Abstract
Background
The cytotoxicity and the rejoining of DNA double-strand breaks induced by γ-rays, H2O2 and neocarzinostatin, were investigated in normal and PARP-1 knockout mouse 3T3 fibroblasts to determine the role of poly(ADP-ribose) polymerase (PARP-1) in DNA double-strand break repair.
Results
PARP-1-/- were considerably more sensitive than PARP-1+/+ 3T3s to induced cell kill by γ-rays and H2O2. However, the two cell lines did not show any significant difference in the susceptibility to neocarzinostatin below 1.5 nM drug. Restoration of PARP-1 expression in PARP-1-/- 3T3s by retroviral transfection of the full PARP-1 cDNA did not induce any change in neocarzinostatin response. Moreover the incidence and the rejoining kinetics of neocarzinostatin-induced DNA double-strand breaks were identical in PARP-1+/+ and PARP-1-/- 3T3s. Poly(ADP-ribose) synthesis following γ-rays and H2O2 was observed in PARP-1-proficient cells only. In contrast neocarzinostatin, even at supra-lethal concentration, was unable to initiate PARP-1 activation yet it induced H2AX histone phosphorylation in both PARP1+/+ and PARP-1-/- 3T3s as efficiently as γ-rays and H2O2.
Conclusions
The results show that PARP-1 is not a major determinant of DNA double-strand break recovery with either strand break rejoining or cell survival as an endpoint. Even though both PARP-1 and ATM activation are major determinants of the cell response to γ-rays and H2O2, data suggest that PARP-1-dependent poly(ADP-ribose) synthesis and ATM-dependent H2AX phosphorylation, are not inter-related in the repair pathway of neocarzinostatin-induced DNA double-strand breaks.
Similar content being viewed by others
Background
Ionizing radiation induces multiple lesions in cell DNA including oxidative base damage, single-strand breaks (SSB) and double-strand breaks (DSB) in proportion to the radiation dose. Among the enzymes that have evolved for the repair of radiation damage, poly(ADP-ribose) polymerase (PARP-1), Ataxia mutated kinase (ATM) and the heterotrimeric DNA-dependent protein kinase (DNA-PK), play a leading role. ATM is rapidly activated by DSB to phosphorylate proteins in chromatin, most notably H2AX histone [1, 2] and the catalytic subunit of the DNA-PK complex (DNA-PKcs). ATM is also essential to coupling of DNA damage detection to NF-κB activation and proper management of the oxidative stress inherent in radiation exposure. DNA-PKcs is recruited by the Ku70-Ku86 complex at sites of DSB [3, 4]. Activated DNA-PKcs phosphorylates a range of protein substrates and, along with XRCC4 and Ligase 4 is essential to V(D)J recombination and DSB repair through the non-homologous end joining (NHEJ) pathway [5].
PARP-1, an ubiquitous 113 kDa enzyme, is also required for the detection and signalization of DNA strand interruptions. It is involved in early DNA damage recognition [6], base excision repair [7, 8] and genome surveillance in a variety of situations (for a review see [9]). Activated PARP-1 carries out synthesis and transfer of long linear or branched ADP-ribose polymers (pADPr) to carboxyl groups in a limited number of nuclear protein acceptors, including PARP-1 itself in a reaction initiated by PARP-1 binding to SSB [10]. In addition, several DNA damage signaling or repair proteins possess high-affinity binding motifs for pADPr, among others XRCC1, DNA ligase III, p21Waf1 and p53 [11, 12]. Two subunits of the DNA-PK heterotrimer, namely, Ku70 and DNA-PKcs also present high affinity motifs for pADPr binding [12], and PARP-1 co-immunoprecipitates with these proteins [13–15]. In vitro, the DNA-PKcs subunit can be ADP-ribosylated and stimulated by PARP-1; PARP-1 can in turn be phosphorylated by DNA-PKcs [16].
Whether these protein complexes and post-translational modifications play a role in repair from radioinduced DSB, has been challenged recently [17, 18]. In the prospect of unravelling this question, we reasoned that radiomimetic compounds acting to produce DSB with high selectivity and efficiency in the DNA of target cells should be used instead of ionizing radiation, since radiation generates oxidative stress response and elicits a large spectrum of lesions located at random in chromatin.
We chose neocarzinostatin (NCS) for this purpose. NCS is the prototype of the "protein antibiotic" family. It is a complex consisting of a dodecadiyne antibiotic (NCSChrom) reversibly bound to a carrier protein [19]. NCS is active in the nanomolar range, and NCSChrom cleaves DNA in the course of a suicide reaction leaving no residual active drug after a few minutes incubation. The major DNA lesions induced by NCSChrom in DNA result from radical attack [20] and consist of a blunt end break bearing a thymidine-5'-aldehyde residue on one strand [21], with an atypical abasic site at two nucleotide interval on the complementary strand [22, 23]. This abasic site is substrate for endonuclease III [24] in such a way that NCS-induced damage is rapidly converted into DSB in living cells [25, 26]. E. coli [27, 28], yeast [29] or mammalian cells [30–37] bearing a defect in DSB repair, are consistently hypersensitive to induced cell kill by NCS.
PARP-1 proficient (PARP-1+/+) and PARP-1 knockout (PARP-1-/-) 3T3 fibroblasts from syngenic mice, were used to investigate the role of PARP-1 in the recovery of NCS-induced DSB. PARP-1-/- 3T3s complemented with the full PARP-1 cDNA, were also used in this assay. The incidence and repair kinetics of DSB were measured in parallel in PARP-1+/+ and PARP-1-/- 3T3s, and the effect of 4-amino-1,8-naphthalimide, a potent PARP-1 inhibitor, was established in PARP-1+/+ 3T3s. The cytotoxic effect of γ-rays and H2O2 was determined for comparison.
Immunofluorescence studies were also performed to assess pADPr synthesis and H2AX histone phosphorylation following exposure to NCS, γ-rays or H2O2. The results show that PARP-1 is likely not to play a crucial role in the repair of DSB, at least in the absence of other types of DNA damage.
Results
Cytotoxicity of NCS, γ-rays and H2O2
PARP-1+/+ and PARP-1-/- 3T3s were exposed to increasing concentrations of NCS and cytotoxicity was determined through a growth assay (see Methods). Below 1.5 nM NCS PARP-1+/+ and PARP-1-/- 3T3s demonstrated exactly the same susceptibility to the lethal effect of NCS (Figure 1A). However, PARP-1+/+ were substantially more sensitive to NCS than PARP-1-/- 3T3s in the high range of NCS concentration.
Effect of PARP-1 knockout on the susceptibility of mouse 3T3 fibroblasts to the lethal effect of NCS. A (top). Cytotoxicity of NCS against PARP-1+/+ and PARP-1-/- 3T3s. The growth assays were performed as described under Methods. For PARP-1-/- 3T3s, the data were fitted to an exponential equation, S = e-α [NCS] where S is the surviving fraction (α = 0.398 ± 0.019 nM-1). For PARP-1+/+ 3T3s, a linear-quadratic equation was used in order to smooth the curve and gave the same value of α. The insert shows the response of PARP-1+/+ and PARP-1-/- 3T3s in the low range of NCS concentration. The amount of NCS (LC37) required to reduce cell survival to 1/e of that in control was 1.79 and 2.48 nM for PARP-1+/+ and PARP-1-/-3T3, respectively. B (bottom). Comparison of the response of PARP-1-/- 3T3s from a unique isolate transduced with a pBabe-puro retroviral vector carrying the full-length PARP-1 cDNA (pBabePARP-1) or the void vector (pBabevoid). Both cell lines showed the same exponentially-dependent response to NCS (α = 0.566 ± 0.044 nM-1).
Pre-treatment with the PARP-1 inhibitor 4-amino-1,8-naphthalimide (ANI) did not alter the susceptibility of PARP-1+/+ 3T3s to NCS (Figure 2A). In contrast, PARP-1-/- were considerably more sensitive than PARP-1+/+ 3T3s to the lethal effect of γ-rays (Figure 2B) and H2O2 (Figure 2C).
Differential susceptibility of PARP-1+/+ and PARP-1-/- 3T3s to γ-rays and H2O2. A (top). Absence of a significant effect of ANI (30 μM) on the cytotoxic response of PARP-1+/+ 3T3s to NCS. The results are expressed as the ratio of the mean lethal NCS concentrations (LC37) determined from survival curves in the same way as in Figure 1. B (middle). γ-Ray survival of PARP-1+/+ and PARP-1-/- 3T3s. A modified form of the linear-quadratic equation,  where D is the radiation dose and (a + b) = 1, was set [6] to take into account the existence of a minor fraction (b) of the cell population experiencing cytolytic cell death at high radiation doses. The values calculated for best fit with the experimental data were: α = 0.134 ± 0.040 Gy-1, β = 0.168 ± 0.030 Gy-2 for PARP-1-/- 3T3; α = 0.0258 ± 0.0323 Gy-1, β = 0.0422 ± 0.0120 Gy-2 for PARP-1+/+ 3T3s. α represents the contribution to radiation-induced cell death of lethal, non-repairable DNA damage. The quadratic parameter, β relates to unrepaired sublethal damage. Though this is still a matter of controversy [66], β is thought to represent the probability of interaction between separate breaks to exchange chromosomal aberrations [67]. The mean lethal radiation doses (D37), i. e., the doses required to reduce cell survival to 1/e of that in control, are given in Table 1. C (bottom). Cytotoxicity of H2O2 against PARP-1+/+ and PARP-1-/- 3T3s. The dose-response curves were fitted to a biexponential equation,
where D is the radiation dose and (a + b) = 1, was set [6] to take into account the existence of a minor fraction (b) of the cell population experiencing cytolytic cell death at high radiation doses. The values calculated for best fit with the experimental data were: α = 0.134 ± 0.040 Gy-1, β = 0.168 ± 0.030 Gy-2 for PARP-1-/- 3T3; α = 0.0258 ± 0.0323 Gy-1, β = 0.0422 ± 0.0120 Gy-2 for PARP-1+/+ 3T3s. α represents the contribution to radiation-induced cell death of lethal, non-repairable DNA damage. The quadratic parameter, β relates to unrepaired sublethal damage. Though this is still a matter of controversy [66], β is thought to represent the probability of interaction between separate breaks to exchange chromosomal aberrations [67]. The mean lethal radiation doses (D37), i. e., the doses required to reduce cell survival to 1/e of that in control, are given in Table 1. C (bottom). Cytotoxicity of H2O2 against PARP-1+/+ and PARP-1-/- 3T3s. The dose-response curves were fitted to a biexponential equation,  with (a + b + c) = 1. c corresponds to the plateau of cell survival at infinite H2O2 concentration (0.48 and 0.046 for PARP-1+/+ and PARP-1-/- 3T3s, respectively). The initial slope of the cells' response to H2O2 was calculated at 2.6 and 25.8 mM-1 for PARP-1+/+ and PARP-1-/- 3T3s, respectively.
with (a + b + c) = 1. c corresponds to the plateau of cell survival at infinite H2O2 concentration (0.48 and 0.046 for PARP-1+/+ and PARP-1-/- 3T3s, respectively). The initial slope of the cells' response to H2O2 was calculated at 2.6 and 25.8 mM-1 for PARP-1+/+ and PARP-1-/- 3T3s, respectively.
The PARP-1+/+ and PARP-1-/- 3T3 clones used in these experiments originated each from independent isolates immortalized at random. There was therefore a possibility that minor differences might exist between both cell lines with regard to cell death mechanism or efficiency at a high level of DSB, and contribute to altered NCS susceptibility in addition to the PARP-1 defect. To settle this question, we used PARP-1-/- 3T3s in which the full PARP-1 cDNA had been re-inserted by retroviral infection with a pBabe construct, allowing complete restoration of the PARP-1 activity [38]. The NCS response of these cells was compared to that of PARP-1-/- 3T3s from the same clonal isolate transfected with the void vector. Both cell lines showed exactly the same susceptibility to NCS, with a purely exponential dose-dependence (Figure 1B).
Incidence of NCS-induced DSB
DSB in PARP-1+/+ and PARP-1-/- 3T3s were measured by PFGE. Briefly, cells were exposed to increasing concentrations of NCS, up to 30 nM for 10-min at 37°C, then chilled in ice, harvested in PBS-EDTA buffer, and inserted into agarose plugs and lysed for PFGE analysis. The effect of NCS was compared to that of γ-rays (up to 60 Gy) on cells irradiated in ice.
The incidence of DSB was found to increase linearly with the γ-ray dose, in agreement with earlier reports [39]. It also grew linearly with the NCS concentration, and incubation with 1 nM NCS yielded the same amount of DSB as 1.21 Gy γ-rays (data not shown). PARP-1+/+ and PARP-1-/- 3T3s did not show any difference in this assay. Taking into account the fact that 1 Gy γ-rays produces 41 DSB per diploid cell nucleus [40], the average incidence of DSB formed by 1 nM NCS in each 3T3 fibroblast was estimated at ca. 50 DSB. With consideration to the number of DSB induced by the treatment, the lethal efficiency of NCS was in the same range as that for γ-rays (Table 1).
Rejoining of NCS-induced DSB
To determine whether the repair of NCS-induced DSB was deficient in PARP-1-/- relative to PARP-1+/+ 3T3s, DSB rejoining in cells treated with 30 nM NCS was measured through PFGE. The data (Figure 3) did not demonstrate any difference in the rejoining kinetics between both cell lines.
Time-dependence of DSB rejoining in PARP-1+/+ and PARP-1-/- 3T3s following exposure to 30 nM NCS. [2-14C]Thymidine-labeled cells were exposed to NCS for 10-min. After drug removal, cells were rapidly chilled in ice at the time indicated, harvested, and embedded into agarose plugs followed by lysis and PFGE (see Methods). The results are expressed as the percentage of remaining damage in Gy.eq. This percentage was determined from the fraction of activity released from the plugs; calibration was made with reference to samples irradiated in ice. The origin of the time scale starts from the introduction of NCS.
Immunofluorescence assays
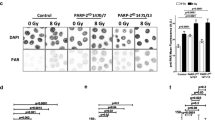
PARP-1+/+ and PARP-1-/- 3T3s were assayed through immunoflurescence for the determination of (i) PARP-1 expression, (ii) PARP-1 activity visualized by pADPr synthesis following γ-ray irradiation, NCS and H2O2, and (iii) ATM-dependent H2AX histone phosphorylation in response to DNA damage induced by γ-rays, H2O2 or NCS. The results are shown in Figure 4.
Immunofluorescent visualization of PARP-1, pADPR synthesis and H2AX phosphorylation in PARP-1+/+ and PARP-1-/- 3T3s exposed to various treatments including H2O2 (1 mM, 10-min) without or with 30 μM ANI, γ-rays (20 Gy, 10-min) or NCS (2 or 30 nM, 10-min). PARP-1-/- 3T3s complemented with the PARP-1 cDNA (pBabePARP-1) are shown for comparison. Cells were grown on coverslips, treated, fixed, incubated with antibodies and counterstained with DAPI. For each preparation the gain of the camera was adjusted relative to DAPI emission. The top part of the figure shows isolated views. The bottom panels show DAPI and immunofluorescence views in coincidence. The bar (top right view) represents 20 μm.
PARP-1 expression was detected in the nucleus of PARP-1+/+ 3T3s and was missing in PARP-1-/- 3T3s, as expected. Also as expected, pADPr synthesis was observed selectively in the nucleus of PARP-1+/+ cells following γ-ray or H2O2 exposure, and was repressed by ANI. Stable transfection with a retroviral vector coding for the full PARP-1 gene, restored PARP-1 expression and function.
In contrast to γ-rays or H2O2, NCS did not induce any measurable pADPr synthesis in PARP-1+/+ 3T3s, even at a concentration of 30 nM representing ca. 20-fold the IC50 value and yielding the same amount of DSB as 36 Gy radiation. However, consistent with induction of DSB in chromosomal DNA, NCS at relatively low concentration (2 nM) initiated rapid phosphorylation of H2AX in the same way as γ-rays or H2O2. In agreement with this observation, MRE11 phosphorylation and focus formation through an ATM- and NBS1-dependent mechanism, has recently been shown to occur after NCS treatment [37].
Discussion
The incidence of NCS-induced DSB was the same in PARP-1+/+ and PARP-1-/- 3T3s, indicating that both cell lines incorporated equal amounts of NCSChrom. Moreover no difference, even minor was found between PARP-1+/+ and PARP-1-/- in the kinetics of DSB rejoining (Figure 3).
Consistent with this, the susceptibility to NCS-induced lethality was the same in PARP-1+/+ and PARP-1-/- 3T3s below 1.5 nM drug. PARP-1+/+ were comparatively more sensitive to NCS in the high range of NCS concentration. This is thought to result from minor differences in the susceptibility to DSB-induced cell death arising between different clonal isolates of 3T3 fibroblasts immortalized at random. As a matter of fact, retrovirus-mediated insertion of the PARP-1 cDNA in a single PARP-1-/- 3T3 clone, did not bear any change in NCS susceptibility (Figure 1).
ANI totally repressed pADPr elongation in response to H2O2 (Figure 4) but did not alter the cytotoxicity of NCS in PARP-1+/+ 3T3s (Figure 2). These results corroborate those of other authors who showed that PARP-1 inhibitors did not affect cell kill by the topoisomerase IIα poison etoposide, another DSB inducer [41–43]. In contrast, and in agreement with earlier reports [6, 44, 45] PARP-1-/- 3T3s were considerably more sensitive than PARP-1+/+ 3T3s to the lethal effect of radiation (Figure 2C). PARP-1-/- 3T3s were also more sensitive than PARP-1+/+ to sub-millimolar concentrations of H2O2, a potent inducer of pADPr synthesis in response to SSB and oxidative base damage [46], highligting the toxic effect of a deficiency in base excision repair in PARP-1 knockout cells [47]. However, for both cell lines, and contrary to CHO cells that exhibited an exponentially concentration-dependent response to H2O2 [46], the survival curves presented a plateau indicating resistance of a major (PARP-1+/+) or minor fraction (PARP-1-/-) of the cell population (Figure 2C). A similar effect was reported by other authors in Chinese hamster V79 cells and in normal and AT human fibroblasts exposed to H2O2 or organic peroxides [48, 49]. Differential expression of catalase, superoxide dismutase or glutathione peroxidase was ruled out [48] and at the moment there is no explanation to this observation.
Other authors using single-cell gel electrophoresis (comet assay) have shown that the rejoining of radiation-, H2O2- or methyl methane sulfonate-induced SSB, is dramatically hampered by PARP-1 knockout, and significantly delayed by PARP-1 inhibitors in L1210 cells [50] and PARP-1+/+ 3T3s [51, 52] yet these inhibitors have poor incidence on the level of H2O2-induced cell death [46, 53]. Ectopic expression of the PARP-1 cDNA in PARP-1-/- 3T3s restored a wild-type phenotype [51], demonstrating unequivocally that the repair capacity of PARP-1-deficient cells was drastically limited. We propose that this effect is relevant to the role of PARP-1 in base excision repair only, as there was no difference between PARP-1+/+ and PARP-1-/- 3T3s with regard to the rejoining kinetics of NCS-induced DSB (Figure 3). Moreover, immunofluorescence assays showed that, even though NCS at supra-lethal concentration did not induce detectable pADPr synthesis, the ATM-dependent phosphorylation of H2AX was activated in response to relatively low levels of NCS, irrespective of whether PARP-1 was defective or not (Figure 4). Bowman et al. [43] also showed that the topoisomerase IIα poison etoposide did not bring about pADPr synthesis in L1210 cells, even at concentrations in excess of those causing significant levels of apoptosis. Likewise, Dziegielewski and Beerman [54] recently showed that a defect in either ATM or DNA-PKcs may not confer cells enhanced susceptibility to the NCS homologue C-1027. On the other hand single-strand nicks in DNA are effective activators for PARP-1, not for DNA-PK [55, 56]. Therefore, PARP-1-dependent pADPr elongation and ATM-dependent H2AX phosphorylation, two major post-translational modifications occurring in response to DNA single- and double-strand breaks, respectively, are likely not to be interrelated or redundant. This may elicit functional synergy between PARP-1 and ATM, consistent with the embryonic lethality observed in the double ATM/PARP-1 knockout [57].
Conclusions
Substantial differences in PARP-1 activation by ionizing radiation and alkylating agents have been reported [58] and our data suggest that PARP-1 is not a major determinant of cell survival to DNA double-strand breaks as long as the level of oxidative stress (as determined from pADPr synthesis) is limited. In consideration of studies with other DNA-damaging agents (reviewed in [59]) and of the comparatively high susceptibility of PARP-1 null 3T3s to H2O2 (Figure 2C), we propose that enhanced radiation sensitivity of PARP-1 null relatively to normal 3T3s, proceeds from defects in base excision repair in the former.
Methods
Antibodies and reagents
Mouse monoclonal antibodies directed against the PARP-1 C-terminal domain (clone 7-D3-6) and pADPr (clone 10 H) were from Becton-Dickinson Biosciences and Alexis Corporation, respectively. Rabbit polyclonal antibody raised against γ-H2AX, was from Trevigen. Alexa-488®-conjugated goat anti-rabbit and anti-mouse secondary antibodies, were purchased from Molecular Probes. HRP-conjugated goat anti-mouse and anti-rabbit IgG came from Jackson Immunoresearch.
[2-14C]Thymidine was purchased from PerkinElmer Life Sciences. 4-amino-1,8-naphthalimide (ANI) came from ACROS Organics. H2O2 was from Sigma-Aldrich. Other chemicals were of the highest purity available and came from VWR International. The products for cell culture were from Invitrogen.
The neocarzinostatin holoprotein, prepared and titrated as described [60], was stored as a sterile 1 mM stock solution in 2 mM sodium formate buffer, pH 4.0, at liquid nitrogen temperature.
Cells and cell cultures
Spontaneously immortalized 3T3 mouse embryo fibroblasts obtained from homozygous wild-type (PARP-1+/+) and knockout (PARP-1-/-) C57BL/6 mice [44], were provided by Dr. Gilbert de Murcia. It should be stressed that the knockout in PARP-1-/- fibroblasts was complete, i. e., no fragment of the functional domain of PARP-1 protein was expressed in these cells. PARP-1-/- 3T3s in which the full PARP-1 gene was reinserted by a retroviral pBabe-puro construct [38] were a generous gift of Dr. Moshe Oren.
3T3 fibroblasts were maintained as exponentially growing monolayers in Dulbecco modified Eagle's minimum essential medium (DMEM), with antibiotics and 10% foetal calf serum as described [6]. The mean doubling time of cells was 27-h and 52-h for PARP-1+/+ and PARP-1-/- 3T3s, respectively.
Cytotoxicity determination
Due to a low cloning efficiency of 3T3 fibroblasts in a feeder layer technique, all cytotoxicity determinations were performed using growth assays. PARP-1+/+ or PARP-1-/- 3T3 fibroblasts were plated in triplicate (105 cells per 25 cm2 vented flasks) and incubated overnight before treatment. After treatment, the flasks were rinsed twice with HBSS and returned to fresh medium for exactly 5 doubling times, with one medium change at day 7 for PARP-1-/- 3T3s. Fibroblasts were harvested by 0.05% trypsin-0.02% EDTA, pelleted, resuspended in medium and counted under microscope in a Malassez cuvette.
Radiation and drug treatment
Irradiation of cells was performed at room temperature in medium equilibrated with 5% CO2 in air, using an IBL-637 (137Cs) γ-ray irradiator (Cis-Biointernational). The dose rate was 0.86 Gy/min or 8.0 Gy/min for survival or immunofluorescence studies, respectively.
Aliquots of NCS were thawed just before use, adjusted to the suitable dilution in ice-cold PBS buffer, pH 6.0, and immediately introduced into culture flasks (25 cm2, 5 ml medium) with gentle agitation. The whole treatment was performed in dim light to avoid photo-induced degradation of the drug. The cytotoxic effect of NCS at fixed concentration was investigated as a function of the length of drug exposure. Cytotoxicity increased steeply with the length of contact and reached completion after 6-min only. More prolonged incubation did not result in increased cell death. Therefore, the length of contact with NCS was 10-min throughout.
For studies of H2O2 response, serial dilutions of concentrated (9.8 M) H2O2 were made in pure water, then in culture medium immediately prior to use. The length of contact with H2O2 was 10-min.
In some experiments ANI was used as a PARP-1 inhibitor [61–63]. When present, ANI (30 μM) from a 3 mM stock solution in pure DMSO was introduced 1-h prior to irradiation or NCS, and removed 1-h later with two HBSS washes. The DMSO concentration in medium was 1% and was kept constant throughout experiments with ANI. Controls were made with DMSO alone at the same concentration.
DNA double-strand break determination
The determination of DSB formation and repair was carried out by pulsed field agarose gel electrophoresis (PFGE) through the clamped homogeneous electric field (CHEF) technique [64, 65]. [2-14C]Thymidine-labeled cells (2–5 106 cells per 25 cm2 flask) were put in ice following treatment, harvested in ice-cold PBS supplemented with 2 mM EDTA by gentle scraping, and collected by sedimentation at 4°C. Pellets from 5 105 cells were resuspended in 150 μl, final volume, of PBS buffer at 37°C, and mixed under mild vortexing with an equal volume of 1.6% w/v low-melting-point agarose (BDH Laboratory) in PBS buffer at 37°C. The suspension (300 μl) was immediately pipetted into pre-chilled 4 × 10 mm moulds and allowed to form plugs on ice for 30-min. Embedded cells were subsequently lysed by immersion of the plugs in 2 ml of a solution containing 2% N-lauroyl sarkosine, 40 mM EDTA, 1 mg/ml proteinase K in PBS, pH 7.8 and incubated, firstly for 2 h in ice, secondly for 24 h at 50°C. The plugs were subsequently washed twice with PBS, and treated for 1-h with 200 μg/ml RNase A. The plugs were finally washed twice with PBS and stored at 4°C overnight prior electrophoresis.
The plugs were inserted into the wells of an 0.8% w/v agarose gel (BioRad, chromosomal grade) made in 0.75 × TAE buffer (40 mM tris-acetate, 2 mM EDTA, pH 8), and the gels submitted to PFGE at 14°C in a CHEF-DR III apparatus (Bio-Rad) with buffer recirculation at 14°C. Migration was for 72 h at 2 V/cm with three switch times, namely, 1200-s, 1500-s and 1800-s with angles of 96°, 100° and 106°, respectively. The molecular weight markers were S. pombe and S. cerevisiae chromosomes (Bio-Rad).
After electrophoresis, the gels were stained with 0.5 μg/ml ethidium bromide under mild agitation (30-min), followed by destaining for 1-h in fresh buffer. DNA fluorescence was visualized over an UV transilluminator with camera recording. The gels were subsequently dried in vacuo over a piece of Whatman paper and analyzed using a Phosphorimager® apparatus (Amersham Pharmacia Biotech), allowing a precise determination of the migration profile of DNA fragments and of the fractional radioactivity released from the plugs (FAR).
Immunofluorescence of PARP-1, pADPr and γ-H2AX
3T3 fibroblasts were grown for 24-h on coverslips and exposed to either γ-rays (5 Gy for γH2AX, 50 Gy for pADPR), H2O2 (1 mM, 10-min contact, 37°C), or graded concentrations of NCS (2 nM, 5 nM or 30 nM, 10-min contact, 37°C), and fixed 10-min after the beginning of treatment. For PARP-1 and pADPr immunofluorescence, fixation was by 4% formaldehyde (10-min, room temperature) in PBS followed by two PBS washes and neutralization of residual formaldehyde by 50 mM NH4Cl (10-min, room temperature). After a further PBS wash (5-min, 4°C), cells were permeabilized by 0.5% Triton X-100 in PBS (5-min, 4°C), and finally rinsed twice with PBS. For γH2AX determination, cells were fixed in a 1:1 v:v acetone:methanol mixe (-20°C, 10-min), dried in air and rehydrated in PBS (15-min, room temperature).
Cell preparations on coverslips were subsequently incubated with 1% bovine serum albumin in PBS (10-min, room temperature) and exposed to anti-pADPr, anti-PARP-1 or anti-γH2AX primary antibody (1/200 dilution, 1-h, 37°C). The coverslips were rinsed thrice with PBS, and incubated with Alexa-488®-conjugated secondary antibody (1/500 dilution, 30-min, 37°C), rinsed thrice with PBS, counterstained with 4,6-diamidino-2-phenylindole (DAPI) and mounted. Immunofluorescence was visualized using a Zeiss Axiophot microscope equipped with a Micromax chilled camera (Princeton Applied Research).
Abbreviations
- ANI:
-
4-amino-1,8-naphthalimide
- ATM:
-
ataxia telangiectasia mutated protein kinase
- CHEF:
-
clamped homogeneous electric field
- DAPI:
-
4,6-diamidino-2-phenylindole
- DNA-PK:
-
DNA-dependent protein kinase
- DSB:
-
double-strand break
- EDTA:
-
ethylene dinitrilo tetraacetate
- HBSS:
-
Hank's balanced salt solution
- NCS:
-
holo-neocarzinostatin
- NCSChrom:
-
NCS chromophore
- pADPr:
-
poly(ADP-ribose)
- PARP-1:
-
poly(ADP-ribose) polymerase
- PBS:
-
phosphate buffered saline
- PFGE:
-
pulsed-field agarose gel electrophoresis
- SSB:
-
single-strand break
- TAE:
-
Tris-acetate-EDTA buffer.
References
Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ: ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001, 276: 42462-42467. 10.1074/jbc.C100466200.
Andegeko Y, Moyal L, Mittelman L, Tsarfaty I, Shiloh Y, Rotman G: Nuclear retention of ATM at sites of DNA double strand breaks. J Biol Chem. 2001, 276: 38224-38230.
Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehman AR, Alt FW, Jackson SP, Jeggo PA: Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994, 265: 1442-1445.
Wu X, Lieber MR: Protein-protein and protein-DNA interaction regions within the DNA end-binding protein Ku70-Ku86. Mol Cell Biol. 1996, 16: 5186-5193.
Jeggo PA, Taccioli GE, Jackson SP: Ménage à trois: double-strand break repair, V(D)J recombination and DNA-PK. Bioessays. 1995, 17: 949-957. 10.1002/bies.950171108.
Fernet M, Ponette V, Deniaud-Alexandre E, Ménissier-de Murcia J, de Murcia G, Giocanti N, Mégnin-Chanet F, Favaudon V: Poly(ADP-ribose) polymerase, a major determinant of early cell response to ionising radiation. Int J Radiat Biol. 2000, 76: 1621-1629. 10.1080/09553000050201118.
Dantzer F, Nasheuer HP, Vonesch JL, de Murcia G, Ménissier-de Murcia J: Functional association of poly(ADP-ribose) polymerase with DNA polymerase α-primase complex: a link between DNA strand break detection and DNA replication. Nucleic Acids Res. 1998, 26: 1891-1898. 10.1093/nar/26.8.1891.
Masson M, Niedergang C, Schreiber V, Müller S, Ménissier-de Murcia J, de Murcia G: XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998, 18: 3563-3571.
de Murcia G, Shall S: From DNA damage and stress signalling to cell death. Poly ADP-ribosylation reactions. 2000, Oxford University Press, New York
de Murcia G, Ménissier-de Murcia J: Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem Sci. 1994, 19: 172-176. 10.1016/0968-0004(94)90280-1.
Wesierska-Gadek J, Bugajska-Schretter A, Cerni C: ADP-ribosylation of p53 tumor suppressor protein: mutant but not wild-type p53 is modified. J Cell Biochem. 1996, 62: 90-101. 10.1002/(SICI)1097-4644(199607)62:1<90::AID-JCB10>3.0.CO;2-J.
Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR: Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000, 275: 40974-40980. 10.1074/jbc.M006520200.
Morrison C, Smith GCM, Stingl L, Jackson SP, Wagner EF, Wang ZQ: Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet. 1997, 17: 479-482. 10.1038/ng1297-479.
Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR: Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998, 273: 14461-14467. 10.1074/jbc.273.23.14461.
Galande S, Kohwi-Shigematsu T: Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999, 274: 20521-20528. 10.1074/jbc.274.29.20521.
Ariumi Y, Masutani M, Copeland TD, Mimori T, Sugimura T, Shimotohno K, Ueda K, Hatanaka M, Noda M: Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene. 1999, 18: 4616-4625. 10.1038/sj.onc.1202823.
Ariumi Y, Ueda K, Masutani M, Copeland TD, Noda M, Hatanaka M, Shimotohno K: In vivo phosphorylation of poly(ADP-ribose) polymerase is independent of its activation. FEBS Lett. 1998, 436: 288-292. 10.1016/S0014-5793(98)01144-2.
Brown ML, Franco D, Burkle A, Chang Y: Role of poly(ADP-ribosyl)ation in DNA-PKcs-independent V(D)J recombination. Proc Natl Acad Sci USA. 2002, 99: 4532-4537. 10.1073/pnas.072495299.
Favaudon V: On the mechanism of reductive activation in the mode of action of some anticancer drugs. Biochimie (Paris). 1982, 64: 457-475.
Dedon PC, Goldberg IH: Free-radical mechanisms involved in the formation of sequence-dependent bistranded DNA lesions by the antitumor antibiotics bleomycin, neocarzinostatin, and calicheamicin. Chem Res Toxicol. 1992, 5: 311-332. 10.1021/tx00027a001.
Kappen LS, Goldberg IH: Deoxyribonucleic acid damage by neocarzinostatin chromophore: strand breaks generated by selective oxidation of C-5' of deoxyribose. Biochemistry. 1983, 22: 4872-4878. 10.1021/bi00290a002.
Favaudon V, Charnas RL, Goldberg IH: Poly(deoxyadenylic-deoxythymidylic acid) damage by radiolytically activated neocarzinostatin. Biochemistry. 1985, 24: 250-259. 10.1021/bi00323a003.
Povirk LF, Houlgrave CW, Han YH: Neocarzinostatin-induced DNA base release accompanied by staggered oxidative cleavage of the complementary strand. J Biol Chem. 1988, 263: 19263-19266.
Hashimoto M, Greenberg MM, Kow YW, Hwang JT, Cunningham RP: The 2-deoxyribonolactone lesion produced in DNA by neocarzinostatin and other damaging agents forms cross-links with the base-excision repair enzyme endonuclease III. J Am Chem Soc. 2001, 123: 3161-3162. 10.1021/ja003354z.
Ohtsuki K, Ishida N: Neocarzinostatin-induced breakdown of deoxyribonucleic acid in HeLa-S3 cells. J Antibiot (Tokyo). 1975, 28: 143-148.
Beerman TA, Goldberg IH: The relationship between DNA strand-scission and DNA synthesis inhibition in HeLa cells treated with neocarzinostatin. Biochim Biophys Acta. 1977, 475: 281-293. 10.1016/0005-2787(77)90019-3.
Tatsumi K, Nishioka H: Effect of DNA repair systems on antibacterial and mutagenic activity of an antitumor protein, neocarzinostatin. Mutat Res. 1977, 48: 195-203. 10.1016/0027-5107(77)90161-0.
Denklau D, Stahl R, Kohnlein W: Effect of neocarzinostatin on E. coli mutants deficient in DNA repair. Z Naturforsch [C]. 1989, 44: 791-796.
Moustacchi E, Favaudon V: Cytotoxic and mutagenic effects of neocarzinostatin in wild-type and repair-deficient yeasts. Mutat Res. 1982, 104: 87-94. 10.1016/0165-7992(82)90125-7.
Shiloh Y, Tabor E, Becker Y: Cellular hypersensitivity to neocarzinostatin in ataxia-telangiectasia skin fibroblasts. Cancer Res. 1982, 42: 2247-2249.
Babilon RW, Soprano KJ, Henderson EE: Hypersensitivity and reduced inhibition of DNA synthesis in ataxia telangiectasia lymphoblasts treated with low levels of neocarzinostatin. Mutat Res. 1985, 146: 79-87. 10.1016/0167-8817(85)90058-6.
Helbig R, Zdzienicka MZ, Speit G: The effect of defective DNA double-strand break repair on mutations and chromosome aberrations in the Chinese hamster cell mutant XR-V15B. Radiat Res. 1995, 143: 151-157.
Povirk LF: DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996, 355: 71-89. 10.1016/0027-5107(96)00023-1.
Muller C, Salles B: Regulation of DNA-dependent protein kinase activity in leukemic cells. Oncogene. 1997, 15: 2343-2348. 10.1038/sj.onc.1201402.
Muller C, Calsou P, Salles B: The activity of the DNA-dependent protein kinase (DNA-PK) complex is determinant in the cellular response to nitrogen mustards. Biochimie (Paris). 2000, 82: 25-28. 10.1016/S0300-9084(00)00341-2.
van Duijn-Goedhart A, Zdzienicka MZ, Sankaranarayanan K, van Buul PP: Differential responses of Chinese hamster mutagen sensitive cell lines to low and high concentrations of calicheamicin and neocarzinostatin. Mutat Res. 2000, 471: 95-105. 10.1016/S1383-5718(00)00122-4.
Yuan SSF, L. CH, Hou MF, Chan TF, Kao YH, Wu YC, Su JH: Neocarzinostatin induces Mre11 phosphorylation and focus formation through an ATM- and NBS1-dependent mechanism. Toxicol. 2002, 177: 123-130. 10.1016/S0300-483X(02)00220-2.
Wang X, Michael D, de Murcia G, Oren M: p53 activation by nitric oxide involves down-regulation of Mdm2. J Biol Chem. 2002, 277: 15697-15702. 10.1074/jbc.M112068200.
Frankenberg-Schwager M: Review of repair kinetics for DNA damage induced in eukaryotic cells in vitro by ionizing radiation. Radiother Oncol. 1989, 14: 307-320. 10.1016/0167-8140(89)90143-6.
Dahm-Daphi J, Dikomey E: Separation of DNA fragments induced by ionizing irradiation using a graded-field gel electrophoresis. Int J Radiat Biol. 1995, 67: 161-168.
Chatterjee S, Cheng MF, Berger RB, Berger SJ, Berger NA: Effect of inhibitors of poly(ADP-ribose) polymerase on the induction of GRP78 and subsequent development of resistance to etoposide. Cancer Res. 1995, 55: 868-873.
Sebestyen A, Mihalik R, Petak I, Kopper L: Modulation of apoptosis signaling in etoposide-treated lymphoma cells. Anticancer Res. 1997, 17: 2609-2614.
Bowman KJ, Newell DR, Calvert AH, Curtin NJ: Differential effects of the poly(ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer. 2001, 84: 106-112. 10.1054/bjoc.2000.1555.
Ménissier-de Murcia J, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G: Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997, 94: 7303-7307. 10.1073/pnas.94.14.7303.
Trucco C, Rolli V, Oliver FJ, Flatter E, Masson M, Dantzer F, Niedergang C, Dutrillaux B, Ménissier-de Murcia J, de Murcia G: A dual approach in the study of poly(ADP-ribose) polymerase: in vitro random mutagenesis and generation of deficient mice. Mol Cell Biochem. 1999, 193: 53-60. 10.1023/A:1006947707713.
Cantoni O, Murray D, Meyn RE: Effect of 3-aminobenzamide on DNA strand-break rejoining and cytotoxicity in CHO cells treated with hydrogen peroxide. Biochim Biophys Acta. 1986, 867: 135-143. 10.1016/0167-4781(86)90073-4.
Dantzer F, de La Rubia G, Ménissier-de Murcia J, Hostomsky Z, de Murcia G, Schreiber V: Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry. 2000, 39: 7559-7569. 10.1021/bi0003442.
Kaneko M, Kodama M, Inoue F: Bimodal pattern of killing of Chinese hamster V79 variant cells by hydrogen peroxide. Free Radic Res. 1994, 20: 229-239.
Shackelford RE, Innes CL, Sieber SO, Heinloth AN, Leadon SA, Paules RS: The Ataxia telangiectasia gene product is required for oxidative stress-induced G(1) and G(2) checkpoint function in human fibroblasts. J Biol Chem. 2001, 276: 21951-21959. 10.1074/jbc.M011303200.
Bowman KJ, White A, Golding BT, Griffin RJ, Curtin NJ: Potentiation of anti-cancer agent cytotoxicity by the potent poly(ADP-ribose) polymerase inhibitors NU1025 and NU1064. Br J Cancer. 1998, 78: 1269-1277.
Trucco C, Oliver FJ, de Murcia G, Ménissier-de Murcia J: DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998, 26: 2644-2649. 10.1093/nar/26.11.2644.
Atorino L, Di Meglio S, Farina B, Jones R, Quesada P: Rat germinal cells require PARP for repair of DNA damage induced by gamma-irradiation and H2O2 treatment. Eur J Cell Biol. 2001, 80: 222-229.
Cantoni O, Cattabeni F, Stocchi V, Meyn RE, Cerutti P, Murray D: Hydrogen peroxide insult in cultured mammalian cells: relationships between DNA single-strand breakage, poly(ADP-ribose) metabolism and cell killing. Biochim Biophys Acta. 1989, 1014: 1-7. 10.1016/0167-4889(89)90234-6.
Dziegielewski J, Beerman TA: Cellular responses to the DNA strand-scission enediyne C-1027 can be independent of ATM, ATR, and DNA-PK kinases. J Biol Chem. 2002, 277: 20549-20554. 10.1074/jbc.M109897200.
Weinfeld M, Chaudhry MA, D'Amours D, Pelletier JD, Poirier GG, Povirk LF, Lees-Miller SP: Interaction of DNA-dependent protein kinase and poly(ADP-ribose) polymerase with radiation-induced DNA strand breaks. Radiat Res. 1997, 148: 22-28.
D'Silva I, Pelletier JD, Lagueux J, D'Amours D, Chaudhry MA, Weinfeld M, Lees-Miller SP, Poirier GG: Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim Biophys Acta. 1999, 1430: 119-126. 10.1016/S0167-4838(98)00278-7.
Ménissier-de Murcia J, Mark M, Wendling O, Wynshaw-Boris A, de Murcia G: Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol Cell Biol. 2001, 21: 1828-1832. 10.1128/MCB.21.5.1828-1832.2001.
Le Rhun Y, Kirkland JB, Shah GM: Cellular responses to DNA damage in the absence of poly(ADP-ribose) polymerase. Biochem Biophys Res Commun. 1998, 245: 1-10. 10.1006/bbrc.1998.8257.
Griffin RJ, Curtin NJ, Newell DR, Golding BT, Durkacz BW, Calvert AH: The role of inhibitors of poly(ADP-ribose) polymerase as resistance-modifying agents in cancer therapy. Biochimie. 1995, 77: 408-422. 10.1016/0300-9084(96)88154-5.
Favaudon V: Gamma-radiolysis study of the reductive activation of neocarzinostatin by the carboxyl radical. Biochimie (Paris). 1983, 65: 593-560.
Banasik M, Komura H, Shimoyama M, Ueda K: Specific inhibitors of poly(ADP-ribose) synthetase and mono(ADP-ribosyl) transferase. J Biol Chem. 1992, 267: 1569-1575.
Bernges F, Zeller WJ: Combination effects of poly(ADP-ribose) polymerase inhibitors and DNA-damaging agents in ovarian tumor cell lines, with special reference to cisplatin. J Cancer Res Clin Oncol. 1996, 122: 665-670. 10.1007/BF01209029.
Schlicker A, Peschke P, Bürkle A, Hahn EW, Kim JH: 4-amino-1,8-naphthalimide: a novel inhibitor of poly(ADP-ribose) polymerase and radiation sensitizer. Int J Radiat Biol. 1999, 75: 91-100. 10.1080/095530099140843.
Blöcher D, Einspenner M, Zajackowski J: CHEF electrophoresis, a sensitive technique for the determination of DNA double-strand breaks. Int J Radiat Biol. 1989, 56: 437-448.
Woudstra EC, Roesink JM, Rosemann M, Brunsting JF, Driessen C, Orta T, Konings AW, Peacock JH, Kampinga HH: Chromatin structure and cellular radiosensitivity: a comparison of two human tumour cell lines. Int J Radiat Biol. 1996, 70: 693-703. 10.1080/095530096144581.
Cornforth MN: Analyzing radiation-induced complex chromosome rearrangements by combinatorial painting. Radiat Res. 2001, 155: 643-659.
Cornforth MN, Bedford JS: A quantitative comparison of potentially lethal damage repair and the rejoining of interphase chromosome breaks in low passage normal human fibroblasts. Radiat Res. 1987, 111: 385-405.
Acknowledgements
Grateful thanks are due to Drs. G. de Murcia and J. Ménissier-de Murcia (UPR 9003 CNRS, ESBS, Illkirch-Graffenstaden, France) for the generous gift of PARP-1-/- 3T3 fibroblasts, and to Dr. Moshe Oren (Dept of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel) who provided PARP-1-/- 3T3s complemented with the PARP-1 cDNA. This work was supported by grants from Electricité de France (RB 2001–02) and the Institut Curie (Protein Remodelling Program), and by financial aid from the Institut National de la Santé et de la Recherche Médicale. M.F. is recipient of a post-doctoral fellowship from the Ligue Nationale Contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' Contributions
GN and NG carried out the survival and DNA repair determinations. MF and FMC performed immunofluorescence analyses. VF conceived the study, and participated in its design and coordination. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Noël, G., Giocanti, N., Fernet, M. et al. Poly(ADP-ribose) polymerase (PARP-1) is not involved in DNA double-strand break recovery. BMC Cell Biol 4, 7 (2003). https://doi.org/10.1186/1471-2121-4-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2121-4-7