Abstract
Background
Bloom syndrome is one of the most cancer-predisposing disorders and is characterized by genomic instability and a high frequency of sister chromatid exchange. The disorder is caused by loss of function of a 3' to 5' RecQ DNA helicase, BLM. The exact role of BLM in maintaining genomic integrity is not known but the helicase has been found to associate with several DNA repair complexes and some DNA replication foci.
Results
Chromatin immunoprecipitation of BLM complexes recovered telomere and ribosomal DNA repeats. The N-terminus of BLM, required for NB localization, is the same as the telomere association domain of BLM. The C-terminus is required for ribosomal DNA localization. BLM localizes primarily to the non-transcribed spacer region of the ribosomal DNA repeat where replication forks initiate. Bloom syndrome cells expressing the deletion alleles lacking the ribosomal DNA and telomere association domains have altered cell cycle populations with increased S or G2/M cells relative to normal.
Conclusion
These results identify telomere and ribosomal DNA repeated sequence elements as chromosomal targets for the BLM DNA helicase during the S/G2 phase of the cell cycle. BLM is localized in nuclear bodies when it associates with telomeric repeats in both telomerase positive and negative cells. The BLM DNA helicase participates in genomic stability at ribosomal DNA repeats and telomeres.
Similar content being viewed by others
Background
Bloom syndrome is caused by loss of function of a DNA helicase
Bloom syndrome (BS) is a rare cancer-prone autosomal recessive disorder characterized by genomic instability, immunodeficiency, infertility and small stature [1, 2]. BS cells have a distinctive genomic instability: a high frequency of sister chromatid exchanges (SCEs) and quadriradial formation. BLM, the gene mutated in BS, encodes a DNA helicase (BLM) of the RecQ family [3]. The 3' to 5' DNA helicase activity of BLM is essential for genomic stability as transfection of the normal BLM cDNA, but not missense alleles lacking helicase activity, into BS cells reduces the frequency of SCEs [4]. Previous work from this laboratory and others [4–8] demonstrated the DNA helicase activity of BLM in vitro on a variety of DNA substrates with a preference for multi-stranded structures [8].
BLM is localized in nuclear bodies and the nucleolus
The BLM DNA helicase is found in two distinct nuclear structures in normal human cells, ND10 or PML nuclear bodies (NBs), and the nucleolus [9, 10]. The NBs are dynamic PML-dependent depots of proteins disrupted upon viral infection and in certain human malignancies [11]. NBs contain multiple transcription regulators and DNA binding proteins, and have been implicated in the regulation of apoptosis and DNA repair [12–14] although the precise function of these nuclear matrix-associated structures [15] is still unknown. BLM is found in alternative or ALT NBs in SV40-transformed cell lines that synthesize telomeres using a recombination-based pathway [10, 16].
BLM helps to maintain genomic stability in the S/G2 phase of the cell cycle and associates with DNA repair proteins
The expression of the BLM DNA helicase is cell cycle regulated, showing a marked increase in S phase and peaking in G2 phase [17, 18]. The increase in BLM mRNA and protein expression coincides with its appearance in the nucleolus and co-localization with WRN [10], the RecQ DNA helicase mutated in Werner syndrome. The localization of BLM in the nucleolus is necessary for the maintenance of genomic stability [19]. BLM and WRN interact in vitro and can be co-localized in NBs in certain cell types [20]. BLM is part of a large BRCA1-containing complex (BASC) containing multiple DNA repair factors and DNA replication components [21]. Studies have documented an interaction between BLM and a number of repair or replication factors: RPA [22], RAD51 [23], TOPIIIα[24], MLH1 [25]. Despite these and other studies of BS cells the precise target sites for the BLM DNA helicase are unknown. This study addresses this issue by using a BLM antibody and chromatin immunoprecipitation to isolate chromosomal target sites for BLM.
Results
Isolation of chromosomal sites where BLM is located
Proteins bound to DNA can be reversibly cross-linked as chromatin complexes by treating cells with formaldehyde [26]. Cross-linked whole cell lysates were sonicated to shear genomic DNA to 200–600 bp fragments and immunoprecipitated with rabbit polyclonal BLM antibody. DNA fragments were isolated from normal lymphoblastoid (GM00103 and GM00536) and fibroblast (AG06814 or WI-38) cell lines as well as a BS fibroblast (GM02932B) cell line. Random DNA fragments from normal lymphoblastoid cells without antibody addition were also purified. DNA fragments from all sources were linkered, amplified, cloned into a plasmid vector, and sequenced. Sequences were analyzed using public databases and websites [27]. Twenty plasmid inserts were sequenced from each source DNA clone library. Two inserts recovered with anti-BLM from the normal lymphoblastoid cell line (GM00536) were readily identified as 435 bp of human telomeric repeat DNA and a region of the rDNA repeat unit (U13369) corresponding to 37501–37980 bp. These two inserts were found only in the GM00536 fragments recovered with anti-BLM. These two fragments recovered in this initial study prompted further studies and are likely not to be the only chromosomal targets of BLM.
BS cells in long term culture have less rDNA and more variation in telomere repeats than normal cells
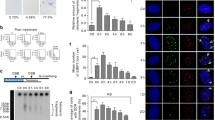
A normal (HG1522) and a BS (HG1525) lymphoblastoid cell line were grown in culture for one year and individual clones were generated as described previously [28] (Figure 1A). Genomic DNA extracted from 24 clones of each parental cell line was analyzed for total rDNA and telomere DNA (Figure 1B). The BS cell lines have significantly less rDNA than the normal clones (Figure 1B, Table 1). In contrast the BS cell lines have a greater variance and somewhat more total telomere DNA (Figure 1B, Table 1). These studies demonstrate that loss of BLM generates diversity in the amount of these two GC-rich repeated sequence elements but does not determine the exact mechanism or location of the variation in these cell lines.
Analysis of rDNA and telomere repeats in normal and BS lymphoblastoid cell lines. A. Diagram of experimental design [28]. B. Relative amounts of total rDNA and telomere DNA in the clones from the normal (HG1522) and BS (HG1525) cell lines using a Perkin-Elmer (ABI) kit for rDNA or rRNA and membrane-transfer hybridization of total genomic DNA for telomere DNA.
Mapping of BLM domains required for telomere association
A series of cell lines expressing GFP-BLM variants (Figure 2A) under tetracycline regulation have been constructed and characterized [19]. ChIP preparations were prepared from each induced cell line expressing the GFP-BLM proteins at approximately the same levels [19] and one expressing EGFP alone. BS cells contain no BLM by western analysis or immunofluorescent microscopy [4]. A measure of the relative amount of total genomic DNA recovered in each preparation was made by quantitation of the Alu-Sb2 family repeats using a molecular beacon assay [29]. The immunoprecipitate from the cell line expressing the GFP-BLM ΔN2 protein contained the most DNA, consistent with the higher level of expression of this protein under standard induction conditions relative to the other BLM alleles [19]. The immunoprecipitate from the cell line expressing EGFP alone contained the least amount of DNA and was 21 fold less than the ΔN2 preparation. Alu-Sb2 equivalent amounts of DNA recovered from each cell line were transferred to a membrane and hybridized with a radiolabeled telomere oligonucleotide [10]. The ΔN2–ΔN4 preparations contained the least amount of total telomere DNA compared to the other BLM proteins (Figure 2B).
BLM mutation map and telomere association domains. A. Map of BLM mutations [19] and domains with assigned sequence identities. NLS indicates the basic nuclear localization signal [60]. The HRD (H elicase and R NAseD) region is common to several members of the RecQ DNA helicase family and structural modeling suggests it is a single-strand nucleic acid binding domain [31]. The red bars indicate regions with a high density of negatively charged amino acids [3]. B. ChIP mapping of the telomere association domains of BLM. Equivalent amounts of DNA recovered from the induced cell lines were denatured and transferred to a nylon membrane. Telomere repeats sequences were quantitated by hybridization with a radiolabeled complementary oligonucleotide [10] and exposure to a phosphorimager screen. Hybridization units were calculated relative to the cell line expressing EGFP alone. Values are the average of two experiments: BLM 8+/-1.4, ΔN1 7+/-0.1, ΔN2 4+/-2.8, ΔN3 1+/-0.1, ΔN4 2+/-1.4, K695T 5+/-2.8, ΔH1 5+/-0.1, ΔC1 6+/-1.4, ΔC2 7+/-1.4.
This observation demonstrates that the region of BLM required for NB localization [19] is the same as the telomere association domain. BLM can be found in ALT NBs at the large heterogeneous telomeres found in telomerase-negative normal SV40-transformed fibroblast cell lines [10, 16]. The BS SV40-transformed cell line (GM0855B) used to construct the tetracycline-inducible cell lines is telomerase-positive (F. B. Johnson and L. Guarente, personal communication, 2003). Therefore BLM in NBs associates with telomere repeats in cell lines with or without telomerase. The BLM alleles that delete the two largest clusters of acidic amino acids (Figure 2A) showed the least association and suggest that these domains may contribute to BLM telomere association.
The C-terminal domain of BLM directs it to two sites in the rDNA repeats
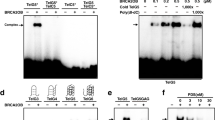
The rDNA genes in human cells have approximately a 15 kb coding region and a 28 kb non-transcribed spacer (NTS) where DNA replication forks initiate [30]. The major sequence features of the rDNA repeat unit are shown (Figure 3A) including the cloned BLM association site and the Sal boxes, an important region where termination of both RNA transcripts and DNA replication forks occur [30]. Equal amounts of DNA (Alu-Sb2 equivalents) from ChIP preparations of each induced cell line were amplified with Taq DNA polymerase using 8 primer sets from the rDNA repeat (Figure 3A). Equivalent volumes from each reaction were transferred to a membrane and hybridized with the radiolabeled forward primer. The recovery of each rDNA region from each cell line was normalized to the cell line expressing EGFP alone (Figure 3B). The region in the NTS around 37 kb proved to be the major BLM association site among the sites tested. BLM was found also in the coding region of the 18S rRNA gene to a lesser degree. BLM was not found at the promoter region or the Sal boxes. Identification of the C-terminal region of BLM as the domain required for association with rDNA repeats is in agreement with the previous study from this laboratory [19] that showed this region is required for nucleolar localization. The ΔC2 BLM protein (Figure 3A) or HRD [31] domain showed the least amount of rDNA localization followed by the ΔC1 and ΔH1 mutations (Figure 3A). The normal and N-terminal deletion proteins showed the greatest rDNA association at both sites in agreement with the previous cell biology study [19]. The finding of BLM primarily in the NTS suggests a role for BLM in the unwinding of multi-stranded tangles that can occur in this region and a potential role in initiation of DNA replication. The presence of multiple Alu elements in this region in opposite orientations can produce complicated intra- and inter-chromosomal structures (Figure 3C). The real substrate for BLM is most likely entangled chromosomes and this cluster of Alu elements may be the most prone to invade a nucleolar neighbor.
Map of sequence elements in the human rDNA repeat unit and identification of the BLM association domains. A. The human rDNA repeat unit (U13369) sequence features and the location of the region isolated with a polyclonal BLM antibody. The orange arrows indicate sites of primer sets. The dotted lines indicate the direction of DNA replication origins [30]. B. ChIP mapping of BLM association with the rDNA repeat and mapping of BLM domains required for rDNA association. Equivalent amounts of DNA recovered from the induced cell lines were amplified with Taq DNA polymerase and one of eight primer sets, denatured and transferred to a nylon membrane. The PCR products were hybridized with the radiolabeled forward primer in each set and quantitated using a phosphorimager. Hybridization units were calculated relative to the cell line expressing EGF alone. C. A simple model for the BLM-binding region of the rDNA repeat.
Purification and assay of GFP-fusion proteins
These studies indicate that deletion of the N3 and N4 domains of BLM have the greatest effect on telomere association, and the C1 and C2 domains are required for BLM to accumulate in the nucleolus. A recombinant baculovirus for each of these GFP-BLM proteins was constructed and expressed in Sf9 cells. The accumulation of the GFP-BLM proteins in the nucleus of Sf9 cells was similar to those seen in human cells (Figure 4A) [19]. The ΔN3 and ΔN4 proteins were diffuse or nucleolar-localized, and the ΔC1 and ΔC2 proteins were aggregated. All proteins were purified using a protocol developed for the normal GFP-BLM fusion protein [19]. The ΔC1 protein was recovered in an amount similar to normal whereas the ΔN3 and ΔN4 were slightly less, and the ΔC2 protein was nearly 10 fold less than normal.
Expression and purification of GFP-BLM alleles in Sf9 cells. A. Fluorescent micrographs of Sf9 cells infected with recombinant baculoviruses expressing GFP-BLM deletion proteins constructed using methods described previously for the full length normal BLM [19]. The ΔN3 image contains a bi-nucleated cell. B. Silver-stained gels of GFP-BLM deletion proteins isolated in the same manner as normal GFP-BLM [19]. Similar volumes of peak fractions are shown to demonstrate the relative recoveries of the different alleles.
Standard helicase assays were performed using all of the purified proteins (Figure 5). The ΔN3 and ΔN4 proteins had DNA-dependent ATPase specific activity nearly identical to normal GFP-BLM (88% and 79%). The ΔC1 and ΔC2 were lower (65% and 60%) but still active. In contrast the ΔC1 and ΔC2 proteins were nearly as active as normal in the unwinding assay (75% and 73%) whereas the ΔN3 and ΔN4 proteins have less specific activity (30% and 39%). All of the deletion proteins have activity in vitro and the lowered unwinding activity of the ΔN3 and ΔN4 proteins may be due to alteration of a DNA-binding domain or destabilization of the helicase domain as these deletions remove amino acids close to it. The deletions may affect interactions with more complex substrates as well. There is a correlation between mis-localization and elevated SCE [19] but not so much with reduced helicase activity. These data suggest that both the localization of BLM and the helicase activity contribute to genomic stability.
Helicase activity of the GFP-BLM deletion proteins. A. Autoradiogram of the unwinding assay gel. The 0 lane contains no enzyme. The Δlane reaction was heated at 95°C for 5 minutes. The C2 reaction contains 1 ng protein and other reactions with the deletion proteins contain 25 ng. The normal BLM reactions contain the amounts shown in the right graph in panel B. Activities are reported relative to 8 units/ng DNA-dependent ATPase activity and 7 units/ng unwinding activity for normal GFP-BLM. B. Graphic representation of helicase activities relative to normal GFP-BLM. Assays were performed as described previously [4]. Quantitation of the autoradiogram using a phosphorimager is shown in the left and right graphs. Relative DNA-dependent ATPase activities are shown in the left graph. The thin layer ATPase assays are not shown.
Cell cycle profiles of BS cells expressing BLM alleles
The cell lines expressing the normal and altered BLM proteins were induced for two days as described before [19] and harvested, fixed and stained for FACS analysis by DNA content. Under these conditions all of the cell lines (except for the ΔH1 cell line) appeared similar to the cell line expressing normal GFP-BLM. There was little difference between the induced and uninduced populations (data not shown). The experiment was repeated except that the cell lines were induced for five days. Among the N-terminal mutations the ΔN3 and ΔN4 cell profiles showed the greatest differences from normal (Figure 6A, Table 2). The ΔN3 profile has an increase in the S and G2/M phase population and the ΔN4 profile has an altered G2/M population. The ΔN1 and ΔN2 cultures show increases in the S phase population as well (Table 2). Analysis of the C-terminal mutations demonstrates an alteration in the S phase population in the ΔC2 cell line as well as an increase in the G2/M population (Figure 6B, Table 2). The ΔC1 cell line has an increase in the G2/M population also (Table 2). Cells expressing BLM proteins with deletions in the telomere and rDNA association domains accumulate cells in the S or G2/M phases of the cell cycle. Therefore BLM function can affect cell cycle progression and BLM localization may be critical for such function.
Cell cycle assay of induced cell lines. Cell lines expressing GFP-BLM alleles described previously [19] were induced for five days with 1 μg/ml doxycycline. Cell cultures were harvested, fixed and stained with PI. Relative DNA content was measured by FACS analysis. A. Comparison of DNA content profiles of the cell lines expressing the normal and N-terminal deletion alleles. B. Comparison of DNA content profiles of the cell lines expressing the normal, helicase and C-terminal mutant alleles.
Formation of quadriradials in BS cells is favoured in gene dense chromosomes
One of the distinctive features found in lymphocytes from BS individuals are quadriradials or recombination figures involving homologs that occur in 1/100 metaphases from lymphocytes [1]. Chromosomes occupy discrete territories within the nucleus and regions of active transcription extend out from the central mass [32]. Recent studies demonstrate that gene dense chromosomes are found at the center of the nucleus and replicate early in S phase relative to low gene density chromosomes that replicate late [33]. This model of nuclear structure suggests that the highly transcribed homologous chromosomes are in closer physical proximity to each other as compared to the chromosomes with lower gene density. This represents an opportunity for entanglement or invasion of one homolog by a single strand from the other during DNA replication or transcription. When the data from a study of 481 quadriradials from BS cells [34] is compared to the predicted gene density for each chromosome from the public database [27] a direct correlation is seen between quadriradial formation and gene density (Figure 7). Some irregularities in the correlation can be due to expression differences in lymphocytes relative to predicted gene density. Related models for quadriradial formation have been previously proposed [35].
Quadriradial formation and gene density. A. Data from a study of 481 quadriradials found in BS lymphocytes [34] is shown as the number of quadriradials observed relative to the length of each chromosome. These values are compared to gene density on each of the 22 human autosomal chromosomes as calculated from the public human genome database [27]. The number of each chromosome is shown above the gene density bar. B-D. Model of origin of quadriradial composed of two acrocentric chromosomes that have replicated in the presence of BrdU and stained for SCE analysis. The rDNA repeats are in close physical proximity, possibly in the nucleolus, and have numerous single-stranded invasions that are not resolved. During DNA replication the sister chromatids can develop entanglements as well. These unresolved invasions combined with the attachments between the sister chromatids lead to mitotic chiasmata. Quadriradial image from Schonberg [61].
Discussion
BLM participates in genomic stability at telomere and rDNA repeats
Aging and cancer are associated with changes in telomeres and rDNA [36, 37]. These two GC-rich repeated sequence elements are concentrated at the ends of chromosomes and are targets for the formation of stable strand invasion events from a sister chromatid or nearby chromosome during DNA replication. These invasion events must be resolved by recombinational repair prior to mitosis or the attached chromosomes will break or assort improperly. The acrocentric chromosomes active in ribosome production in the nucleolus are especially vulnerable due to their close spatial proximity and high rate of transcription, an activity that can lead to invasive single-strands as well. Failure to resolve tangles in the rDNA repeat or the telomeres of these chromosomes could produce the satellite associations seen in the cells of persons with BS [38], leading to aneuploidy or breaks in the acrocentric chromosomes. This study demonstrates the localization of the RecQ family DNA helicase BLM at telomeres and at two sites in the human rDNA repeat, and provides the first evidence for specific genomic targets for the BLM helicase.
BLM in NBs associates with telomeres
Telomeres have specialized chromatin with several well characterized binding proteins and potentially form a variety of three-dimensional structures including G quartets and D-loops [39, 40]. Studies on BLM from several laboratories have documented the localization of BLM in ALT NBs in telomerase-negative SV40-transformed cell lines and interaction or co-localization with telomere-binding proteins [10, 41, 42]. This study maps the telomere-association domain of BLM to the same region required for its localization in NBs in a telomerase-positive cell line. Therefore BLM associates with telomeres from within NBs regardless of the expression of telomerase. Telomeres and PML-containing NBs are both tethered to the nuclear matrix [13, 15, 43]. This may facilitate co-localization of telomeres and NBs containing BLM.
Helicases are required for many DNA manipulation mechanisms and are deleterious when deregulated or mislocalized. The packaging of BLM into the NBs can regulate its activity as well as coordinate its arrival to sites undergoing DNA repair [44]. Many DNA repair or damage surveillance proteins are found in distinct focal complexes in the nucleus at sites of DNA damage where they co-localize and co-purify [21]. The observation that the MRE11/RAD50/NBS complex associates with telomeres in a cell cycle dependent manner [45] as well as co-localization with PML in nuclear bodies [14], suggests that repair surveillance is an essential component of telomere maintenance. BS cells show alterations in formation of MRE11/RAD50/NBS foci [46] consistent with a mechanism of coordination of DNA response pathways. The two cell lines most deficient in BLM telomere association, ΔN3 and ΔN4, show an accumulation of cells in the S or G2/M phase of the cell cycle. These proteins in vitro have nearly normal DNA-dependent ATPase activity but both show lowered unwinding activity as well as a reduced efficiency in correction of the frequency of SCEs [19]. These data suggest that reduced BLM activity at telomeres triggers cell cycle checkpoints that delay progression until an alternative resolution activity involving SCEs completes the chromosome duplication program.
Comparison of normal and BS lymphoblastoid cell lines demonstrates that the BS cell lines have a greater variance in the amount of telomeric DNA relative to the normal. This observation suggests that BLM is not required for telomere synthesis or function but rather helps to limit the amount of telomeric DNA. A simple model for BLM or other RecQ family DNA helicases at telomeres would be to process the 3' end of the telomere tail sequestered in the D-loop structure as has been suggested for the MRE11/RAD50/NBS complex [45] to prevent rolling circle replication of the telomere. Loss of BLM activates SCEs that could resolve telomere structures in an unequal manner whereas normal cells would resolve invasion events without crossing-over or amplification.
BLM is found where forks originate in the rDNA repeat and may participate in the initiation of DNA replication in the rDNA
The ribosomal DNA repeat in eukaryotic cells is replicated in a unique manner. Replication forks initiate at multiple sites in the NTS region and terminate at the same region as rRNA transcripts such that forks do not oppose transcription activity [30]. Transcription affects the topology of this DNA region, as RNA polymerase is itself a DNA helicase. Movement through the DNA helix produces positive supercoils in front of the polymerase, making the DNA helix more difficult to open and negative supercoils behind the enzyme facilitating DNA helix opening [47]. Accumulation of negative supercoils in the NTS region behind the promoter site may facilitate loading of the replication origin complex by allowing access of the replication machinery, as well as creating an opportunity for formation of complex DNA structures involving the many repeated sequence elements in the NTS. BLM could facilitate DNA replication by unwinding complex structures that would impede fork formation or movement. Regions of non-coding DNA could act as traps for superhelical tension generated in the coding regions. GC-rich regions such as telomeres and clusters of Alu elements could use RecQ DNA helicases plus topoisomerase partners to help fork passage. Superhelical tension in non-coding regions could contribute to the initiation of DNA replication by facilitating origin recognition or opening of condensed regions.
DNA replication, recombination and repair pathways work together to maintain genomic integrity
Chromosomes replicate in a mosaic manner with euchromatin replicating early and heterochromatin replicating late in general [48, 49]. Early forks in open chromatin regions may stall upon entering silent heterochromatin [50]. Therefore restarting stalled forks using a recombination-based mechanism [51–53] may be a normal process of chromosomal DNA replication initiation in heterochromatin. Studies on the RecQ DNA helicases in model organisms have demonstrated a role for E. coli RecQ in restarting stalled forks [54], Xenopus BLM in DNA replication [55], Drosophila BLM in a single-strand annealing recombination pathway [56] and yeast Sgs1p in maintaining telomeres using recombination [57] and limiting homeologous recombination [58]. Recent work has shown that hBLM complements both replication and repair defects of yeast dna2 mutants [59]. This work demonstrates a role for human BLM in maintenance of rDNA and telomere repeats.
Conclusions
This study isolates telomere and rDNA repeats by chromatin immunoprecipitation with anti-BLM. Hybridization experiments using deletion alleles of GFP-BLM demonstrate that the N-terminal (NB localization) domain of BLM is the telomere association domain, and the C-terminal domain (nucleolar localization) domain is the rDNA association domain, directing BLM primarily to the NTS. BS cells in long-term culture have less rDNA and more variability in telomere repeats than normal cells. Cell lines expressing BLM proteins deleted in the telomere and rDNA association regions have altered cell cycle profiles. BLM localized in NBs associates with telomere repeats in telomerase positive or negative cells. These studies suggest a role for BLM-containing complexes to untangle rDNA repeats in the nucleolus in S phase and at telomere repeats in S/G2/M as a participant in NB-mediated DNA repair. These studies suggest that the localization of BLM as well as its enzymatic activity affects genomic stability.
Methods
Cell lines
Fibroblast and lymphoblastoid cell lines used for ChIP were obtained from the Coriell Cell Repositories: Normal human lung fibroblasts AG06814 (WI-38), BS skin fibroblasts GM02932B (blmAsh/blmAsh), normal lymphoblastoid cell lines GM00103 and GM00536. The Tet-responsive SV40-transformed BS cell line HG2551a and cell lines expressing GFP-BLM fusion genes were described before [19]. HG1522 and HG1525 cell lines were isolated and described previously [28].
Chromatin immunoprecipitation
Fibroblast cell lines were grown as described before [4, 19] in T-75 flasks. Normal and BS fibroblasts were split and seeded out 24 hours before fixation. The medium was removed by aspiration, followed by the addition of PBS + 1% formaldehyde (Sigma) to the flasks (5 mls) After 10 minutes of incubation at room temperature glycine pH 7.0 was added to 0.125 M and the flasks were further incubated for 5 minutes. The fixed cells were washed with PBS twice and harvested by scraping with 1 ml cold lysis buffer (RIPA buffer (1% SDS, 0.1% NP40, 1% triton X-100, 0.5% sodium deoxycholate, 1 mM EDTA) + 1 mM PMSF + 1 Roche Biomedical Complete protease inhibitor tablet). Lymphoblastoid cell lines were harvested by centrifugation and fixed in 5 mls of PBS + 1% formaldehyde. After washing with PBS the cells were lysated on ice by addition of 1 ml of the cold lysis buffer. The lysate was collected on ice and sonicated with a Braun sonicator fitted with a microtip for 5 times for 20 seconds at the maximum setting for the microtip. The lysate was centrifuged at 13 k rpm in a Sorval SS-34 rotor for 10 minutes. The final lysate preparation was aliquoted into siliconized sterile 1 ml microfuge tubes and stored at -70°C. The protein concentration of the lysates and the affinity-purified BLM antibody [4] were determined using the Bradford reagent from BioRad. Immunoprecipitations were performed by mixing 1 mg of lysate with 10 μg of polyclonal BLM antibody and overnight incubation at 4°C on a Clay Adams Nutator platform. Protein A beads (Pierce) were equilibrated with the lysis buffer and 50 μl of a 50% suspension was added to the immunoprecipitate and mixed on the Nutator at 4°C for 1 hour. The beads were collected by spinning at 10 k rpm in an Eppendorf microfuge for 15 seconds, washed twice in lysis buffer and resuspended in 50 μl 2% SDS. The eluted chromatin fragments were processed as described [26] using the linkers listed below and PCR amplification with one primer [26] using the Promega PCR Master Mix. Qiagen PCR purification kits were used to purify the DNA fragments before and after amplification as described [26].
DNA cloning, sequence analysis and websites
The purified DNA fragments were visualized on a 1% agarose gel to verify recovery of 200–600 bp products and cloned into the pCR 2.1 TOPO vector using the TOPO TA Cloning kit (Invitrogen). Plasmid inserts were sequenced directly or after PCR amplification with the M13 primers supplied in the cloning kit using an ABI BigDye Terminator v3.0 kit and an ABI377 sequencer.
Sequences were screened for repeated sequence elements using Repeatmasker2 http://repeatmasker.genome.washington.edu/. Sequences were compared to the human ribosomal DNA repeat unit (U13369) using Pairwise BLAST and to the public database of the human genome using BLAST http://www.ncbi.nlm.nih.gov with high and low stringency settings. Sequences that were not identified using these programs were analyzed for sequence motifs using MEME version 3.0 http://meme.sdsc.edu/meme/website/meme.html and for folding patterns using the DNA fold server http://bioinfo.rpi.edu.
Linkers and primers
Invitrogen synthesized oligonucleotides. Linkers for ChIP clones:
5'-TACGTGCGATAACGAGCGAGTATCGTC-3'/5'-GACGATACTCGC-3' or 5'-ATGCACGCTATTGCTCGCTCATAGCAG-3' / 5'-CTGCTATGAGCG-3'. Primers for amplifying plasmid inserts:
5'-CAGCAAACAGCTATCACC-3' / 5'-TGTAAAACGACGGCCAGT-3'.
Ribosomal DNA primers:
1 kb 5'-CTCTGCCGCGATCCTTCT-3'/5'-GGGAGCCGGAAGCATTTT-3',
16 kb 5'-ATGTATCGAAGCCCCATTTC-3'/5'-GGCCCCAAAAGAAAAGAAAA-3',
19 kb 5'-GTGTGTTGGGGCTGAAAACT-3'/5'-CCTCCTCAAACGCAAGAAAG-3',
27 kb 5'-TGTTGGCTTGTTTTGTTTCG-3'/5'-AAACATCAACCGGCTCTCAC-3',
34 kb 5'-GACAACTCACGCCCTAATGC-3'/5'-ATCCTCCGAAAGAGGTTGCT-3',
37 kb 5'-AGGAGTCCCCTGGTCTGTCT-3'/5'-GTCAAGGTCCAAACCGAAAA-3',
42 kb 5'-GATGGTGGCGTTTTTGGG-3'/5'-CGACTCGGAGCGAAAGATATA-3'
4 kb primers were supplied by Perkin-Elmer and included in the kit for rRNA quantitation (see below). Oligonucleotide primers for PCR were chosen using the primer3 Website at MIT http://www.genome.wi.mit.edu/.
Hybridizations
The molecular beacon and primers for quantitating the recovery of genomic DNA using the Alu-Sb2 elements were made by Research Genetics and used as described [27]. The relative amounts of rDNA were measured from 200 ng of total genomic DNA prepared previously [28] isolated from each cell line using a kit from Perkin-Elmer (ABI TaqMan Ribosomal RNA Control Reagents) containing primers and a VIC probe to the 18S rRNA coding region in an ABI 7700. Membrane-transfer hybridizations were performed using Hybond N+ membranes (Amersham-Pharmacia) and a dot blot transfer apparatus and protocols supplied by the manufacturer (Schleicher and Schuell Minifold I Spot Blot System). Radiolabeled oligonucleotide probes were made as described [10]. Relative hybridization units were measured using a Molecular Dynamics Storm Phosphorimager and Imagequant. Relative total rDNA and telomere DNA are reported by normalizing values to the cell line with the lowest amount.
Baculovirus Constructions
BLM alleles were constructed by replacing fragments of the full-length normal cDNA construction with the deleted ones described before [19]. Co-transfection of the GFP-fusion plasmids and linear Bac-N-Blue AcMNPV DNA into Sf9 cells was according the to supplier's recommendations (Invitrogen). Recombinant baculoviruses were isolated by limiting dilution and inspection by immunofluorescent microscopy to confirm nuclear localization [19, 59].
Purification of GFP-fusion proteins and helicase assays
Full length and deleted BLM proteins were expressed and purified as before [19] except that a 5 ml Hi-trap heparin column (Amersham-Pharmacia) was substituted for the ion-exchange step. Standard helicase assays were performed as previously [4].
Cell cycle analysis
SV40-transformed BS fibroblasts containing different BLM alleles [19] were grown for 5 days in DMEM + 10% fetal bovine serum (Clontech tetracycline-free) + 0.2 mg/ml G418 (Geneticin, Invitrogen) + 0.1 mg/ml hygromycin (Roche Molecular Biochemicals) and 5% CO2 with 1 μg/ml doxycycline. Cells were harvested and fixed in PBS + 50% ethanol on ice and stored at 4°C. Fixed cells were resuspended in PBS + PI according to a protocol supplied by Becton-Dickinson. Cell cycle populations were calibrated using control cells and software purchased from Becton-Dickinson (FACSCalibur flow cytometer and CellQuest).
Abbreviations
- ALT:
-
alternative telomere synthesis
- BS:
-
Bloom syndrome
- ChIP:
-
chromatin immunoprecipitation
- FACS:
-
fluorescence activated cell sorter
- HRD:
-
helicase and RNAseD
- NB:
-
ND10 or PML nuclear body
- NTS:
-
non-transcribed spacer
- PBS:
-
phosphate buffered saline
- PI:
-
propidium iodide
- rDNA:
-
ribosomal DNA
- SCE:
-
sister chromatid exchange
- SDS:
-
sodium dodecylsulfate
References
German J: Bloom syndrome: a Mendelian prototype of somatic mutational disease. Medicine. 1993, 72: 393-406.
German J, Ellis NA: Bloom syndrome. In The Genetic Basis of Human Cancer. Edited by: Vogelstein B, Kinzler KW. 1998, New York: McGraw-Hill, 301-315.
Ellis NA, Groden J, Ye TZ, Straughen J, Lennon D, Ciocci S, Proytcheva M, German J: The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995, 83: 655-666.
Neff NF, Ellis NA, Ye TZ, Noonan J, Huang K, Sanz M, Proytcheva M: The DNA helicase activity of BLM is necessary for the correction of the genomic instability of Bloom syndrome cells. Mol Biol Cell. 1999, 10: 665-676.
Sun H, Karow JK, Hickson ID, Maizels N: The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem. 1998, 273: 27587-27592. 10.1074/jbc.273.42.27587.
Karow JK, Constantinou A, Li JL, West SC, Hickson ID: The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc Natl Acad Sci U S A. 2000, 97: 6504-6508. 10.1073/pnas.100448097.
van Brabant AJ, Ye TZ, Sanz M, German J, Ellis NA, Holloman WK: Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000, 39: 14617-14625. 10.1021/bi0018640.
Mohaghegh P, Karow JK, Brosh RM, Bohr VA, Hickson ID: The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001, 29: 2843-2849. 10.1093/nar/29.13.2843.
Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff NF, Kamitani T, Yeh ETH, Strauss JF, Maul GG: PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999, 147: 221-233. 10.1083/jcb.147.2.221.
Yankiwski V, Marciniak RA, Guarente L, Neff NF: Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci USA. 2000, 97: 5214-5219. 10.1073/pnas.090525897.
Hodges M, Tissot C, Howe K, Grimwade D, Freemont P: Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am J Hum Genet. 1998, 63: 297-304. 10.1086/301991.
Borden KL: Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol Cell Biol. 2002, 22: 5259-5269. 10.1128/MCB.22.15.5259-5269.2002.
Maul GG, Negorev D, Bell P, Ishov AM: Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000, 129: 278-287. 10.1006/jsbi.2000.4239.
Lombard DB, Guarente L: Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 2000, 60: 2331-2334.
Tse WT, Tang J, Jin O, Korsgren C, John KM, Kung AL, Gwynn B, Peters LL, Lux SE: A new spectrin, beta IV, has a major truncated isoform that associates with promyelocytic leukemia protein nuclear bodies and the nuclear matrix. J Biol Chem. 2001, 276: 23974-23985. 10.1074/jbc.M009307200.
Henson JD, Neumann AA, Yeager TR, Reddel RR: Alternative lengthening of telomeres in mammalian cells. Oncogene. 2001, 20: 7234-7242. 10.1038/sj.onc.1204764.
Gharibyan V, Youssoufian H: Localization of the Bloom syndrome helicase to punctate nuclear structures and the nuclear matrix and regulation during the cell cycle: comparison with the Werner's syndrome helicase. Mol Carcinog. 1999, 26: 261-273. 10.1002/(SICI)1098-2744(199912)26:4<261::AID-MC5>3.0.CO;2-A.
Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M: Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene. 2000, 19: 2731-2738. 10.1038/sj.onc.1203595.
Yankiwski V, Noonan J, Neff NF: The C-terminal domain of the Bloom syndrome DNA helicase is essential for genomic stability. BMC Cell Biology. 2001, 2: 11-10.1186/1471-2121-2-11.
von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng X, Brosh RM, Hickson ID, Bohr VA: Colocalization, physical and functional interaction between Werner and Bloom syndrome proteins. J Biol Chem. 2002, 277: 22035-22044. 10.1074/jbc.M200914200.
Wang Y, Cortez D, Yazdi P, Neff NF, Elledge SJ, Qin J: BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000, 14: 927-939.
Brosh RM, Li J, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA: Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem. 2000, 275: 23500-23508. 10.1074/jbc.M001557200.
Wu L, Davies SL, Levitt NC, Hickson ID: Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001, 276: 19375-19381. 10.1074/jbc.M009471200.
Johnson FB, Lombard DB, Neff NF, Mastrangelo M, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L: Association of the Bloom syndrome protein with topoisomerase IIIα in somatic and meiotic cells. Can Res. 2000, 60: 1162-1167.
Langland G, Kordich J, Creaney J, Goss KH, Lillard-Wetherell K, Bebenek K, Kunkel TA, Groden J: The Bloom's syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J Biol Chem. 2001, 276: 30031-30035. 10.1074/jbc.M009664200.
Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA: Genome-wide location and function of DNA binding proteins. Science. 2000, 290: 2306-2309. 10.1126/science.290.5500.2306.
International Human Genome Sequencing Consortium: Initial sequencing and analysis of the human genome. Nature. 2001, 469: 860-921.
Groden J, Nakamura Y, German J: Molecular evidence that homologous recombination occurs in proliferating human somatic cells. Proc Nat Acad Sci USA. 1990, 87: 4315-4319.
Lee DH, Andrus L, Prince AM: Stabilized viral nucleic acids in plasma as an alternative shipping method for NAT. Transfusion. 2002, 42: 409-413. 10.1046/j.1525-1438.2002.00068.x.
Little RD, Platt TH, Schildkraut CL: Initiation and termination of DNA replication in human rDNA genes. Mol Cell Biol. 1993, 13: 6600-6613.
Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M: The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure Fold Des. 1999, 7: 1557-1566. 10.1016/S0969-2126(00)88346-X.
Cremer T, Cremer C: Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001, 2: 292-301. 10.1038/35066075.
Croft JA, Bridges JM, Boyle S, Perry P, Teague P, Bickmore WA: Difference in the localization and morphology of chromsomes in the human nucleus. J Cell Biol. 1999, 145: 1119-1131. 10.1083/jcb.145.6.1119.
Kuhn EM: Localization by Q-banding of mitotic chiasmata in cases of Bloom's syndrome. Chromosoma. 1976, 57: 1-11.
Kuhn EM, Therman E, Denniston C: Mitotic chiasmata, gene density and oncogenes. Hum Genet. 1985, 70: 1-5.
Harley CB, Villeponteau B: Telomeres and telomerase in aging and cancer. Curr Opin Genet Dev. 1995, 5: 249-255. 10.1016/0959-437X(95)80016-6.
Johnson FB, Marciniak RA, Guarente L: Telomeres, the nucleolus and aging. Curr Opin Cell Biol. 1998, 10: 332-338. 10.1016/S0955-0674(98)80008-2.
Therman E, Otto PG, Shahidi NT: Mitotic recombination and segregation of satellites in Bloom's syndrome. Chromosoma. 82: 627-636.
de Lange T: Protection of mammalian telomeres. Oncogene. 2002, 21: 532-540. 10.1038/sj.onc.1205080.
Chan SW, Blackburn EH: New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002, 21: 553-563. 10.1038/sj.onc.1205082.
Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS: The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet. 2002, 11: 3135-3144. 10.1093/hmg/11.25.3135.
Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA: Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002, 277: 41110-41119. 10.1074/jbc.M205396200.
Lunderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T: Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996, 135: 867-681.
Bischof O, Kim SH, J Irving J, Beresten S, Ellis NA, Campisi J: Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001, 153: 367-80. 10.1083/jcb.153.2.367.
Zhu XD, Kuster B, Mann M, Petrini JH, Lange T: Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000, 25: 347-352. 10.1038/77139.
Franchitto A, Pichierri P: Bloom's syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J Cell Biol. 2002, 157: 19-30. 10.1083/jcb.200110009.
Kornberg RD, Lorch Y: Chromatin structure and transcription. Annu Rev Cell Biol. 1992, 8: 563-587.
Stubblefield E: Analysis of the replication pattern of Chinese hamster chromosomes using 5-bromo-deoxyuridine suppression of 33258 Hoechst fluorescence. Chromosoma. 1975, 53: 209-221.
Lima-de-Faria A, Jaworska H: Late DNA synthesis in heterochromatin. Nature. 1968, 217: 138-142.
Fuss J, Linn S: Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J Biol Chem. 2002, 277: 8658-8666. 10.1074/jbc.M110615200.
Jones CE, Mueser TC, Dudas KC, Kreuzer KN, Nossal NG: Bacteriophage T4 gene 41 helicase and gene 59 helicase-loading protein: a versatile couple with roles in replication and recombination. Proc Natl Acad Sci USA. 2001, 98: 8312-8338. 10.1073/pnas.121009398.
Haber JE: DNA recombination: the replication connection. Trends Biochem Sci. 1999, 24: 271-275. 10.1016/S0968-0004(99)01413-9.
Cox MM: The nonmutagenic repair of broken replication forks via recombination. Mut Res. 2002, 510: 107-210. 10.1016/S0027-5107(02)00256-7.
Courcelle J, Hanawalt PC: RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999, 262: 543-551. 10.1007/s004380051116.
Liao S, Graham J, Yan H: The function of xenopus Bloom's syndrome protein homolog (xBLM) in DNA replication. Genes Dev. 2000, 14: 2570-2575. 10.1101/gad.822400.
Adams MD, McVey M, Sekelsky JJ: Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003, 299: 265-267. 10.1126/science.1077198.
Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L: The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001, 20: 905-913. 10.1093/emboj/20.4.905.
Myung KA, Datta A, Chen C, Kolodner RD: SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001, 27: 113-116. 10.1038/83673.
Imamura O, Campbell JL: The human Bloom syndrome gene suppresses the DNA replication and repair defects of yeast dna2 mutants. Proc Natl Acad Sci USA. 2003, 100: 8193-8198. 10.1073/pnas.1431624100.
Kaneko H, Orii KO, Matsui E, Shimozawa N, Fukao T, Matsumoto T, Shimamoto A, Furuichi Y, Hayakawa S, Kasahara K, Kondo K: BLM (the causative gene of Bloom syndrome) protein translocation into the nucleus by a nuclear localization signal. Biochem Biophys Res Comm. 1997, 240: 348-353. 10.1006/bbrc.1997.7648.
Schonberg SA: Chromatid exchange in Bloom's Syndrome cells. Thesis (Ph.D.) Cornell University Graduate School of Medical Sciences, New York, NY. 1980
Acknowledgements
The authors thank Alfred Prince for use of the ABI7700, Dong-Hun Lee for help with the TaqMan assays and Linda Andrus for the Alu-Sb2 primers and beacon. Susana Albano Maraia and Pablo Rubinstein provided assistance with the DNA sequencing. Jill Storry provided advice and reagents for the cloning. Ruth Croson-Lowney and Karina Yazdanbakhsh helped us with the FACS study. We gratefully acknowledge the suggestion of a reviewer of our previous BMC Cell Biology publication to look at the cell cycle profiles of the cell lines. The Elsa U. Pardee Foundation and NYBC provided funding for this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
Author 1 JS performed the FACS analysis, the DNA cloning and sequencing of the lymphoblastoid fragments, expression and purification of the GFP-BLM fusion proteins and participated in the helicase assays. Author 2 EP carried out the chromatin immunoprecipitation, DNA cloning and sequencing of the fibroblast fragments, the PCR analyses, and performed ChIP on the Tet-on cell lines. Author 3 NN participated in the isolation of the recombinant baculoviruses, protein purification, and helicase assays as well as performing the hybridizations and drafting the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Schawalder, J., Paric, E. & Neff, N.F. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol 4, 15 (2003). https://doi.org/10.1186/1471-2121-4-15
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2121-4-15