphological (including fossil) and molecular data [2–33]. Some of the issues have been well resolved including the monophyly of Cetacea [5, 12–17, 19–22] and its sister relationship with Hippoptamidae [10, 12, 13, 22–24]. However, these studies left unresolved issues: 1) the phylogenetic relationships of some major cetacean lineages; 2) the systematic status and phylogenetic position of some taxa such as the Ganges River dolphin or susu (Platanista gangetica) and the now nearly extinct Yangtze river dolphin or Baiji (Lipotes vexillifer), as well as those between the three delphinoid families: Monodontidae (narwhals and belugas), Phocoenidae (porpoises) and Delphinidae (dolphins) (Figure 1). The phylogenetic relationships among the various river dolphin genera (Inia, Pontoporia, Platanista, Lipotes) remain controversial, despite that a variety of studies have been conducted using a diverse array of systematic markers [12, 17, 31, 33], even in large concatenations of data [10]. The now nearly extinct Lipotes has been difficult to classify especially with respect to Inia and Pontoporia [12, 31]. Additionally, the position of Platanista at the base of Odontoceti was unstable, with conflicting evidence coming from morphology, mtDNA, and nuclear DNA (reviewed in [10]). In addition to these conflicts, previous phylogenetic hypotheses disagreed with one another in revealing relationships and diversity of the species within Delphinidae, especially within the Sousa-Delphinus-Tursiops-Stenella complex (Figure 2). In this complex, Tursiops truncatus (bottlenose dolphin) was long considered as the single species in the genus Tursiops, but recently two species, T. truncatus and T. aduncus, have been recognized as valid for this genus [34–36]. LeDuc et al. [34] suggested that T. aduncus was more closely related to the striped dolphin (Stenella coeruleoalba) than to the congener T. truncatus based on cytochrome b analysis. This is contrasted with morphological and other molecular evidence supporting Tursiops and Stenella as monophyletic genera [10, 11, 35].
SINEs (short interspersed elements) have been proposed as perfect molecular markers for studies of systematics, phylogenetics, evolution, and population biology, etc. [16, 22, 23, 31, 32, 37–47]. They have been successfully applied to resolve phylogenetic relationships among various groups at different taxonomic ranks [31, 32, 37, 39–42, 44]. SINEs are one of the major classes of retroposons that are dispersed throughout eukaryotic genomes. They are nonautonomous retroposons lacking the machinery to replicate themselves and they propagate in the genome via cDNA intermediating and are reintegrated into the host genome by retroposition [48–51]. Integration of a SINE sequence at a specific site in the genome is irreversible, and its target site is chosen almost at random [52]. To date, no mechanism has been described for the reversal of retroposon integration, and it is highly unlikely that the same type of retroposon would be integrated into the same genomic locus independently in different lineages [53]. SINEs, which are shared by some taxa but missing from the genomes of others, are ideal shared, derived phylogenetic characters at the molecular level [22, 31, 32, 37–47, 51–56]. Thus, a SINE sequence found at an orthologous locus in two or more lineages can be regarded as evidence for synapomorphy.
Nikaido and his colleagues pioneered the use of SINE insertions to address the relationships among cetaceans and other orders of mammals as well as to address relationships among both mysticetes and odontocetes [16, 22, 31, 32, 44]. For example, they examined 25 informative SINE insertions to support the monophyly of toothed whales and the paraphyly of river dolphins [31]. However, the interrelationships among some cetacean lineages, especially three families within Delphinoidea (i.e. Delphinidae, Phocoenidae and Mondontidae), were not well resolved with SINE markers, although their analysis of the SINE-flanking sequences supported the sister group relationship of Monodontidae and Phocoenidae with the exclusion of Delphinidae.
Thus, the main objectives of the present study are to: 1) address some of the remaining problematic areas of the cetacean phylogenetic tree through the analysis of additional SINE insertions and flanking sequences, and 2) utilize flanking sequences of 12 retroposed elements to estimate divergence times associated with the cetacean radiation. Identifying additional SINE elements that resolve the relationships within superfamily Delphinoidea and family Delphinidae will be important steps forward in completely resolving cetacean phylogenetic relationships in the future.
Results
Phylogenetic relationships
A total of 219 insertion loci were identified from random sequencing of genomic DNA from the Indo-Pacific bottlenose dolphin, screening genomic libraries from five species (i.e. long-beaked common dolphin, striped dolphin, Indo-Pacific humpbacked dolphin, Risso's dolphin, and finless porpoise), and screening the genome sequence of the common bottlenose dolphin. After eliminating loci that failed to amplify in all taxa (118 loci), were difficult to decipher (1 locus), and were present in all taxa (36 loci), 64 loci proved phylogenetically informative (Additional file 1 and 2).
Figure 3 shows the PCR patterns of 15 representative SINE loci in cetacean clades of A-J. Eight newly isolated SINE loci are present in all cetaceans but not in the hippopotamus, supporting the monophyly of the order Cetacea (clade A in Figures 3 and 4 and Additional file 1). Clade B represented the monophyly of the suborder Odontocete (toothed whales), which was supported by one independent locus Neop28 (Figures 3 and 4 and Additional file 1). Furthermore, we also elucidated the order from which toothed whales diverged. The sister relationship between sperm whales and the other toothed whales was supported by one SINE insertion Neop28 (Figures 3 and 4 and Additional file 1). The Ganges River dolphins and the remaining toothed whales formed a monophyletic group supported by the presence of four SINE insertions (clade C in Figures 3 and 4 and Additional file 1). The sister relationship between beaked whales and Yangtze River dolphin + Delphinoidea (Delphinidae + Phocoenidae +Mondontidae), as well as a sister relationship of the latter two families were supported by ten and thirteen SINE loci respectively (clade D and E in Figures 3 and 4 and Additional file 1). Finally, the monophyly of the superfamily Delphinoidea was supported by eleven informative loci (clade F in Figures 3 and 4 and Additional file 1). Within the superfamily Delphinoidea, the differentiation between Delphinidae and other two families was clearly suggested with four SINE insertions (clade G in Figures 3 and 4 and Additional file 1). Four SINE insertions indicate clades from H to J (Figures 3 and 4, Additional file 1). For example, the locus Plag35 and Plag113 indicated two species-specific integrations for the Ganges River dolphins, whereas the locus Turt127 indicated a species-specific insertion for the Common bottlenose dolphin.
Electrophoretic gel patterns of PCR products for 15 representative SINE loci. All loci analyzed in this study are shown in Additional file 1. Bands indicating the presence of the SINE are shown by black arrowheads, whereas gray arrowheads show those that indicate SINE absence. Loci are assigned alphabetically from A to J according to the clade on the phylogenetic tree shown in Figure 4. The species are numbered as follows: 1, Striped dolphin; 2, Risso's dolphin; 3, Indo-Pacific bottlenose dolphin; 4, Common bottlenose dolphin; 5, Long-beaked common dolphin; 6, Chinese white dolphin; 7, Pantropical spotted dolphin; 8, Beluga; 9, Finless porpoise; 10, Yangtze River dolphin; 11, Ginkgo-toothed beaked whale; 12, Ganges River dolphin; 13, Pygmy sperm whale; 14, Omura's whale; 15, Common minke whale; 16, hippopotamus.
Phylogenetic relationships of the major lineages of Cetaceans reconstructed using retroposon insertion data shown in Additional file 2. Closed vertical arrowheads denote insertions of retroposons into each lineage. All loci mapped onto the tree were newly isolated and characterized in the present study. Each clade is named alphabetically from A to J. Cetacean families are delimited by vertical lines to the right of the tree along with representative members.
Figure 5 shows the cetacean relationships inferred from Bayesian analysis of the 3, 974 sites of SINE-flanking sequences. The topology supported the monophyly of Odontoceti (toothed whales), with a posterior probability of 1.00. The basal divergence within odontocetes is between the physeteroids (with the pygmy sperm whale as the representative) and a clade (PP = 1.0) of remaining odontocete species. The sister relationship between Platanistidae (Indian River dolphins) and other dolphins and porpoises was weakly supported (PP = 0.59), whereas the relationship between Ziphiidae (beaked whales) and Lipotidae (Yangtze River dolphin) + Delphinoidea (Delphinidae + Phocoenidae +Mondontidae) was well supported with PP = 1.0, and the support for the sister relationship of the latter two families was significant (PP = 1.0). The oceanic dolphins and porpoises formed a clade (PP = 1.0), with a basal divergence between monophyletic Delphinidae (PP = 1.0) and a sister relationship of Phocoenidae (porpoises) and Monodontidae (narwhals and belugas) (PP = 1.0). Within the Delphinidae, the Risso's dolphin (G. griseus) was the sister group to the remaining delphinids, whereas the remaining delphinids were subdivided into two clades: one well supported clade T. aduncus + D. capensis (Figure 5, clade K; PP = 1.0), and the other weakly supported clade ((Sousa chinensis + St. coeruleoalba) + (T. truncatus + St. attenuata)) (Figure 5, clade L; PP = 0.83). As revealed in previous studies, two species of Tursiops (T. truncates and T. aduncus) and two species of Stenella (St. coeruleoalba and St. attenuata) did not form respective monophyletic clades, which suggested that both genera are not monophyletic.
Time-calibrated cetacean phylogeny derived from BEAST using the flanking regions of 12 retroposed elements. Numbers above the clades represent Bayesian posterior probabilities. Clade letters are identical to those in Table 1. Red boxes indicate nodes for which a prior calibration constraint distribution was used and blue boxes indicate divergence dates estimated without prior calibration constraints for that node. The bounds of the boxes correspond to the 95% highest posterior density (HPD) of each node.
Divergence time estimation
All estimated divergence dates for nodes with labels from A to N in Figure 5 were presented in Table 1. The split between Mysticeti and Odontoceti was estimated to have occurred in the Late Eocene, shortly before the appearance of the first documented fossil mysticete Llanocetus denticrenatus (~34.2 MYA) (Figure 5). Radiation of the major clades of Odontocetes (Physeteroidea, Platanistidae, Ziphiidea, Lipotidae, Delphinoidea) dated from 15.55 to 29.05 MYA (Figure 5 and Table 1), suggesting a rapid early radiation within the major Odontocete lineages. These estimates are close to and at some degree later than previous estimates which were primarily based on mitochondrial DNA sequences and other markers [10, 12, 19, 31]. The divergence of the three extant delphinoid families took place in the Middle Miocene, whereas the radiation of the crown Delphinid lineages appeared to occur in the Middle Miocene, while the Sousa-Delphinus-Tursiops-Stenella complex may have a recent divergence in the Middle-Late Miocene and Pliocene.
Anomalous PCR amplification patterns of retroposon insertions in cetaceans
Although the vast majority of SINE insertions in our study supported a single most parsimonious tree, two anomalies in the present SINE analysis of phylogenetics remain noteworthy. At the locus Stec35, it was present in the Ganges River dolphins, based on preliminary analysis of the agarose gel electrophoresis. However, further analysis of the DNA sequences indicated that a different SINE insertion has occurred near the insertion Stec35 locus (68-bp distance between the two loci) (Figure 6A and Additional file 3). This indicated that the locus Stec35 was absent in the genome of the Ganges River dolphins, instead, there was a novel species-specific insertion and we tentatively named it Plag35, owing to its discovery only in Platanista gangetica.
Potential confounding SINE insertions. Two samples of incongruent loci are shown. Picture 6A shows a near-parallel insertion event occurring at locus Stec35. Picture 6B is the agarose gel electrophoresis result of Turt164 from 15 cetacean samples. It is polymorphic in the two species of the genus Stenella. The species are numbered as in Figure 3.
The second anomaly came from the locus Turt164. St. attenuate and St. coeruleoalba exhibited the typical heterozygous profile consisting of the insertion amplicon (band A) and the lack of insertion PCR product (band B) (Figure 6B) at this locus, while nearly all other species examined (exclusive of T. truncatus and T. aduncus) amplified a single amplicon of band B corresponding to the lack of insertion allele (Figure 6B). In contrast, T. truncatus and T. aduncus generated the usual single band A of homozygote for the insertion allele. To confirm this polymorphic amplification, four more T. truncatus and T. aduncus individuals were examined and they all generated the same single band A. In order to investigate this interesting scenario, different amplicon types (i.e., band A and band B) were isolated, cloned and sequenced (see Methods). As shown in Additional file 4, the only difference between the sequence of amplicon A and B in both St. attenuate and St. coeruleoalba is the lack of a SINE element in B.
Discussion
Phylogeny of Odontoceti and its Oligocene radiation
Relationships among odontocete families obtained in the present study were broadly congruent with most previous molecular and morphological hypotheses [5, 7, 8, 12, 13, 19, 20, 29, 31, 33, 57–65]. For example, the monophyly of Odontoceti and the sister relationship of Physeteroidea to all other extant odontocetes (Figures 4 and 5), supported the SINE analysis of Nikaido et al. [31] and was compatible with the morphological evidence [29]. The grouping of Ziphiidae (beaked whales) with Delphinida to the exclusion of Platanistidae and Physeteroidea (clade D in Figures 3 and 4 and Additional file 1), was concordant with previous SINE insertion analyses [31], as well as the SINE-flanking sequence analysis in the present study (Figure 5).
The grouping of the four genera of 'river dolphins' in family Platanistidae or superfamily Platanistoidea [66] has long been challenged by both morphologists and molecular systematists [5, 7, 19, 31, 33, 60, 61, 67–69], and instead conflicting relationships of the four major river dolphin clades have been proposed (Figure 1). Although the lack of Inia and Pontoporia in the present study made it difficult to discern the phylogeny of river dolphins, the present finding that Platanista has no direct affinity with Lipotes clarifies that river dolphins are an artificial rather than a natural group, which is consistent with many previous molecular studies [7, 31, 33, 63, 69, 70].
Our estimates of divergence times suggested that the common ancestors of extant cetaceans occurred in the Late Eocene Epoch, prior to approximately 34.40 (33.52-36.09; 95% highest posterior density) MYA (Figure 5 and Table 1), slightly younger than several previous estimates [10, 11, 19, 31, 71], but conflicted dramatically with the Early Eocene split around 50 MYA proposed by Cassens et al. [33] based on only one delphinid calibration. The present estimate accorded closely with the earliest known fossil crown cetacean, the archaic mysticete Llanocetus denticrenatus (~34.2) [72]. In addition, the present study estimates divergence of the major Odontocete lineages such as Physeteroidea, Platanistidae, Ziphiidea, Lipotidae and Delphinoidea occurred primarily in the Early Oligocene and Early Miocene (Figure 5 and Table 1). Climate change from greenhouse to icehouse which occurred in the Late Eocene to Early Oligocene [73, 74] might have played an important role in the cetacean radiation. During that period, atmospheric CO2 level decreased, and the polar ice caps expanded rapidly, Southern Ocean upwelling and ocean productivity increased [75–78], which may explain the radiation of cetaceans [10–12, 31]. Early representatives of cetacean fossils including Ferecetotherium, Waipatia and Kentriodontidae were present in the Late Oligocene, demonstrating that these lineages were diverged during this time period [79–82].
Interrelationship within Delphinoidea and rapid divergence of Delphinidae
The interrelationships among the three families within Delphinoidea were disputed and several alternative branching patterns were proposed [2, 5, 7, 17, 19, 20, 29, 31, 33, 64, 65, 70, 83]. While several morphological and molecular studies agreed that a close relationship existed between Delphinidae and Phocoenidae [2, 7, 17, 29], other molecular analyses supported the sister relationship of Monodontidae and Phocoenidae [12, 13, 19, 20, 33, 65, 70, 83]. Besides these hypotheses, an unresolved relationship between the three families was mentioned in some studies [5, 29, 31, 64]. In the present study, the differentiation between Delphinidae and other two families was suggested with four SINE insertions, while no SINE was found to suggest the divergence between Phocoenidae and Mondontidae. However, SINE flanking-sequences analysis here further resolved the relationship among three Delphinoidea families (Delphinidae + (Monodontidae + Phocoenidae)), which was the same as those revealed in Waddell et al. [83], Nishida et al. [8, 65] and May-Collado and Agnarsson [70]. Within Delphinoidea, the divergence between Phocoenidae and Monodontidae was estimated at 11.39 (10.02-14.02; 95% highest posterior density) MYA (Figure 5 and Table 1), which are close to and at some degree later than previous analyses [10, 12, 19, 84], but are much younger than Nikaido et al. [31], which predicted the divergence at 20 (17-23) MYA on the basis of SINE flanking sequences using the calibration date (55 Myr) for the separation of Cetacean from the hippopotamus based on the relaxed clock of cytochrome b data (lacking fossil calibration). Our result is consistent with the age of the oldest representative fossil, the late Miocene phocoenid Salumiphocaena stocktoni [80].
Of the Delphinidae species examined, the sister relationship of Grampus griseus and Sousa-Delphinus-Tursiops-Stenella complex [10–13, 34, 70] was confirmed by SINE-flanking sequences analysis with a posterior probability of 1.00 (Figure 5). Within Sousa-Delphinus-Tursiops-Stenella complex, it was supported the closest affinity between Sousa and Stenella coeruleoalba, with T. truncatus and S. attenuata as their sister clades, then they cluster with a clade of D. capensis and T. aduncus. This is in contrast with Caballero et al. 's [9] and McGowen et al. 's [10] suggestion of the basal position of Sousa among delphinine, the alliance of Sousa with Stenella and Delphinus [11], or the alliance of Sousa with Steno [85]. Further, the sister relationship of T. aduncus and D. capensis obtained in the present study with a posterior probability of 1.00 was well congruent with the studies based on Mt genomes [12]. Obviously, the present SINE flanking sequence data rejected the monophyly of genera Tursiops and Stenella [79, 84, 86–88], although some branches were not supported by high posterior probability (Figure 5). The delphinids was estimated to radiate in the Middle-Late Miocene and Pliocene, with branch events taking place within a narrow time frame (3-6 MYA) (Figure 5 and Table 1). Unfortunately, no SINE insertion was identified to solve the relationship within Delphinidae and especially within Sousa-Delphinus-Tursiops-Stenella complex, and more SINEs are necessary to solve this problem.
Anomalous events in SINE-based phylogenetic analysis
Several potential anomalous SINE intertion events were revealed in the present study (Figure 6). These anomalies may have been brought about through near-parallel insertions, lineage sorting, and paralogous insertions, as discussed in previous studies [47].
A. Parallel insertion
According to Ray et al. [47], near-parallel insertion meant that a secondary independent SINE was inserted into a site near the insertion originally being studied. To detect whether this is the case in cetaceans, we sequenced and analyzed the insertions. At locus Stec35, the original insertion was not found in the Ganges River dolphins, while an additional independent insertion was found to occur near the first insertion (68-bp interval between them) (as shown in Additional file 3).
B. Anomalous PCR amplification patterns of Turt164: Paralogous insertion, incomplete lineage sorting, or introgressive hybridization?
Turt164 is another interesting SINE that appeared to be polymorphic (Figure 6B). For example, St. attenuata and St. coeruleoalba exhibited the typical heterozygous profile consisting of the insertion amplicon (band A) and the lack of insertion PCR product (band B), whereas a single PCR amplicon (band A or band B) was found in other representative species examined in the present study (Figure 6B).
Paralogous insertion [47, 89] might be a potential interpretation of this anomalous phenomenon. Only one band (band A) was amplified from the genus Tursiops, a scenario that can be interpreted as segmental duplications occurred around the locus Turt164 of genus Stenella. Further studies including more samples of Stenella species should be performed to confirm this interpretation.
Incomplete lineage sorting [44, 90] may also be an alternative cause. Rapid speciation might occur in the common ancestor of genera Tursiops and Stenella [10–13, 79] and Turt164 inserted into their genome during a short period. This insertion might have been fixed in genus Tursiops, but not in genus Stenella because of incomplete lineage sorting. However, because only a small number of Tursiops individuals were examined in this study, further studies including more samples of the two Tursiops species should be performed in the future to confirm this.
Introgression could be the third explanation for the anomalous PCR amplification pattern. Numerous cases of dolphin hybridization both in captivity and in the wild [91–94] have been reported. It is reasonable that insertion might have occurred only in the genome of Tursiops, however introgression between Tursiops and Stenella may have taken place at some time, which may explain the unexpected polymorphism of Turt164 between them (Figure 6B).
Conclusions
A series of additional SINEs were identified to support the monophyly of the order Cetacea as well as Odontoceti, Delphinoidea, and Delphinidae. Especially, several novel SINEs were found to differentiate Delphinidae with other two Delphinoidea families (i.e. Monodontidae and Phocoenidae), whereas the sister group relationship of Monodontidae and Phocoenidae with exclusion of Delphinidae was revealed by the SINE-flanking sequences. Furthermore, members of classical river dolphins and the genera Tursiops and Stenella were found to be paraphyletic. Estimates of divergence times based on the flanking regions of 12 retroposed elements using a relaxed-clock Bayesian approach furthered our understanding of the rapid radiation events in cetacean evolution. Interestingly, potential ancestral SINE polymorphisms and incomplete lineage sorting in Delphinidae were detected. Although a few loci are potentially anomalous, this study still demonstrated that SINE-based approach is a powerful tool in phylogenetic studies. Identifying additional novel SINE elements that resolve the relationships in the superfamily Delphinoidea and family Delphinidae will be important steps forward in completely resolving the cetacean phylogenetic relationships in the future.
Methods
DNA samples and location
Fifteen cetacean species (13 odontocetes and 2 mysticetes, Table 2.) were examined in this study, using hippopotamus as an outgroup. Because all the muscle tissues used in our study were collected from the incidentally killed or stranded dead individuals, no ethical approval is necessary in such cases. All tissue samples were subsequently frozen at -20°C. The voucher specimens were preserved at Nanjing Normal University. Total genomic DNA from muscle tissues was extracted with a standard phenol/chloroform procedure followed by ethanol precipitation [95]. For blood, we used the DNAeasy Blood Extraction Kit (Qiagen) in a separate laboratory facility.
Strategies to identify novel SINE elements
Three different procedures were applied to isolate and characterize novel phylogenetically informative SINEs from cetaceans.
Strategy 1
Considering that typical SINEs are often present in numbers that exceed 104 copies per genome, a sufficient amount of SINE sequences can usually be gained with 60 kbp genomic sequence data. In order to identify novel SINEs in the Indo-Pacific bottlenose dolphin, we used the strategy suggested by Okada et al. [96]. Genomic libraries were constructed for T. aduncus (Indo-Pacific bottlenose dolphin). Genomic DNA was first digested by HindIII, and then DNA fragments with the size of 1.5-2.5 kb were cut out of the gel and purified using QIAquick Gel Extraction Kit (QIAGEN). The purified DNA fragments were ligated into the plasmid vector pUC118 HindIII/BAP (TaKaRa) at 16°C overnight. Aliquots of the ligation reactions were transformed into Escherichia coli Top10 competent cells and plated for blue/white selection on media containing X-gal and IPTG. White clones were chosen, isolated, purified, and the inserts were then sequenced and analyzed employing an ABI PRISM 310 Automated Genetic Analyzer (Applied Biosystems, Foster City, CA) with universal (forward and reverse) M13 primers under the instruction of the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). 62 kb of genomic sequence data of the Indo-Pacific bottlenose dolphin were randomly sequenced. To find SINEs among these sequences, we aligned these sequences using CLUSTAL X [97] and performed a RepeatMasker search using the RepeatMasker software (Smit & Green, Repeat Masker at http://ftp.genome.Washington.edu/RM/RepeatMasker.html). As most SINEs are derived from tRNA genes, we also performed a local Blast search against all published tRNA-genes. Using this procedure, we discovered 12 copies of tRNA-derived SINEs.
Strategy 2
In order to further identify novel SINEs in the genome of cetaceans, we used the strategy suggested by Chen and Yang [98]. The genomic libraries were constructed for long-beaked common dolphin, striped dolphin, Indo-Pacific humpbacked dolphin, Risso's dolphin and finless porpoise. About three thousands colonies were screened for each species. Clones identified by nonradioactive southern blotting based on digoxigenin-labeling system were sequenced. With this strategy, 25 informative SINEs that inserted into unique genomic loci during evolution were isolated and characterized.
Strategy 3
To extract potential novel SINEs from GenBank entries, we downloaded sequence data of about 1.8 million bases for the common bottlenose dolphin from the National Institutes of Health Intramural Sequencing Center at http://asia.ensembl.org/Tursiops_truncatus/Info/Index. To identify SINEs from these sequences, we developed a computer-based search profile in the C programming language that extracts sequences of 100 to 500 nt flanked by 8-nt to 25-nt perfect repeats. About 501 corresponding sequences could be extracted from the common bottlenose dolphin sequences. We subsequently used the local version of RepeatMasker (Smit & Green, Repeat Masker at http://ftp.genome.Washington.edu/RM/RepeatMasker.html) containing a specific library comprising all CHR-1 and CHR-2 subfamily consensus sequences to scan for novel SINEs. We also performed a local Blast search against all published SINEs isolated from the cetacean genomes. In the end, we found 182 novel copies of tRNA-derived SINE element flanked with perfect direct repeats (DRs).
PCR amplification
To examine the presence or absence of a SINE unit at orthologous in various species, we designed and synthesized a pair of primers that flanked the unit based on the novel SINE loci (Additional file 5). PCR was performed with these primer sets for each SINE locus using cetacean and hippopotamus DNAs as templates. All amplification reactions were conducted on a BioRAD PTC-200 using 2×EasyTaq PCR SuperMix (TransGen Biotech) under the profile: 30 cycles at 93°C for 5 min, 93°C for 1 min, 53°C-59°C for 1 min, and 72°C for 1 min, followed by a 10-min extension at 72°C. The PCR products were electrophoresed in a 1.5% agarose gel and visualized under UV irradiation. Longer products indicated the presence of the SINE, whereas shorter products indicated the absence of the SINE. To confirm the presence or absence of a SINE at the loci, PCR products were sequenced employing an ABI PRISM 310 or 3700 system with bi-directional primers.
Sequence alignment and phylogenetic analyses
All amplified sequences were analyzed and compared with the GenBank-NCBI database using the BLAST network service (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignments were performed by using CLUSTAL X [97] and manually adjusted in GeneDoc. For phylogenetic analysis, the SINE insertion data were compiled into the data matrix, in which SINE absence was coded as 0, and SINE presence was coded as 1 (see Additional file 2). In case where a PCR band was invisible or PCR was not performed, the character state was coded as missing (denoted with '?'). The resultant data matrix were applied to PAUP* (ver. 4. 0b10; [99]) for reconstruction of a strict consensus parsimony tree. The analysis was carried out under ''IRREV.UP'' option, regarding '0' as the ancestral state. Newly obtained sequences data have been deposited in GenBank database (accession numbers JN120481-JN120757).
In addition, for phylogenetic reconstructions using the flanking regions of 12 retroposed elements, the retroposed elements were entirely removed from the concatenation to make subsequent phylogenetic inferences fully independent of the retroposed insertions, excluding ambiguously aligned sites and highly gapped regions (Figure 7). Bayesian phylogenetic analyses of the concatenated SINE flanking sequence data set (3771 nucleotides in total for each species) were implemented using MrBayes 3.1.2 [100]. Two concurrent runs of one cold and 3 heated Metropolis-coupled Markov chains Monte Carlo (MCMCMC) were launched from random starting points. For DNA sequence alignments, Modeltest 3.7 [101] was employed to choose optimal models for the partition according to the AIC [102]. The 4 MCMCMC were simultaneously run for 20, 000, 000 generations using the program default parameters and trees were sampled every 1000 generations, and the stationarity of the likelihood scores of sampled trees was checked in Tracer 1.4 [103]. Bayesian posterior probabilities (PP) were obtained from the 50% majority-rule consensus of the post burn-in trees sampled at stationarity after removing the first 10% of trees as the "burn-in" stage.
Concatenations of parts of sequences of the 10 representative SINE loci. The name of the SINE family as well as its subfamily is indicated in a bold box (CD, Cetacean deletions; CDO, Cetacean deletion Odontoceti; MDI, Middle deletion type I) [16]. The dots indicate nucleotides identical to the consensus sequence at the top. Putative flanking direct repeats are underlined.
Molecular divergence estimates
Although SINE insertions allow one to construct tree topologies, they cannot be used for reliable calculation of relative branch lengths without the potential to model amplification rates of SINE markers over time. SINE-flanking sequences, however, may potentially be used for dating historical retropositional events that diagnose common ancestry, because of the probable neutral nature of evolution in nonfunctional regions of the genome [104]. Here, estimation of divergence times was conducted using the flanking regions of 12 retroposed elements with uncorrelated lognormal model, as implemented in BEAST v 1.6 [105]. Age estimates were obtained using the lognormal distribution, with the following fossils as calibration age constraints. The age of the Cetacea-Hippopotamidae split was calibrated using the Ypresian (Eocene: 55.8-48.6 Ma) fossil Pakicetus [24, 106] with standard deviation (SD) = 1.2. Crown Cetacea was calibrated based on the earliest record of mysticete from the Eocene/Oligocene boundary [79] (33.5-40 Ma, 1.138 SD). The age of the basal of the crown Odontoceti was calibrated using the oldest physeterid: the late Oligocene Ferecetotherium [107] (23.7-30 Ma, 1.119 SD). And the age of Phocenidae-Monodontidae split was established based on the oldest Phocoenid, Salumiphocaena stocktoni [1982] (10-11.2 Ma, 1.138 SD). The BEAST analysis was executed for 20, 000, 000 generations with a random starting tree, birth-death default priors sampled every 1000 generations. Results were examined using Tracer 1.4 [103] to evaluate stationarity, and the first 10% of trees were discarded as burn-in.
References
Jefferson TA, Webber MA, Pitman RL: Marine Mammals of the World: a Comprehensive Guide to their Identification. 2008, San Diego: Academic Press
Milinkovitch MC, Ortí G, Meyer A: Revised phylogeny of whales suggested by mitochondrial ribosomal DNA sequences. Nature. 1993, 361: 346-348. 10.1038/361346a0.
Rosel PE, Haygood MG, Perrin WF: Phylogenetic relationships among the true porpoises (Cetacea: Phocoenidae). Mol Phylogenet Evol. 1995, 4: 463-474. 10.1006/mpev.1995.1043.
Montgelard C, Catzeflis FM, Douzery E: Phylogenetic relationships of artiodactyls and cetaceans as deduced from the comparison of cytochrome b and 12S rRNA mitochondrial sequences. Mol Biol Evol. 1997, 14: 550-559.
Messenger SL, McGuire JA: Morphology, molecules, and the phylogenetics of cetaceans. Syst Biol. 1998, 47: 90-124. 10.1080/106351598261058.
Rychel AL, Reeder TW, Berta A: Phylogeny of mysticete whales based on mitochondrial and nuclear data. Mol Phylogenet Evol. 2004, 32: 892-901. 10.1016/j.ympev.2004.02.020.
Yan J, Zhou K, Yang G: Molecular phylogenetics of 'river dolphins' and the baiji mitochondrial genome. Mol Phylogenet Evol. 2005, 37: 743-750. 10.1016/j.ympev.2005.06.009.
Nishida S, Goto M, Pastene LA, Kanda N, Koike H: Phylogenetic relationships among cetaceans revealed by Y-chromosome sequences. Zool Sci. 2007, 24: 723-732. 10.2108/zsj.24.723.
Caballero S, Jackson J, Mignucci-Giannoni AA, Barrios-Garrido H, Beltrán-Pedreros S, Montiel-Villalobos MG, Robertson KM, Baker CS: Molecular systematics of South American dolphins Sotalia: sister taxa determination and phylogenetic relationships, with insights into a multilocus phylogeny of the Delphinidae. Mol Phylogenet Evol. 2008, 46: 252-268. 10.1016/j.ympev.2007.10.015.
McGowen MR, Spaulding M, Gatesy J: Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol Phylogenet Evol. 2009, 53: 891-906. 10.1016/j.ympev.2009.08.018.
Steeman ME, Hebsgaard MB, Fordyce RE, Ho SYW, Rabosky DL, Nielsen R, Rahberk C, Glenner H, Sorensen MV, Willerslev E: Radiation of extant cetaceans driven by restructuring of the oceans. Syt Biol. 2009, 58: 573-585. 10.1093/sysbio/syp060.
Xiong Y, Brandley MC, Xu S, Zhou K, Yang G: Seven new dolphin mitochondrial genomes and a time-calibrated phylogeny of whales. BMC Evol Biol. 2009, 9: 20-10.1186/1471-2148-9-20.
Zhou X, Xu S, Yang Y, Zhou K, Yang G: Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol Phylogenet Evol. 2011, 61: 255-264. 10.1016/j.ympev.2011.02.009.
Geisler JH, McGowen MR, Yang G, Gatesy J: A supermatrix analysis of genomic, morphological, and paleontological data from crown Cetacea. BMC Evol Biol. 2011, 11: 112-10.1186/1471-2148-11-112.
Gatesy J, Milinkovitch M, Waddell V, Stanhope M: Stability of cladistic relationships between Cetacea and higher-level artiodactyl taxa. Syst Biol. 1999, 48: 6-20. 10.1080/106351599260409.
Nikaido M, Matsuno F, Abe H, Shimamura M, Hamilton H, Matsubayashi H, Okada N: Evolution of CHR-2 SINEs in cetartiodactyl genomes: possible evidence for the monophyletic origin of toothed whales. Mamm Genome. 2001, 12: 909-915. 10.1007/s0033501-1015-4.
Geisler JH, Sanders AE: Morphological evidence for the phylogeny of Cetacea. J Mamm Evol. 2003, 10: 23-129. 10.1023/A:1025552007291.
Zhou X, Xu S, Xu J, Chen B, Zhou K, Yang G: Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian mammals. Syt Biol.
Arnason U, Gullberg A, Janke A: Mitogenomic analyses provide new insights into cetacean origin and evolution. Gene. 2004, 333: 27-34.
Agnarsson I, May-Collado LJ: The phylogeny of Cetartiodactyla: the importance of dense taxon sampling, missing data, and the remarkable promise of cytochrome b to provide reliable species-level phylogenies. Mol Phylogenet Evol. 2008, 48: 964-985. 10.1016/j.ympev.2008.05.046.
O'Leary MA, Gatesy J: Impact of increased character sampling on the phylogeny of Cetartiodactyla (Mammalia): combined analysis including fossils. Cladistics. 2008, 24: 397-442. 10.1111/j.1096-0031.2007.00187.x.
Nikaido M, Rooney AP, Okada N: Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: hippopotamuses are the closest extant relatives of whales. Proc Natl Acad Sci USA. 1999, 96: 10261-10266. 10.1073/pnas.96.18.10261.
Lum JK, Nikaido M, Shimamura M, Shimodaira H, Shedlock AM, Okada N, Hasegawa M: Consistency of SINE Insertion Topology and Flanking Sequence Tree: Quantifying Relationships Among Cetartiodactyls. Mol Biol Evol. 2000, 17: 1417-1424.
Thewissen JG, Williams EM, Roe LJ, Hussain ST: Skeletons of terrestrial cetaceans and the relationship of whales to artiodactyls. Nature. 2001, 413: 277-281. 10.1038/35095005.
Milinkovitch MC: Molecular phylogeny of cetaceans prompts revision of morphological transformations. Trends Ecol Evol. 1995, 10: 328-334. 10.1016/S0169-5347(00)89120-X.
Milinkovitch MC, LeDuc RG, Adachi J, Farnir F, Georges M, Hasegawa M: Effects of character weighting and species sampling on phylogeny reconstruction: a case study based on DNA sequence data in cetaceans. Genetics. 1996, 144: 1817-1833.
Milinkovitch MC, Meyer A, Powell JR: Phylogeny of all major groups of cetaceans based on DNA sequences from three mitochondrial genes. Mol Biol Evol. 1994, 11: 939-948.
Smith MR, Shivji MS, Waddell VG, Stanhope MJ: Phylogenetic evidence from the IRBP gene for the paraphyly of toothed whales, with mixed support for Cetacea as a suborder of Artiodactyla. Mol Biol Evol. 1996, 13: 918-922.
Heyning JE: Sperm whale phylogeny revisited: analysis of the morphological evidence. Mar Mammal Sci. 1997, 13: 596-613. 10.1111/j.1748-7692.1997.tb00086.x.
Gatesy J: Molecular evidence for the phylogenetic affinities of Cetacea. The Emergence of Whales. Edited by: Thewissen J. 1998, New York: Plenum, 63-111.
Nikaido M, Matsuno F, Hamilton H, Brownell JRL, Cao Y, Wang D, Zhu ZY, Shedlock AM, Fordyce RE, Hasegawa M, Okada N: Retroposon analysis of major cetacean lineages: the monophyly of toothed whales and the paraphyly of river dolphins. Proc Natl Acad Sci USA. 2001, 98: 7384-7389. 10.1073/pnas.121139198.
Nikaido M, Piskurek O, Okada N: Toothed whale monophyly reassessed by SINE insertion analysis: the absence of lineage sorting effects suggests a small population of a common ancestral species. Mol Phylogenet Evol. 2007, 43: 216-224. 10.1016/j.ympev.2006.08.005.
Cassens I, Vicario S, Waddell VG, Balchowsky H, Belle DV, Wang D, Chen F, Mohan RSL, Simões-Lopes PC, Bastida R, Meyer A, Stanhope MJ, Milinkovitch MC: Independent adaptation to riverine habitats allowed survival of ancient cetacean lineages. Proc Natl Acad Sci USA. 2000, 97: 11343-11347.
Leduc RG, Perrin WF, Dizon AE: Phylogenetic relationships among the delphinid cetaceans based on full cytochrome b sequences. Mar Mammal Sci. 1999, 15: 619-648. 10.1111/j.1748-7692.1999.tb00833.x.
Wang J, Chou L, White BN: Osteological differences between two sympatric forms of bottlenose dolphins (genus Tursiops) in Chinese waters. J Zool Lond. 2000, 252: 147-162. 10.1111/j.1469-7998.2000.tb00611.x.
Kakuda T, Tajima Y, Arai K, Kogi K, Hishii T, Yamada KT: On the Resident "Bottlenose Dolphins" from Mikura Water. Mem Nat Sci Mus Tokyo. 2002, 38: 255-272.
Nishihara H, Terai Y, Okada N: Characterization of novel Alu- and tRNA-related SINEs from the Tree Shrew and evolutionary implications of their origins. Mol Biol Evol. 2002, 19: 1964-1972.
Piskurek O, Nikaido M, Boeadi BM, Okada N: Unique mammalian tRNA-derived repetitive elements in dermopterans: the t-SINE family and its retrotransposition through multiple sources. Mol Biol Evol. 2003, 20: 1659-1668. 10.1093/molbev/msg187.
Nikaido M, Nishihara H, Hukumoto Y, Okada N: Ancient SINEs from African endemic mammals. Mol Biol Evol. 2003, 20: 522-527. 10.1093/molbev/msg052.
Sasaki T, Takahashi K, Nikaido M, Miura S, Yasukawa Y, Okada N: First application of the SINE (Short interspersed repetitive element) method to infer phylogenetic relationships in reptiles: an example from the turtle superfamily Testudinoidea. Mol Biol Evol. 2004, 21: 705-715. 10.1093/molbev/msh069.
Nishihara H, Satta Y, Nikaido M, Thewissen JGM, Stanhope MJ, Okada N: A retroposon analysis of Afrotherian phylogeny. Mol Biol Evol. 2005, 22: 1823-1833. 10.1093/molbev/msi179.
Nishihara H, Hasegawa M, Okada N: Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci USA. 2006, 103: 9929-9934. 10.1073/pnas.0603797103.
Nishihara H, Smit AF, Okada N: Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006, 16: 864-874. 10.1101/gr.5255506.
Nikaido M, Hamilton H, Makino H, Sasaki T, Takahashi K, Goto M, Kanda N, Pastene LA, Okada N: Baleen whale phylogeny and a past extensive radiation event revealed by SINE insertion analysis. Mol Biol Evol. 2006, 23: 866-873. 10.1093/molbev/msj071.
Matveev V, Nishihara H, Okada N: Novel SINE families from Salmons validate Parahucho (Salmonidae) as a distinct genus and give evidence that SINEs can incorporate LINE-related 3'-tails of other SINEs. Mol Biol Evol. 2007, 24: 1656-1666. 10.1093/molbev/msm083.
Nishihara H, Kuno S, Nikaido M, Okada N: MyrSINEs: A novel SINE family in the anteater genomes. Gene. 2007, 400: 98-103. 10.1016/j.gene.2007.06.003.
Ray DA, Xing J, Salem AH, Batzer MA: SINEs of a nearly perfect character. Syst Biol. 2006, 55: 928-935. 10.1080/10635150600865419.
Rogers JH: The origin and evolution of retroposons. Int Rev Cytol. 1985, 93: 187-279.
Weiner AM, Deininger PL, Efstratiadis A: Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986, 55: 631-661. 10.1146/annurev.bi.55.070186.003215.
Okada N: SINEs. Curr Opin Genet Dev. 1991, 1: 498-504. 10.1016/S0959-437X(05)80198-4.
Okada N: SINEs: short interspersed repeated elements of the eukaryotic genome. Trends Ecol Evol. 1991, 6: 358-361. 10.1016/0169-5347(91)90226-N.
Okada N, Hamada M, Ogiwara I, Ohshima K: SINEs and LINEs share common 3' sequences: a review. Gene. 1997, 205: 229-243. 10.1016/S0378-1119(97)00409-5.
Shedlock AM, Okada N: SINE insertions: powerful tools for molecular systematics. BioEssays. 2000, 22: 148-160. 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z.
Salem AH, Ray DA, Xing J, Callinan PA, Myers JS, Hedges DJ, Garber RK, Witherspoon DJ, Jorde LB, Batzer MA: Alu elements and hominid phylogenetics. Proc Natl Acad Sci USA. 2003, 100: 12787-12791. 10.1073/pnas.2133766100.
Roos C, Schmitz J, Zischler H: Primate jumping genes elucidate strepsirrhine phylogeny. Proc Natl Acad Sci USA. 2004, 101: 10650-10654. 10.1073/pnas.0403852101.
Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J: Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006, 4: e91-10.1371/journal.pbio.0040091.
Sasaki T, Nikaido M, Hamilton H, Goto M, Kato H, Kanda N, Pastene LA, Cao Y, Fordyce RE, Hasegawa M, Okada N: Mitochondrial phylogenetics and the evolution of mysticete whales. Syst Biol. 2005, 54: 77-90. 10.1080/10635150590905939.
Yang X: Baysian inference of cetacean phylogeny based on mitochondrial genome. Biologia, Bratislava Section Cellular and Molecular Biology. 2009, 64: 811-818.
Gatesy J, Hayashi C, Cronin MA, Arctander P: Evidence from milk casein genes that cetaceans are close relatives of hippopotamid artiodactyls. Mol Biol Evol. 1996, 13: 954-963.
Verma SN, Sinha RK, Singh L: Phylogetic position of Platanista gangetica: insights from the mitochondrial cytochrome b and nuclear interphotoreceptor retinoid-binding protein gene sequences. Mol Phylogenet Evol. 2004, 33: 280-288. 10.1016/j.ympev.2004.06.018.
Heyning JE: Comparative facial anatomy of beaked whales (Ziphiidae) and a systematic revision among the families of extant Odontoceti. Contrib Sci Nat Hist Mus Los Angeles. 1989, 405: 1-64.
Fordyce RE, Barnes LG: The evolutionary history of whales and dolphins. Annu Rev Earth Planet Science. 1994, 22: 419-455. 10.1146/annurev.ea.22.050194.002223.
Arnason U, Gullberg A: Cytochrome b nucleotide sequences and the identification of five primary lineages of extant cetaceans. Mol Biol Evol. 1996, 13: 407-417.
Hamilton H, Caballero S, Collins AG, Brownell JRL: Evolution of river dolphins. Proc R Soc B. 2001, 268: 549-556. 10.1098/rspb.2000.1385.
Nishida S, Pastene LA, Goto M, Koike H: SRY gene structure and phylogeny in the cetacean species. Mammal Study. 2003, 28: 57-66. 10.3106/mammalstudy.28.57.
Simpson GG: The principles of classification and a new classification of mammals. Bull Am Mus Nat Hist. 1945, 85: 1-350.
de Muizon Cde: Les relations phyloge'ne'tiques des Delphinida (Cetacea, Mammalia). Annales de pale'ontologie. 1988, 74: 159-227.
de Muizon Cde: A new Ziphiidae from the Early Miocene of Washington state and a phylogenetic analysis of the major group of odontocetes. Bull Mus Natl Hist Nat Paris. 1991, 3-4: 279-326.
Yang G, Zhou K: A study on the molecular phylogeny of river dolphins. Acta Theriol Sin. 1999, 19: 1-9.
May-Collado L, Agnarsson I: Cytochrome b and Bayesian inference of whale phylogeny. Mol Phylogenet Evol. 2006, 38: 344-354. 10.1016/j.ympev.2005.09.019.
Sasaki T, Nikaido M, Hamilton H, Goto M, Kato H, Kanda N, Pastene LA, Cao Y, Fordyce RE, Hasegawa M, Okada N: Mitochondrial phylogenetics and the evolution of mysticete whales. Syst Biol. 2005, 54: 77-90. 10.1080/10635150590905939.
Fordyce RE, de Muizon C: Evolutionary history of cetaceans: a review. Secondary Adaptation of Tetrapods to Life in Water. Edited by: Mazin JM, de Buffrenil V. 2001, Verlag Dr. Friedrich Pfeil Munich, 169-233.
Coxall HK, Pearson PN: The Eocene-Oligocene transition. Deep time perspectives on climate change: marrying the signal from computer models and biological processes. Edited by: Williams M, Haywood AM, Gregory FJ, Schmidt DN. 2008, London: Geological Society London, 351-387.
Lear CH, Bailey TR, Pearson PN, Coxall HK, Rosenthal Y: Cooling and ice growth across the Eocene-Oligocene transition. Geology. 2008, 36: 251-254. 10.1130/G24584A.1.
Scher HD, Martin EE: Timing and climatic consequences of the opening of Drake Passage. Science. 2006, 312: 428-430. 10.1126/science.1120044.
Livermore R, Hillenbrand CD, Meredith M, Eagles G: Drake Passage and Cenozoic climate: an open and shut case?. Geochem Geophys Geosyst. 2007, 8: 1-11.
Whitfield P: Encyclopedia of Animals. Edited by: Simon, Schuster. 1998, New York, 184-193.
Coxall HK, Wilson PA, Palike H, Lear CH, Backman J: Rapid stepwise onset of Antarctic glaciation and deeper calcite compensation in the Pacific Ocean. Nature. 2005, 433: 53-57. 10.1038/nature03135.
Rice DW: Marine mammals of the world: Systematics and distribution. The Society for Marine Mammalogy. Special Publication No.4. 1998, 1-231.
Barnes LG: Evolution, taxonomy and antitropical distributions of the porpoises (Phocoenidae, Mammalia). Mar Mammal Sci. 1985, 1: 149-165. 10.1111/j.1748-7692.1985.tb00003.x.
Fordyce RE: Cetacean evolution and Eocene-Oligocene oceans revisited. From Greenhouse to Icehouse: the Marine Eocene-Oligocene Transition. Edited by: Prothero DR, Ivany LC, Nesbitt EA. 2003, New York: Columbia University Press, 154-170.
Fordyce RE: Cetacean fossil record. Encyclopedia of Marine Mammals. Edited by: Perrin W, Würsig B, Thewissen JGM. 2009, San Diego: Academic Press, 201-207. 2
Waddell VG, Milinkovitch MC, Bérubé M, Stanhope MJ: Examination of the Delphinoidea trichotomy: congruent evidence from three nuclear loci indicates that porpoises (Phocoenidae) share a more recent common ancestry with white whales (Monodontidae) than they do with true dolphins (Delphinidae). Mol Phylogenet Evol. 2000, 15: 314-318. 10.1006/mpev.1999.0751.
True FW: Contributions to the natural history of the cetaceans, a review of the family Delphinidae. US Natl Mus Bull. 1889, 36: 1-191.
Perrin WF: Dolphins, porpoises, and whales. An action plan for the conservation of biological diversity: 1988-1992. 1989, Gland Switzerland: IUCN, 2
Hershkovitz P: Catalog of living whales. US Natl Mus Bull. 1966, 246: 1-259.
Perrin WF, Mitchell ED, Mead JG, Caldwell DK, van Bree PJH: Stenella clymene, a rediscovered tropical dolphin of the Atlantic. J Mammal. 1981, 62: 583-598. 10.2307/1380405.
Ross GJB, Cockcroft VG: The bottlenose dolphin. Comments on Australian bottlenose dolphins and the taxonomic status of Tursiops aduncus (Ehrenberg, 1832). Edited by: Leatherwood S, Reeves RR. 1990, San Diego: Academic Press, 101-128.
Luis JR, Terreros MC, Martinez L, Rojas D, Herrera RJ: Two problematic human polymorphic Alu insertions. Electrophoresis. 2003, 24: 2290-2294. 10.1002/elps.200305451.
Nei M, Kumar S: Molecular evolution and phylogenetics. 2000, New York: Oxford University Press
Fraser FC: Three anomalous dolphins from Blacksod Bay, Ireland. Proc Roy Irish Acad. 1940, 45: 413-462.
Shallenberger EW, King I: Dolphin birth at Sea Life Park. Breeding dolphins. Present status, suggestions for the future. Edited by: Ridgway SM, Benirschke K. 1977, Washington: US Marine Mammal Commission, 77-84.
Nishiwaki M: Remarks on the systematics of Cetacea. New result of cetacean and pinniped research. Acad Sci USSR Severtsov Memorial Inst Evolutional Morpholo and Ecolo Animals Nauka. 1978, 9-19.
Nishiwaki M, Tobayama T: Morphological study on the hybrid between Tursiops and Pseudorca. Sci Rep Whales Res Inst. 1982, 34: 109-121.
Sambrook J, Russell DW: Molecular Cloning: a laboratory manual. 2001, New York: Cold Spring Harbor Laboratory Press, 3
Okada N, Shedlock AM, Nikaido M: Retroposon mapping in molecular systematics. Methods in Molecular Biology: Mobile Genetic Element. Edited by: Miller WJ, Capy P. 2004, Totowa NJ: Humana Press, 189-226.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG: The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 24: 4876-4882.
Chen Z, Yang G: Novel CHR-2 subfamilies and t-SINEs identified in cetaceans using nonradioactive Southern blotting. Genes Genom. 2010, 32: 345-352. 10.1007/s13258-010-0044-3.
SwoVord DL: PAUP*: Phylogenetic Analysis Using Parsimony (Sinauer, Sunderland, MA), Version 4.0. 1998
Ronquist F, Huelsenbeck JP: MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003, 19: 1572-1574. 10.1093/bioinformatics/btg180.
Posada D, Crandall KA: Modeltest: testing the model of DNA substitution. Bioinformatics. 1998, 14: 817-818. 10.1093/bioinformatics/14.9.817.
Posada D, Buckley TR: Model selection and model averaging in phylogenetics: advantages of Akaike Information Criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004, 53: 793-808. 10.1080/10635150490522304.
Rambaut A, Drummond AJ: Tracer v1.4. Distributed by the Authors. 2007, [http://beast.bio.ed.ac.uk/Tracer]
Lum JK, Nikaido M, Shimamura M, Shimodaira H, Shedlock AM, Okada N, Hasegawa M: Consistency of SINE insertion topology and flanking sequence tree: quantifying relationships among Cetartiodactyls. Mol Biol Evol. 2000, 17: 1417-1424.
Drummond AJ, Rambaut A: BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Bio. 2007, l7: 214-
Gingerich PD, Russel DE: Pakicetus inachus, a new archaeocete (Mammalia, Cetacea) from the Early-Middle Eocene Kuldana Formation of Kohat (Pakistan). Contributions from the Museum of Paleontology University of Michigan. 1981, 28: 1-20.
Mchedlidze GA: Nekotorye obshie cherty istorii kitoobraznykh. (Some general characteristics of the evolution of cetaceans). 1970, Metsniereba Press, Institut Paleobiologii Akademia Nauk, Gruzinskoi SSR, Tbilisi, Georgia, 1-112.
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (NSFC) key project grant no. 30830016, the Program for New Century Excellent Talents in University (NCET-07-0445), the Ministry of Education of China, the major project of the Natural Science Foundation of the Jiangsu Higher Education Institutions of Jiangsu Province, China (07KJA18016), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). We are grateful to Mr Anli Gao, Xinrong Xu, and Bingyao Chen for their contribution to collecting samples. Section Editor and three anonymous reviewers provided constructive comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
GY and ZC designed the study. ZC carried out the experiments, performed the data analyses and prepared the draft of the manuscript. SX helped to perform the analyses and improve the manuscript. KZ helped to improve the manuscript. GY helped to perform the data analyses and improve the manuscript. All authors read and approved the final manuscript.
Electronic supplementary material
12862_2011_1919_MOESM1_ESM.PPT
Additional file 1: Electrophoretic gel patterns of PCR products for the SINE loci analyzed in this study. Bands indicating the presence of the SINE are shown by black arrowheads, whereas gray arrowheads show those that indicate SINE absence. Loci are assigned alphabetically from A to J according to the clade on the phylogenetic tree shown in Figure 4. The species are numbered as follows: 1, Striped dolphin; 2, Risso's dolphin; 3, Indo-Pacific bottlenose dolphin; 4, Common bottlenose dolphin; 5, Long-beaked common dolphin; 6, Chinese white dolphin; 7, Pantropical spotted dolphin; 8, Beluga; 9, Finless porpoise; 10, Yangtze River dolphin; 11, Ginkgo-toothed beaked whale; 12, Ganges River dolphin; 13, Pygmy sperm whale; 14, Omura's whale; 15, Common minke whale; 16, hippopotamus. (PPT 2 MB)
12862_2011_1919_MOESM2_ESM.DOC
Additional file 2: Data matrix showing the character states for the loci isolated in the present study. 0 = absence, 1 = presence,? = missing. The descriptions of each locus and taxa analyzed in this study are shown in the boxes. (DOC 57 KB)
12862_2011_1919_MOESM3_ESM.JPEG
Additional file 3: Alignments of sequences for loci Stec35 (A) and the two different SINE insertions (B). Dots indicate nucleotides identical to the consensus sequence at the top. The name of the SINE family as well as the two different SINEs are indicated in a bold box. The line above the sequences represents the tRNA-related region of the SINE. Box A and Box B promoters for RNA Polymerase III are boxed and highlighted. Putative flanking direct repeats are underlined. (JPEG 458 KB)
12862_2011_1919_MOESM4_ESM.JPEG
Additional file 4: Alignments of sequences for loci Turt164 (A) (including Band A and Band B) and the four SINE insertions amplified in the four species in this study (B). Dots indicate nucleotides identical to the consensus sequence at the top. The name of the SINE family as well as its subfamily is indicated in a bold box. The line above the sequences represents the tRNA-related region of the SINE. Box A and Box B promoters for RNA Polymerase III are boxed and highlighted. Putative flanking direct repeats are underlined. (JPEG 625 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Chen, Z., Xu, S., Zhou, K. et al. Whale phylogeny and rapid radiation events revealed using novel retroposed elements and their flanking sequences. BMC Evol Biol 11, 314 (2011). https://doi.org/10.1186/1471-2148-11-314
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2148-11-314