Abstract
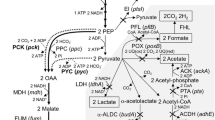
The glutamate decarboxylase (Gad) system is an important amino acid-dependent acid resistance system commonly found in microorganisms. Actinobacillus succinogenes is one of the best natural producers of succinic acid (SA) but lacks glutamate decarboxylase. This study assessed the effects of Gad system introduction into A. succinogenes. The recombinant strains gadB-SW and gadBC-SW were constructed by heterologous expression of gadB alone, or gadB together with gadC, respectively. After 1.0 and 1.5 h of acid stress at pH 4.6, cell survival of gadBC-SW was greater than gadB-SW. The growth of gadB-SW and gadBC-SW was both affected by the expression of heterologous proteins and by γ-aminobutyric acid, with gadBC-SW growth reduced at a neutral pH. SA production in acidic conditions was evaluated by a shake flask and by 3-L bioreactor fermentation. The results showed gadBC-SW to increase SA production by 8.4% in shake flask compared to the parent strain, SW. For a 3-L bioreactor batch fermentation under acidic environment, the highest conversion rate of sugar to SA was observed for gadBC-SW, reaching 96%. However, SA concentration by gadBC-SW was only 47 g/L and 31 g/L at pH 6.5 and pH 6.0, respectively. In summary, the introduction of heterologous gadB and gadC into A. succinogenes not only improved acid tolerance but also influenced the synthesis of SA and added a metabolic burden.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Sauer M, Porro D, Mattanovich D, Branduardi P. Microbial production of organic acids: expanding the markets. Trends Biotechnol. 2008;26:100–8. https://doi.org/10.1016/j.tibtech.2007.11.006.
Kover A, Kraljić D, Marinaro R, Rene ER. Processes for the valorization of food and agricultural wastes to value-added products: recent practices and perspectives. Syst Microbiol Biomanuf. 2021. https://doi.org/10.1007/s43393-021-00042-y.
Jansen ML, Gulik WMV. Towards large scale fermentative production of succinic acid. Curr Opin Biotechnol. 2014;30:190–7. https://doi.org/10.1016/j.copbio.2014.07.003.
Zhang W, Qiao Y, Wu M, et al. Metabolic regulation of organic acid biosynthesis in Actinobacillus succinogenes. Front Bioeng Biotech. 2019. https://doi.org/10.3389/fbioe.2019.00216.
McKinlay JB, Laivenieks M, Schindler BD, Mckinlay AA, Siddaramappa S, Challacombe JF, et al. A genomic perspective on the potential of Actinobacillus succinogenes for industrial succinate production. BMC Genomics. 2010;11:680. https://doi.org/10.1186/1471-2164-11-680.
Guettler MV, Jain MK, Rumler D (1996) Method for making succinic acid, bacterial variants for use in the process, and methods for obtaining variants. U. S. patent 5573931 A
Guettler MV, Rumler D, Jain MK. Actinobacillus succinogenes sp. nov., a novel succinic-acid-producing strain from the bovine rumen. Int J Syst Bacteriol. 1999;49:207–16. https://doi.org/10.1099/00207713-49-1-207.
Corona-Gonzalez RI, Bories A, González-Álvarez V, Snell-Castro R, Toriz-González G, Pelayo-Ortiz C. Succinic acid production with Actinobacillus succinogenes, zt-130 in the presence of succinic acid. Curr Microbiol. 2010;60:71–7. https://doi.org/10.1007/s00284-009-9504-x.
Tag A, Madcs A, Erd B, Los C, Rma A, Rg A. Optimization of anaerobic fermentation of Actinobacillus succinogenes for increase the succinic acid production. Biocatal Agric Biotechnol. 2020. https://doi.org/10.1016/j.bcab.2020.101718.
Liu YP, et al. Strategies of pH control and glucose-fed batch fermentation for production of succinic acid by Actinobacillus succinogenes CGMCC1593. J Chem Technol Biotechnol. 2008;83:722–9. https://doi.org/10.1002/jctb.1862.
Wang CC, Zhu LW, Li HM, et al. Performance analyses of a neutralizing agent combination strategy for the production of succinic acid by Actinobacillus succinogenes ATCC 55618. Bioproc Biosyst Eng. 2012;35(4):659–64. https://doi.org/10.1007/s00449-011-0644-6.
Dai ZX, Guo F, Zhang SJ, Zhang WM, Yang Q, Dong WL, Jiang M, et al. Bio-based succinic acid: an overview of strain development, substrate utilization, and downstream purification. Biofuel Bioprod Bior. 2019. https://doi.org/10.1002/bbb.2063.
Chong LA, Klo B, Zc C, Zsa D, Xl B, Rdp B. Promising advancement in fermentative succinic acid production by yeast hosts. J Hazard Mater. 2020. https://doi.org/10.1016/j.jhazmat.2020.123414.
Zhang Q, Chen PC, Zheng P. Physiological and transcriptional responses of Actinobacillus succigenes to acid stress. J Microbiol. 2017. https://doi.org/10.13343/j.cnki.wsxb.20170400.
Guarnieri MT, Chou YC, Salvachúa D, Mohagheghi A, Beckham GT. Metabolic engineering of Actinobacillus succinogenes provides insights into succinic acid biosynthesis. Appl Environ Microbiol. 2017. https://doi.org/10.1128/AEM.00996-17.
Hu S, You Y, Xia F, Liu J, Dai W, Liu J. Genome shuffling improved acid-tolerance and succinic acid production of Actinobacillus succinogenes. Food Sci Biotechnol. 2019;28(3):817–22. https://doi.org/10.1007/s10068-018-0505-z.
Liu X, Zheng P, Ni Y, Dong JQ, Sun ZH. Breeding Actinobacillus succinogenes with acid-tolerance by genome shuffling. Microbiol. 2009. https://doi.org/10.13344/j.microbiol.china.2009.11.008.
Zhang WM, Tao YX, Wu M, Xi FX, Dong WL, Zhou J, Gu JC, Ma JF, Jiang M, et al. Adaptive evolution improves acid tolerance and succinic acid production in Actinobacillus succinogenes. Process Biochem. 2020;98:76–82. https://doi.org/10.1016/j.procbio.2020.08.003.
Tang HZ, Zhang L, Liu YP, et al. Mechanisms of acid tolerance in bacteria and prospects in biotechnology and bioremediation. Biotechnol Adv. 2015. https://doi.org/10.1016/j.biotechadv.2015.06.001.
Feehily C, Karatzas KAG. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114:11–24. https://doi.org/10.1111/j.1365-2672.2012.05434.x.
Li Q, Tao QY, Teixeira JS, Su SWM, Ganzle MG. Contribution of glutaminases to glutamine metabolism and acid resistance in Lactobacillus reuteri and other vertebrate host adapted lactobacilli. Food Microbiol. 2020;86:103343. https://doi.org/10.1016/j.fm.2019.103343.
Cotter PD, O’Reilly K, Hill C. Role of the glutamate decarboxylase acid resistance system in the survival of Listeria monocytogenes LO28 in low pH foods. J Food Prot. 2001;64:1362–8. https://doi.org/10.4315/0362-028x-64.9.1362.
Lu P, Ma D, Chen YL, Guo YY, Chen GQ, Deng HT, Shi YG, et al. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013;23:635–44. https://doi.org/10.1038/cr.2013.13.
Zhao H, Feng Z, Quan X, Cao Z, Jie L. The soluble transhydrogenase UdhA affecting the glutamate-dependent acid resistance system of Escherichia coli under acetate stress. Biol Open. 2018. https://doi.org/10.1242/bio.031856.
Mancini A, Carafa I, Franciosi E, Nardin T, Tuohy KM. In vitro probiotic characterization of high GABA producing strain Lactobacilluas brevis DSM 32386 isolated from traditional “wild” Alpine cheese. Ann Microbiol. 2019;69:1435–43. https://doi.org/10.1007/s13213-019-01527-x.
Krammer EM, Prevost M. Function and regulation of acid resistance antiporters. J Membr Biol. 2019;252:465–81. https://doi.org/10.1007/s00232-019-00073-6.
Ma D, Lu P, Shi Y. Substrate selectivity of the acid-activated glutamate/γ-aminobutyric acid (GABA) antiporter GadC from Escherichia coli. J Biol Chem. 2013;288:15148–53.
Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–41. https://doi.org/10.1128/JB.186.18.6032-6041.2004.
Cozzani I, Barsacchi R, Dibenedetto G, et al. Regulation of breakdown and synthesis of L-glutamate decarboxylase in Clostridium perfringens. J Bacteriol. 1975;123(3):1115–23. https://doi.org/10.1128/JB.123.3.1115-1123.1975.
Krulwich T, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–43. https://doi.org/10.1038/nrmicro2549.
Acknowledgements
The authors are grateful for the financial support from the National First-class Discipline Program of Light Industry Technology and Engineering (Grant No. LITE2018-04) and the Topnotch Academic Programs Project of Jiangsu Higher Education Institutions (TAPP).
Author information
Authors and Affiliations
Contributions
CCM had a major contribution to this work and wrote the manuscript. ZQ and QJZ did some supplementary experiments and analyzed the data. WD and CPC revised the manuscript. ZP conceived and supervised the study and contributed to the writing of the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, C., Zhang, Q., Qian, J. et al. Effect of the Gad system on Actinobacillus succinogenes during acid stress. Syst Microbiol and Biomanuf 2, 177–185 (2022). https://doi.org/10.1007/s43393-021-00054-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-021-00054-8