Abstract
The vitamin D prophylaxis of rickets in pregnant women and newborns may play a role in early allergic sensitization. We now asked if an already diseased population may have inherited genetic variants in the vitamin D turnover or signalling pathway.
Serum levels of calcidiol (25-OH-D3) and calcitriol (1,25-(OH)2-D3) were retrospectively assessed in 872 partipants of the German Asthma Family Study. 96 DNA single base variants in 13 different genes were genotyped with MALDI-TOF and a bead array system. At least one positive SNP with a TDT of p < 0.05 for asthma or total IgE and calcidiol or calcitriol was seen in IL10, GC, IL12B, CYP2R1, IL4R, and CYP24A1. Consistent strong genotypic association could not be observed. Haplotype association were found only for CYP24A1, the main calcidiol degrading enzyme, where a frequent 5-point-haplotype was associated with asthma (p = 0,00063), total IgE (p = 0,0014), calcidiol (p = 0,0043) and calcitriol (p = 0,0046).
Genetic analysis of biological pathways seem to be a promising approach where this may be a first entry point into effects of a polygenic inherited vitamin D sensitivity that may affect also other metabolic, immunological and cancerous diseases.
Similar content being viewed by others
Background
Asthma is a chronic inflammatory condition of the airways, variable airway obstruction and elevated serum IgE levels of unclear pathogenesis [1]. A hypothesis relating early vitamin D supplementation and induction of later allergy has initially been postulated as the main cholecalciferol metabolite calcitriol may suppresses dendritic cell maturation and consecutive development of Th1 cells [2] which is now supported by in vitro, animal and human studies [3, 4].
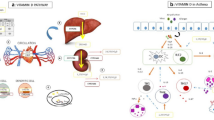
Exposure studies in humans, however, are difficult as nearly all newborns in Western countries are now being exposed in utero or during the first year of life to vitamin supplements [5, 6]. We now asked if there are DNA sequence variants that are associated with higher or lower levels of vitamin D metabolites. As it is unlikely that any complex disease is determined by variants in a single gene we tested the main genes that code for enzymes in the metabolic pathway of vitamin D conversion (Figure 1).
Methods
Study population
The German Asthma Family Study collected affected sib pairs in 26 paediatric centres in Germany and Sweden for a two-stage genome-wide linkage scan [7, 8]. In these families at least two children were required with confirmed clinical asthma, while prematurity or low birth weight of the children were excluded, along with any other severe pulmonary disease. All affected children over age 3 had a history of at least 3 years of recurrent wheezing and with no other airway disease diagnosed. Unaffected siblings were also sampled if they were at least 6 years old and eligible for pulmonary function testing. Each study participant signed a consent form. All study methods were approved by the ethics commission of "Ärztekammer Nordrhein-Westfalen".
A complete pedigree of the family was drawn and information collected in a questionnaire. Participants were examined for several closely associated phenotypes. Pulmonary function tests were performed by forced flow volume tests and bronchial challenge was done by methacholine (discontinued in the second stage of the study) as reported earlier [7, 8].
The current analysis differs from previous publications [7, 8]. We excluded here all families with at least one member of non white skin colour (families 2, 14, 16, 19 to 21 and 27 to 32) as these individuals had considerable lower levels of 25-OH-D3 (data not shown) compared to all other participants (Figure 2).
Total IgE was determined with an ELISA (Pharmacia Diagnostics, Uppsala, Sweden). 25-OH-D3 was determined with an enzymatic immunoassay (OCTEIA 25-Hydroxy Vitamin D kit, Immunodiagnostic Systems IDS, Frankfurt, Germany) that has a working range of 6–360 nmol/L, an intra-assay of 8% and inter-assay variation of 10% with a 100% specificity for 25-OH-D3 and 75% specificity for 25-OH-D2 according to the manufacturer. 1,25-OH2-D3 was determined by immunoextraction followed by an enzyme-immunoassay (OCTEIA 1,25-Hydroxy Vitamin D kit, Immunodiagnostic Systems IDS, Frankfurt, Germany) that has a working range of 6–500 pmol/L, a 100% specificity for 1,25-OH2-D3 and 0,009% specificity for 25-OH-D2. 25-OH-D3 values reported are the mean of a duplicate analysis while due to limited serum availability only single assays have been performed for 1,25-OH2-D3.
Control population
191 anonymized DNAs were selected randomly from the ECRHS II study [9] to fill in remaining slots on the genotyping plates. These DNA samples served as population-based controls to test if the parents of the famillies had different allele spectrum.
DNA preparation and genotyping
DNA was isolated from peripheral white blood cells using Qiamp (Qiagen, Germany) or Puregene isolation kits (Gentra Systems, Minneapolis, MN, USA).
Genes were selected as coding either for key enzymes in the vitamin D conversion pathway or being regulated by vitamin D metabolites [10]. SNPs were being picked more or less randomly either for tagging haplotypes or being functional relevant [11]. Most SNPs were genotyped using MALDI-TOF mass spectrometry of allele-specific primer extension products generated from amplified DNA sequences (MassARRAY, SEQUENOM Inc., San Diego, CA, USA). A few SNPs were also genotyped at Illumina (San Diego CA, USA) by use of the Sentrix bead arrays. VDR [12] and IL4R [13] SNP results have been published earlier and are reanalysed here for the vitamin D levels. The following SNPs were analyzed (genotyping details upon request): rs3024498 (IL10), rs3024493 (IL10), rs1518111 (IL10), rs10000076 (IL10), rs1800872 (IL10), Il10-571CA (IL10), rs1800895 (IL10), rs1800894 (IL10), rs1800896 (IL10), rs1800893 (IL10), rs705120 (GC), rs222040 (GC), rs7041 (GC), rs4752 (GC), rs222011 (GC), rs221999 (GC), rs6811536 (SPP1), rs4754 (SPP1), rs1042714 (ADRB2), rs1800888 (ADRB2), rs1368439 (IL12B), rs3212227 (IL12B), rs2853697 (IL12B), rs3213119 (IL12B), rs2853696 (IL12B), rs2853694 (IL12B), rs2288831 (IL12B), rs3213096 (IL12B), rs2569254 (IL12B), rs3181216 (IL12B), rs3212220 (IL12B), rs3212218 (IL12B), rs1433048 (IL12B), rs2546890 (IL12B), rs3132299 (RXRA), rs877954 (RXRA), rs1045570 (RXRA), rs10500804 (CYP2R1), rs1562902 (CYP2R1), rs10766197 (CYP2R1), rs2853563 (VDR), rs731236 (VDR), rs7975232 (VDR), rs1544410 (VDR), rs2239185 (VDR), rs987849 (VDR), rs1540339 (VDR), rs3819545 (VDR), rs3782905 (VDR), rs2239186 (VDR), rs2228570 (VDR), rs1989969 (VDR), rs2853564 (VDR), hCV2880804 (VDR), rs238532 (CYP27B1), rs2072052 (CYP27B1), rs1048691 (CYP27B1), rs4646537 (CYP27B1), rs4646536 (CYP27B1), rs8176345 (CYP24A1), rs703842 (CYP27B1), I50V (IL4R), rs2234897 (IL4R), rs1805011 (IL4R), C406R (IL4R), rs1805015 (IL4R), Q551R (IL4R), rs1805016 (IL4R), rs10000306 (CARD15), rs2076753 (CARD15), rs2066842 (CARD15), rs2066843 (CARD15), rs2076756 (CARD15), rs10000331 (CARD15), rs3135499 (CARD15), rs3135500 (CARD15), rs375947 (IL12RB1), rs447009 (IL12RB1), rs436857 (IL12RB1), rs2045387 (IL12RB1), rs8118441 (CYP24A1), rs751089 (CYP24A1), rs6068816 (CYP24A1), rs4809958 (CYP24A1), rs2244719 (CYP24A1), rs2296241 (CYP24A1), rs17219266 (CYP24A1), rs6022999 (CYP24A1), rs17219315 (CYP24A1), rs11699278 (CYP24A1), rs2762942 (CYP24A1), rs2248137 (CYP24A1), rs2762943 (CYP24A1), rs2585427 (CYP24A1), rs2248359 (CYP24A1) and rs2426496 (CYP24A1).
Data handling and statistical analysis
Clinical data and genotypes were all transferred to a SQL 2000 database and checked for completeness, paternity, and Hardy-Weinberg equilibrium. Further analyses were performed using R 2.0 statistical software[14]. Linkage disequilibrium was determined by Haploview [15] using the Gabriel method for block definition. TDT association for quantitative and qualitative traits was done with SIBPAIR [16] using the TDT option for qualitative and the Haseman-Elston regression for quantitative traits. Family-based haplotype association analysis was performed by FBAT [17] using a dominant model.
Bioinformatics
SNP information was obtained from dbSNP [18], Innate Immunity PGA [19] and UCSC genome browser [20]. SNP selection was done with the help of Perlegen [21] and Hapmap [22] data. Sequence context annotation was done by SNPper [23], PUPA [24], TAMAL [25] and SNPPi [26]).
Results
The total sample consisted of 947 individuals from 224 families where 872 serum measurements of 25-OH-D3, 876 1,25- OH2-D3 and 934 total IgE measurements could be performed. After exclusion of non-white families 903 individuals from 201 families remained under analysis with 812 assays of 25-OH-D3, 807 1,25- OH2-D3 and 903 total IgE.
Clinical details of the families are given in table 1. Mean 25-OH-D3 level in children was 68 nmol/l (s.d. 38 nmol/l). 50% of values fell below and 17% above the Merck manual reference range of 62.4 to 99.8 nmol/l. Mean 1,25- OH2-D3 in children was 102 pmol/l (s.d. 38 nmol/l). 3% of values fell below and 40% above the Merck manual reference range of 48.4 to 108 pmol/l. The highest measured value was 257 pmol/l in two children from unrelated families.
There were no major differences in serum levels between children and parents. There was also no major influence by sex or age. An important factor, however, was found with month of examination representing seasonal sun exposure in mid Europe (Figure 2). Even after serum storage of 10 years, the individual 25-OH-D3 levels followed a clear time course with a major peak in August. The hormonal form 1,25-OH2-D3 did not vary over the course of the year, as the conversion rate decreased with higher levels of 25-OH-D3 (Figure 3).
The overall heritability index H2 for 25-OH-D3 was 80.3% while the H2 for 1,25-OH2-D3 was only 30.0% [27]. There was neither an association of 25-OH-D3 and total IgE nor an association of 1,25-OH2-D3 and total IgE levels.
13 genes were selected for genotyping (IL10, GC, SPP, ADRB2, IL12B, RXRA, CYP2R1, VDR, CYP27B1, IL4R, CARD15, IL12RB1, CYP24A1) and could be successfully completed for 96 SNPs. 4 of these SNPs were not in Hardy-Weinberg equilibrium: rs221999 (GC, P = 0,0299), rs10500804 (CYP2R1, P = 0,0498), rs10766197 (CYP2R1, P = 0,0100) and rs2248359 (CYP24A1, P = 0,0299). SNP rs221999 was also not in Hardy-Weinberg equilibrium in controls. The population-based controls showed similar allele frequencies compared to the family samples except for SNPs rs4754, rs2288831 and rs3819545.
SNP allele transmission in 7 of the tested 13 genes showed a p-value of less than 0.05 when testing for 25-OH-D3 levels (IL10, GC, ADRB2, CYP2R1, IL4R, IL12RB1 and CYP24A1, see table 2) while only 3 showed transmission disequilibrium with 1,25-OH2-D3 (IL10, IL12B and CYP24A1). SNPs in 5 genes showed a p-value < 0.05 with asthma (IL10, IL12B, VDR, CARD15 and CYP24A1). Most significances, however, were weak. For 96 SNPs we would expect 4.8 tests to be positive for each trait which was exceeded by testing asthma (N = 8), total IgE (N = 13), 25-OH-D3 (N = 8) but not 1,25-OH2-D3(N = 3). Only 2 SNP showed an effect with both traits, one in CYP2R1 (rs10766197) and one in IL4R (rs1805011). rs10766197 is situated in the CYP2R1 promotor; while rs1805011 is leading to an Ala- > Glu amino acid exchange in the IL4 receptor.
In a next step we performed multivariate regression in the parental dataset while adjusting for age, sex, and month of examination (table 3). This confirmed 11 SNPs already found in the family based-aproach; again, association results were weak. Some CARD15 variants had an asthma protective effect while IL12B SNPs carried risk alleles.
Haplotypes were constructed from all significantly associated SNPs (table2). No significant association was found in any of the 13 genes except for CYP24A1 where a 5-point frequent haplotype (rs2296241:rs17219315:rs2762942:rs2248137:rs2248359) spanning both LD blocks of CYP24A1 was associated with a diagnosis of asthma (p = 0.001), total IgE (p = 0.001), 25-OH2-D3 (p = 0.004) and 1,25-OH2-D3 serum level (p = 0.005, table 4).
Discussion
We have shown that serum 25-OH-D3 (calcidiol) levels -although highly influenced by environmental sunlight exposure- is a heritable trait in asthma families. In contrast, a major genetic influence on 1,25-OH2-D3 (calcitriol) levels could not be found, a finding that requires replication in further family and population-based studies.
The reason for this discrepancy is not fully clear as the conversion of 25-OH-D3 to 1,25-OH2-D3 is closely regulated by a direct feedback loop. It is generally agreed, however, that 25-OH-D3 reflects best the current vitamin D status [28]. Unfortunately standardized reference values for this age group are not available but values for 25-OH-D3 in children seem to be in the upper normal range [29]. An explanation therefore could be that a delayed downstream metabolism is leading to an (unintended) afflux or -also possible- that an increased peripheral demand needs a larger reservoir.
We observed a number of positive associations with single nucleotide polymorphisms. Although the selection of candidate genes was rather subjective, it turned out that some of the tested candidate genes are associated with both allergy and vitamin D metabolites. Statistical significance, however, was weak, and varied even with different analysis strategies and software packages (unpublished own observation). There was also no fully consistent pattern when comparing the family transmission and the case-control approach which makes it unlikely that any of the tested SNPs is already an important functional variant. The new associations may instead indicate the effects of physically closely related variants in these genes (which is also supported by the haplotype results of CYP24A1).
Genomic organization of CYP24A1 gene, location of genotyped SNPs, linkage disequilibrium between SNPs (with R2 indicated by bullet size) and LD block structure (highlighted by red boxes; rs2248359 was excluded from LD calculations for not being in HWE). SNPs indicated by ¶ were used to build haplotypes.
The associated candidate genes are of particular interest. CYP24A1 is the major enzyme of the calcitriol degradation pathway that showed nearly 100-fold increase after vitamin D treatment of rats [30]. Previous studies also suggest that CYP24A1 null mice cannot clear calcitriol efficiently [31] which would support the above mentioned afflux hypothesis. An alternative splicing variant in CYP24A1 has been described recently [32] leading to a truncated and catalytically dysfunctional protein while it is unclear if any of our tested SNPs will have functional relationship to this protein variant. Dark skinned Asian Indians seem to have increased 24-hydroxylase activity compared to white skinned Caucasians [33] whereas both skin colour and metabolic capacity seem to be adapted to less sun light exposure in Caucasians.
The evidence that the human CYP2R1 is a key vitamin D 25-hydoxylase is rather new [34] where the identity of the hepatic 25-hydroxylase has remained unclear for several decades. At least six CYPs can catalyze this step where the most viable candidates are CYP27A1 and CYP2R1 [34] with the renal enzyme responsible for 1-α-hydroxylation being CYP27B1. A loss of function mutation in CYP2R1 has also been described [34] and deserves further testing.
Variants in CYP2R1, CYP27B1 and CYP24A1 or other genes in the metabolic pathway of vitamin D have not been tested so far with asthma or allergy but several of the VDR-controlled genes tested here already have been associated with asthma and allergy. These include IL12B [35–37], IL12RB [38, 39], IL10 [40], VDR [41, 42, 12], GC [43], ADRB2 [44], CARD15 [45] and IL4R [46].
Of these, IL12B is a particular interesting cytokines. Macrophage engulfed microorganism are leading to IL12p70 production, a heterodimer of IL12p40 (IL12B) and IL12p35 (IL12A), which is a primary inducer of Th1 cell development and a critical factor in the development of allergy [47]. Also IL10 seems to be important where production in circulating T cells from atopic asthmatics is maximally stimulated [48]; allergen specific IL10 producing T regulatory cells can inhibit allergen specific effector cells and represent an important line of defense in the allergic reaction [49]. Functional variants in these genes leading to human disease are not known so far.
The many positive but weak associations represent a common dilemma in complex disease. In asthma more than 75 genes have now been claimed to be associated [50] but none of them has been shown to contribute to risk in all populations studied [51]. Obviously there are only small genetic effects and a large heterogeneity; sometimes there is unidentified population stratification and there might be phenotyping and genotyping errors. Most likely, however, not the "center" SNPs have been choosen [11]. The current pathway based approach seems to be an alternative in particular when an environmental trait can be included. It is likely that some of the genes identified here are acting in concert to determine the overall vitamin D sensitivity.
Besides increasing sample size and testing additional populations, further work may concentrate on monitoring vitamin D supplementation by immunological readouts and the identification of contributing functional genetic elements. The present rediscovery of a genetic vitamin D sensitivity [52] may be an important step in allergy induction and also surmount many other diseases including type 1 diabetes, osteoporosis, tuberculosis, rheumatoid arthritis, multiple sclerosis, inflammatory bowel diseases, and prostate cancer where adequate vitamin D support has been found to be beneficial.
Abbreviations
- SNP:
-
single nucleotide polymorphism
- D3 :
-
cholecalciferol, vitamin D
- 25-OH-D3 :
-
calcidiol
- 1,25-OH2-D3 :
-
calcitriol
- CYP24A1:
-
cytochrome P450, family 24, subfamily A, polypeptide 1
- VDR:
-
nuclear vitamin D receptor
- IL12B:
-
interleukin 12 B (cytotoxic lymphocyte maturation factor 2, p40)
- RXRA:
-
retinoid X receptor α
- IL4R:
-
interleukin 4 receptor
- ADRB2:
-
ß2 adrenergic receptor
- IL12RB1:
-
interleukin 12 receptor, ß1
- IL10:
-
interleukin 10
- GC:
-
group-specific component (vitamin D binding protein)
- CYP2R1:
-
cytochrome P450, family 2, subfamily R, polypeptide 1
- CARD15:
-
caspase recruitment domain family, member 15 (NOD2)
- SPP1:
-
secreted phosphoprotein 1, osteopontin (OPN, ETA-1, BNSP,...)
- CYP27B1:
-
cytochrome P450, family 27, subfamily B, polypeptide 1
References
Holt PG, Macaubas C, Stumbles PA, Sly PD: The role of allergy in the development of asthma. Nature 1999, 402:B12–17.
Adorini L: Tolerogenic dendritic cells induced by vitamin D receptor ligands enhance regulatory T cells inhibiting autoimmune diabetes. Ann N Y Acad Sci 2003, 987:258–261.
Wjst M, Dold S: Genes, factor X, and allergens: what causes allergic diseases? Allergy 1999, 54:757–759.
Wjst M: The triple T allergy hypothesis. Clin Dev Immunol 2004, 11:175.
Hyppönen E, Sovio U, Wjst M, Patel S, Pekkanen J, Hartikainen AL, Järvelin MR: Vitamin D supplementation in infancy and the risk of allergies in adulthood: a birth cohort study. Ann Am Acad Sci 2004, 1037:84–95.
Milner JD, Stein DM, McCarter R, Moon RY: Early infant multivitamin supplementation is associated with increased risk for food allergy and asthma. Pediatrics 2004, 114:27–32.
Wjst M, Fischer G, Immervoll T, Jung M, Saar K, Rüschendorf F, Reis A, Ulbrecht M, Gomolka M, Weiss EH, Jäger L, Nickel R, Richter K, Kjellman NI, Griese M, von Berg A, Gappa M, Riedel F, Boehle M, van Koningsbruggen S, Schoberth P, Szczepanski R, Dorsch W, Silbermann M, Wichmann HE: A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics 1999, 58:1–8.
Altmuller J, Seidel C, Lee YA, Loesgen S, Bulle D, Friedrichs F, Jellouschek H, Kelber J, Keller A, Schuster A, Silbermann M, Wahlen W, Wolff P, Schlenvoigt G, Rüschendorf F, Nürnberg P, Wjst M: Phenotypic and genetic heterogeneity in a genome-wide linkage study of asthma families. BMC Pulm Med 2005, 5:1.
Werner M, Topp R, Wimmer K, Richter K, Bischof W, Wjst M, Heinrich J: TLR4 gene variants modify endotoxin effects on asthma. J Allergy Clin Immunol 2003, 112:323–330.
Wang TT, Tavera-Mendoza LE, Laperriere D, Libby E, MacLeod NB, Nagai Y, Bourdeau V, Konstorum A, Lallemant B, Zhang R, Mader S, White JH: Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol 2005, 19:2685–2695.
Wjst M: Target SNP selection in complex disease association studies. BMC Bioinformatics 2004, 5:92.
Wjst M: Genetic variants in the vitamin D receptor and childhood asthma. BMC Genetics 2005, 6:2.
Wjst M, Kruse S, Illig T, Deichmann K: Asthma and IL-4 receptor alpha gene variants. Eur J Immunogenet 2002, 29:263–268.
Rproject [http://www.r-project.org]
Haploview [http://www.broad.mit.edu/mpg/haploview]
PGA II [http://innateimmunity.net]
Browser UG [http://genome.ucsc.edu]
Perlegen [http://www.perlegen.com]
Hapmap [http://www.hapmap.org]
SNPper [http://snpper.chip.org]
TAMAL [http://neoref.ils.unc.edu/tamal]
SNPPI [http://cooke.gsf.de]/wjst/paper/2004BMCBioinformaticsSuppl/snppi.cfm.
Wjst M: Maternal vitamin D status and childhood bone mass. Lancet 2006, in press.
Vieth R: Vitamin D supplementation, 25-hydroxyvitamin D concentrations, and safety. Am J Clin Nutr 1999, 69:842–856.
Greenway A, Zacharin M: Vitamin D status of chronically ill or disabled children in Victoria. J Paediatr Child Health 2003, 39:543–547.
Kutuzova GD, Deluca HF: Gene expression profiles in rat intestine identify pathways for 1,25-dihydroxyvitamin D(3) stimulated calcium absorption and clarify its immunomodulatory properties. Arch Biochem Biophys 2004, 432:152–166.
Masuda S, Byford V, Arabian A, Sakai Y, Demay MB, St-Arnaud R, Jones G: Altered pharmacokinetics of 1alpha,25-dihydroxyvitamin D3 and 25-hydroxyvitamin D3 in the blood and tissues of the 25-hydroxyvitamin D-24-hydroxylase (Cyp24a1) null mouse. Endocrinology 2005, 146:825–834.
Ren S, Nguyen L, Wu S, Encinas C, Adams JS, Hewison M: Alternative splicing of vitamin D-24-hydroxylase: a novel mechanism for the regulation of extrarenal 1,25-dihydroxyvitamin D synthesis. J Biol Chem 2005, 280:20604–20611.
Awumey EM, Mitra DA, Hollis BW, Kumar R, Bell NH: Vitamin D metabolism is altered in Asian Indians in the southern United States: a clinical research center study. J Clin Endocrinol Metab 1998, 83:169–173.
Cheng JB, Levine MA, Bell NH, Mangelsdorf DJ, Russell DW: Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase. Proc Natl Acad Sci U S A 2004, 101:7711–7715.
Randolph AG, Lange C, Silverman EK, Lazarus R, Silverman ES, Raby B, Brown A, Ozonoff A, Richter B, Weiss ST: The IL12B gene is associated with asthma. Am J Hum Genet 2004, 75:709–715.
Morahan G, Huang D, Wu M, Holt BJ, White GP, Kendall GE, Sly PD, Holt PG: Association of IL12B promoter polymorphism with severity of atopic and non-atopic asthma in children. Lancet 2002, 360:455–459.
Hirota T, Suzuki Y, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, Cheng L, Takahashi N, Shimizu M, Doi S, Fujita K, Enomoto T, Ebisawa M, Yoshihara S, Nakamura Y, Kishi F, Shirakawa T, Tamari M: Functional haplotypes of IL-12B are associated with childhood atopic asthma. J Allergy Clin Immunol 2005, 116:789–795.
Bassuny WM, Ihara K, Kimura J, Ichikawa S, Kuromaru R, Miyako K, Kusuhara K, Sasaki Y, Kohno H, Matsuura N, Nishima S, Hara T: Association study between interleukin-12 receptor beta1/beta2 genes and type 1 diabetes or asthma in the Japanese population. Immunogenetics 2003, 55:189–192.
Takahashi N, Akahoshi M, Matsuda A, Ebe K, Inomata N, Obara K, Hirota T, Nakashima K, Shimizu M, Tamari M, Doi S, Miyatake A, Enomoto T, Nakashima H, Ikezawa Z, Shirakawa T: Association of the IL12RB1 promoter polymorphisms with increased risk of atopic dermatitis and other allergic phenotypes. Hum Mol Genet 2005, 14:3149–3159.
Lyon H, Lange C, Lake S, Silverman EK, Randolph AG, Kwiatkowski D, Raby BA, Lazarus R, Weiland KM, Laird N, Weiss ST: IL10 gene polymorphisms are associated with asthma phenotypes in children. Genet Epidemiol 2004, 26:155–165.
Poon AH, Laprise C, Lemire M, Montpetit A, Sinnett D, Schurr E, Hudson TJ: Association of vitamin D receptor genetic variants with susceptibility to asthma and atopy. Am J Respir Crit Care Med 2004, 170:967–973.
Raby BA, Lazarus R, Silverman EK, Lake S, Lange C, Wjst M, Weiss ST: Association of vitamin D receptor gene polymorphisms with childhood and adult asthma. Am J Respir Crit Care Med 2004, 170:1057–1065.
Laufs J, Andrason H, Sigvaldason A, Halapi E, Thorsteinsson L, Jonasson K, Soebech E, Gislason T, Gulcher JR, Stefansson K, Hakonarson H: Association of vitamin D binding protein variants with chronic mucus hypersecretion in Iceland. Am J Pharmacogenomics 2004, 4:63–68.
Dewar JC, Wilkinson J, Wheatley A, Thomas NS, Doull I, Morton N, Lio P, Harvey JF, Liggett SB, Holgate ST, Hall IP: The glutamine 27 beta2-adrenoceptor polymorphism is associated with elevated IgE levels in asthmatic families. J Allergy Clin Immunol 1997, 100:261–265.
Weidinger S, Klopp N, Rummler L, Wagenpfeil S, Baurecht HJ, Gauger A, Darsow U, Jakob T, Novak N, Schäfer T, Heinrich J, Behrendt H, Wichmann HE, Ring J, Illig T: Association of CARD15 polymorphisms with atopy-related traits in a population-based cohort of Caucasian adults. Clin Exp Allergy 2005, 35:866–872.
Mitsuyasu H, Izuhara K, Mao XQ, Gao PS, Arinobu Y, Enomoto T, Kawai M, Sasaki S, Dake Y, Hamasaki N, Shirakawa T, Hopkin JM: Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet 1998, 19:119–120.
Yoshida M, Watson RM, Rerecich T, O'Byrne PM: Different profiles of T-cell IFN-gamma and IL-12 in allergen-induced early and dual responders with asthma. J Allergy Clin Immunol 2005, 115:1004–1009.
Matsumoto K, Narita S, Rerecich T, Snider DP, O'Byrne PM: Different profile of interleukin-10 production in circulating T cells from atopic asthmatics compared with healthy subjects. Can Respir J 2004, 11:33–38.
Akdis M, Blaser K, Akdis CA: T regulatory cells in allergy: novel concepts in the pathogenesis, prevention, and treatment of allergic diseases. J Allergy Clin Immunol 2005, 116:961–968. quiz 969
Ober C, Hoffjan S: Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun 2006.
Ober C, Thompson EE: Rethinking genetic models of asthma: the role of environmental modifiers. Curr Opin Immunol 2005, 17:670–678.
Schulze E: Überempfindlichkeit gegen Vitamin D. Dtsch Med Wochenschr 1956, 81:1364–1365.
Acknowledgements
We thank the participating families and clinical centers for their help: R. Nickel, K. Beyer, R. Kehrt, U.Wahn (Berlin), K. Richter, H. Janiki, R. Joerres, H. Magnussen (Grosshansdorf), I. M. Sandberg, L. Lindell, N.I.M. Kjellman (Linkoeping), C. Frye, G. Woehlke, I. Meyer, O. Manuwald (Erfurt), A. Demirsoy, M. Griese, D. Reinhardt (München), G. Oepen, A. Martin, A. von Berg, D. Berdel (Wesel), Y. Guesewell, M. Gappa, H. von der Hardt (Hannover), J. Tuecke, F. Riedel (Bochum), M. Boehle, G. Kusenbach [+], H. Jellouschek, M. Barker, G. Heimann (Aachen), S. van Koningsbruggen, E. Rietschel (Köln), P. Schoberth (Köln), G. Damm, R. Szczepanski, T. Lob-Corzilius (Osnabrück), L. Schmid, W. Dorsch (München), M. Skiba, C.Seidel, M. Silbermann (Berlin), A. Schuster (Düsseldorf), J. Seidenberg (Oldenburg), W. Leupold, J. Kelber (Dresden), W. Wahlen (Homburg), F. Friedrichs, K. Zima (Aachen), P. Wolff (Pfullendorf), D. Bulle (Ravensburg), W. Rebien, A.Keller (Hamburg) and M. Tiedgen (Hamburg). M. Hoeltzenbein organized the first part of the study, G. Schlenvoigt and L.Jaeger did the IgE determination and G. Fischer supervised data entry. T. Illig (former T.Immervoll), P. Lichtner and J. Heinrich supported the project during various stages; B. Wunderlich for excellent laboratory work during set up of the family study. We wish to thank also Amelie Elsaesser for programming the Jonkheere-Terpstra trend test and Michelle Emfinger for proof-reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
M.W. initiated the study, applied for funding, developed protocols, trained investigators, planned laboratory analysis, did statistical analysis and wrote the report. J.A. did the clinical survey, C.B. did the SNP analysis, M.B. built serum and DNA bank and did the vitamin D assays together with E.A. who supervised also laboratory work and did functional assays. T.F-K. participated in the data analysis. All authors critically revised the paper.
Conflicts of Interest
The author(s) declare that they have no competing interests.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Wjst, M., Altmüller, J., Faus-Kessler, T. et al. Asthma families show transmission disequilibrium of gene variants in the vitamin D metabolism and signalling pathway. Respir Res 7, 60 (2006). https://doi.org/10.1186/1465-9921-7-60
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1465-9921-7-60