Abstract
The global emergence of antimicrobial resistance (AMR) has become a serious threat to human and animal health. Recent studies have shown that synanthropic animals can act as reservoirs and disseminators of pathogens and resistant bacteria. The aim of this study was to evaluate the frequency, distribution, and antimicrobial susceptibility of staphylococcal species and Clostridioides difficile isolated from the feces of free-living rodents and marsupials from two urban parks in Belo Horizonte, Brazil. During a 12-month period, fecal samples from 159 free-living animals, including 136 rodents and 23 marsupials, were collected from two urban parks in Belo Horizonte, Minas Gerais, Brazil. Staphylococcus spp. were more likely to be isolated from rodents than marsupials (p = 0.0164). Eight different staphylococcal species were isolated from 36 (26.5%) rodents and one marsupial (4.3%). S. saprophyticus (48.6%) was the most frequently isolated species, and almost a quarter of the isolates (24.3%) were resistant to at least one antimicrobial agent, four (10.8%) of which were multi-drug resistant (MDR). Two (5.4%) strains were resistant to cefoxitin and were then classified as methicillin-resistant staphylococci, and one also tested positive for the mecA gene. C. difficile was isolated from two rodents (1.5%), and one strain was toxigenic and classified as ribotype 064. One isolate was resistant to rifampicin, but both strains were susceptible to all other antimicrobials tested, including metronidazole and vancomycin. All C. difficile isolates and all staphylococcal strains resistant to antimicrobials were recovered from the same park. The present study suggests that free-living rodents in Belo Horizonte (Brazil) are mainly colonized by S. saprophyticus and may act as reservoirs of antimicrobial-resistant Staphylococcus spp. and C. difficile strains. This is the first study to evaluate the presence of staphylococci and C. difficile from free-living opossums and suggest a low fecal shedding of these organisms by these mammals.
Similar content being viewed by others
Introduction
The global emergence of antimicrobial resistance (AMR) has become a serious threat to human and animal health due to the widespread use of antimicrobials. However, there has been a significant increase in reports of AMR in bacteria isolated from environments and animals that have not been exposed to direct selective pressure from these agents [1]. Horizontal gene transfer between bacteria has been identified as an explanation for this phenomenon [2] and, in this context, staphylococci are highlighted as excellent carriers and transferors of resistance genes [3, 4] causing a wide variety of diseases in humans and animals [5,6,7].
Over the years, staphylococci and AMR have been extensively investigated in humans and domestic animals, but there are limited studies on wild and pest species, particularly in developing countries [8, 9]. It is known that some pest species can act as reservoirs for antimicrobial-resistant bacteria, transmitting and disseminating these microorganisms by different routes, including feces and urine [10].
Clostridioides (prev. Clostridium) difficile is an emergent pathogen responsible for antimicrobial-associated diarrhea in humans. In the last decade, animals and the environment have been suggested as possible reservoirs of C. difficile strains [11]. Recent studies in Canada and European countries have shown that rats and mice are sources of C. difficile in both urban and farm environments [12,13,14,15]. The role of other peridomestic rodents, including Cerradomys and Necromys spp., as reservoirs of C. difficile strains, has not been addressed.
Given the notable lack of data on the carriage and antimicrobial resistance profile of potentially pathogenic staphylococci and C. difficile in wild animals living in urban areas in Brazil, this study evaluated the frequency, distribution, and antimicrobial susceptibility patterns of staphylococcal species and C. difficile isolated from the feces of free-living rodents and marsupials from two urban parks in Belo Horizonte, Brazil.
Material and methods
Capture sites
The capture was conducted in two urban parks in Belo Horizonte (Minas Gerais, Brazil). Park 1 was “Jacques Cousteau Municipal Park” (19°58′ S and 43°59′ W), with a total area of 335 thousand square meters. Park 2, “Mangabeiras Municipal Park” (19°56′ S and 43°54′ W), has a total area of 2.4 million square meters (Fig. 1). Previous studies on C. difficile, Vaccinia virus, and different ectoparasites and endoparasites in free-living South American coatis (Nasua nasua) have been performed in park 2 [16,17,18,19]. In park 1, currently only one study on the abundance and diversity of amphibians has been conducted [20].
Mapping of parks 1 and 2 located in the city of Belo Horizonte, Minas Gerais, Brazil. (A) Map of Brazil with the location of the Minas Gerais state. (B) Map of Minas Gerais state with the location of Belo Horizonte. (C1) Jacques Cousteau Municipal Park (park 1). (C2) Mangabeiras Municipal Park (park 2). Yellow and blue dots represent the locations where animals were captured in transects 1 and 2, respectively.
These parks serve as recreation centers and leisure areas for the population and usually receive a large number of visitors every day throughout the year. Park 1 is located completely inside the city. The space functioned as a landfill for Belo Horizonte for 20 years (1951–1971), and then transformed into a park and horticultural garden for the production of tree and plant seedlings used for city landscaping. The site has springs and perennial watercourses that are impacted by sewage effluent from the city [21]. Park 2 is considered one of the largest urban parks in Brazil with approximately fifteen thousand visitors per month. It is located in an urban area in contact with some of the city's neighborhoods, but it is surrounded by native vegetation and other environmental preservation areas. It has water springs around and throughout, but the courses are not impacted by wastewater [22].
Animals sampled
A total of 159 free-living animals, including 136 rodents and 23 marsupials, were sampled between April 2018 and March 2019 (Table 1). For the capture, two transects were established for park 1 and three transects for park 2. Each transect had fifteen collection stations 20 m apart from each other. The stations contained a Sherman trap for capturing small rodents and marsupials, and a cage trap with suspended bait for capturing larger animals, totaling thirty traps on each transect. Each transect was surveyed once per day. The bait used was a mixture of sardines, peanuts, bananas, and corn bran. The traps were baited at the time of the survey, in the morning, between 08:00 and 09:00, on each collection day. The collections occurred for 5 consecutive days, every 2 months, totaling six campaigns in each park over a period of 1 year.
After capture, the animals were weighed. They were then anesthetized with a combination of 2% xylazine (rodents, 10 mg/kg, IP; marsupials, 5 mg/kg, IM) and ketamine hydrochloride (rodents, 100 mg/kg, IP; marsupials, 25 mg/kg, IM). After sedation, fecal samples were collected directly from the rectal ampoule of the marsupials. These animals were marked with ear tags to prevent multiple samples from the same animal, and released at the same capture site. The rodents were euthanized with an overdose of propofol (10 mg/kg) via the intracardiac route, and intestinal contents were collected from the rectum (feces) during necropsy.
The fecal samples were placed in a sterile microtube, stored in a transport box with ice packs, and transported to the Bacteriosis and Research Laboratory of the Veterinary School of the Federal University of Minas Gerais (UFMG), where they were stored at − 80 °C until laboratory processing. This study was approved by the Ethical Committee on Animal Use (CEUA) of UFMG under protocol 306/2017 and by Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) under protocol 12,989–2.
Staphylococci isolation and antimicrobial susceptibility
For Staphylococcus spp. isolation, fecal samples were suspended in 0.85% saline solution and 100 μL was streaked onto mannitol salt agar (MSA; Difco Laboratories Inc., USA) that was then incubated at 37 °C for 24 h [23]. Colonies were subcultured on brain heart infusion agar (BHI, Difco Laboratories Inc., USA) and identified by matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics, Germany). The cutoff log score of 2 was used to validate identification at the species level, as recommended by the manufacturer. The strains were then subjected to DNA extraction [24] and methicillin-resistant staphylococci were investigated by detection of the mecA gene [25]. Antimicrobial susceptibility tests were performed using disk diffusion in agar, according to the Clinical and Laboratory Standards Institute (CLSI) documents M100-S30 [26] and VET08 [27]. The following antimicrobials were tested: cefoxitin (30 μg), penicillin (10 units), tetracycline (30 μg), trimethoprim/sulfamethoxazole (25 μg), chloramphenicol (30 μg), erythromycin (15 μg), clindamycin (2 μg), gentamicin (10 μg), and ciprofloxacin (5 μg) (DME, BRA). Staphylococcus aureus ATCC 25,923 was used as a control. Isolates were considered multidrug-resistant (MDR) when resistant to three or more classes of antimicrobial agents [28].
Clostridioides difficile isolation and antimicrobial susceptibility
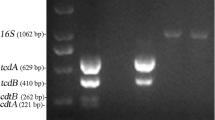
For C. difficile isolation, fecal samples were incubated in 96% ethanol for 30 min (1:1) and aliquots of 10 μL were plated on cycloserine-cefoxitin fructose agar (CCFA) supplemented with 7% horse blood and 0.1% sodium taurocholate (Sigma, USA) [29]. After incubation in an anaerobic atmosphere at 37 °C for 96 h, C. difficile colonies (flat, irregular, and with ground-glass appearance) were subjected to a multiplex PCR to identify the housekeeping gene (tpi) and the virulence genes of toxin A (tcdA), toxin B (tcdB), and binary toxin (cdtB) [30]. Toxigenic C. difficile isolates were also subjected to PCR ribotyping, as previously described [31]. The minimal inhibitory concentrations (MIC) of metronidazole, vancomycin, clindamycin, moxifloxacin, ciprofloxacin, erythromycin, rifampicin, and tetracycline were determined using Etest strips (bioMerieux, Marcy I'Etoile, France) in Brucella agar (Oxoid, USA) with 5% lysed blood, supplemented with hemin (Difco Laboratories, USA) and vitamin K (Sigma-Aldrich Co., USA). The MIC values were interpreted according to the clinical breakpoints of the CLSI and EUCAST guidelines [32,33,34].
Statistical analysis
The association between phenotypic resistance and Staphylococcus species was evaluated using the chi-square or Fisher’s exact tests. The chi-square test for adherence was used to evaluate the distribution of variables. All statistical analyses were performed using GraphPad Prism v.8 (GraphPad Software, San Diego, CA, USA). Differences were considered significant at p < 0.05.
Results
Marsupial and rodent species captured
A total of 159 free-living animals, including 136 rodents and 23 marsupials, were sampled over a 1-year period (Table 1). The number of animals sampled in park 1 (76.1%) was approximately three times higher than that in park 2 (23.9%) (p < 0.001). In park 1, rodents were captured more frequently (97.5%) than marsupials (2.5%), with black rats (Rattus rattus) the most commonly collected (31.4%) (Table 1). Among marsupials, only the white-eared opossum (Didelphis albiventris) was trapped. In park 2, the capture frequency was 52.6% for marsupials and 47.4% for rodents. The main representatives of marsupials and rodents in this park were white-eared opossum (Didelphis albiventris) (45%) and the genus Cerradomys (55.5%), respectively (Table 1). For marsupials (Table 1), the frequency of capture of these animals was higher (52.6%) and more species diversity was observed in park 2 than in park 1 (2.5%) (p < 0.001).
Staphylococcal isolation and identification
Overall, staphylococci were isolated from 36 out of 136 (26.5%) tested rodents, with one animal presenting two isolates with different Staphylococcus species (Table 1S). Among the 37 staphylococci isolates, 35 (94.5%) were from park 1, and only two were from park 2. There was no statistically significant difference in staphylococcal carriage by rodent species (Table 2). There was also no difference in the frequency of isolation in different seasonal periods (rain and drought) (Table 1S). Only one isolate (4.3%) was recovered from marsupials, specifically from a black-eared opossum (Didelphis aurita) captured in park 2. Staphylococci were more frequently isolated from rodents than from marsupials (p = 0.0164).
Eight different staphylococcal species were detected in rodents, with S. saprophyticus (48.6%) being isolated significantly more frequently than the other species (p < 0.001) (Table 2). S. aureus was isolated from three animals (8.1%). For marsupials, the only strain isolated was identified as S. saprophyticus, which was susceptible to all tested antimicrobials.
Staphylococcus spp. antimicrobial susceptibility
Nine (6.6%) rodents harbored antimicrobial-resistant staphylococci, all from park 1, and of the 37 isolates, nine (24.3%) were resistant to at least one antimicrobial agent. Four (10.8%) were classified as MDR, two (5.4%) of which were resistant to cefoxitin, and were classified as methicillin-resistant staphylococci. One of these isolates was positive for mecA (Table 3). Penicillin G had the highest frequency of resistance (24.3%), followed by erythromycin (8.1%), cefoxitin (5.4%), and clindamycin (5.4%). Resistance to penicillin G (cefoxitin/clindamycin: p = 0.04; others: p = 0.002) was significantly higher than that of the other tested antimicrobial agents, except for erythromycin (p = 0.11). However, no significant differences were found in resistance to erythromycin and other antimicrobials. All isolates were susceptible to chloramphenicol, ciprofloxacin, gentamicin, tetracycline, and trimethoprim/sulfamethoxazole. All rodent species in the present study showed at least one antimicrobial resistant isolate, and no statistical difference was found between these species (p = 0.15). In addition, there was no difference in the frequency of resistant isolates among transects in each park (p = 0.24) or between the parks (p = 0.6).
Clostridioides difficile isolation and antimicrobial susceptibility
C. difficile was isolated from two (1.7%) animals, both rodents from park 1. No association was observed between rodent species and the isolation of C. difficile. One strain was toxigenic (A + B + CDT-) and was classified as ribotype 064, while the other isolate was non-toxigenic (A-B-CDT-). The non-toxigenic C. difficile isolated in the present study was resistant to rifampicin (MIC 3.0 mg/mL). The two isolates were susceptible to all other seven antimicrobials tested.
Discussion
Studies have suggested that wild animals living closer to humans and domestic animals may become a threat to public health because they harbor and disseminate pathogens and MDR microorganisms, including Staphylococcus spp. and C. difficile [10, 35]. However, there are few investigations on the role of rodents and marsupials in urban areas, especially in Brazil. Thus, the present study evaluated the presence of C. difficile and MDR staphylococci among rodents and marsupials from two urban parks in Belo Horizonte, Brazil.
Two parks were used to trap and sample rodents and marsupials. Several differences were observed in the animals sampled from each park. First, the number of animals sampled in park 1 was almost three times higher than that in park 2. In addition, differences in the species captured were also observed; black rats were the most common rodent trapped in park 1, but this animal was not captured in park 2. In contrast, the frequency and diversity of marsupials were higher in park 2 than in park 1. Ecological aspects are the main hypotheses for these differences, since park 1 is substantially smaller (335 thousand square meters versus 2.4 million square meters), is completely surrounded by urban environment (Fig. 1), has experiencing a rapidly growing of vertical urbanization, and has sewage effluent present in its waterways [21]. This environment seems more attractive to synanthropic rodents, including black rats, while the more conserved area observed in park 2 might favor the trapping of marsupials [36,37,38].
S. saprophyticus was the most frequently recovered species from rodents in the present study (Table 2). Staphylococcal species in rodents vary considerably between studies, with S. xylosus, S. succinus, and S. sciuri being the most frequently noted [39,40,41]. Interestingly, S. saprophyticus was only reported in a few animals in one study on bank voles (Myodes glareolus) conducted in Poland [39]. In a public health context, it is also important to remember that S. saprophyticus is the second highest cause of urinary tract infections in women, including in Brazil, and is typically classified as a human colonizer [42,43,44,45,46,47]. Although less frequently, S. aureus (8.1%) and S. epidermidis (5.4%) were also detected in the present study. These two species are commonly found in human microbiota and are well-known opportunistic pathogens that can cause serious infections in humans and animals [48,49,50].
Overall, the incidence of staphylococci in rodents in this study (27.2%) was much lower than that reported by other authors, which is generally more than 75% [39, 41]. Differences in rodent species were observed in these studies, and differences in host ecology, such as food, geographical location, and contact with different anthropogenic sources may have contributed to this large variation in carriage rate [51, 52].
Only one isolate of S. saprophyticus (4.3%) was recovered from marsupials, and rodents appeared to be more prone to staphylococcal colonization than marsupials (p = 0.01). The only study published to date that evaluated the distribution of staphylococci in marsupials, specifically in the nasal swabs of Australian wallabies (Petrogale xanthopus, Petrogale lateralis, and Macropus eugenii), reported a much higher isolation rate than this study, reaching 90.8%. In addition, fourteen species of staphylococci were recovered, with S. delphini and S. succinus being the predominant species. S. saprophyticus, also isolated in the present study, was recovered from 4.5% of the animals evaluated [53]. In that study, 70% of the wallabies lived in captivity, and the authors attributed the high frequency of isolation and diversity of staphylococcal species to environmental selection pressure and anthropogenic activity.
Staphylococci are known for their capacity to carry and disseminate antimicrobial resistance determinants, which contribute to their pathogenic potential [4, 54, 55]. In the present study, 24.3% of the rodent isolates, all from park 1, were resistant to at least one tested antimicrobial agent, including three isolates classified as MDR, one of which was also classified as methicillin-resistant Staphylococcus. Penicillin G and erythromycin, two drugs widely used in human and veterinary medicine, had the highest resistance rate. These results demonstrate that these rodents, although not directly exposed to antimicrobial agents, can harbor and disseminate resistant bacteria. Only animals from park 1, which is more anthropized and contains sewage effluent, showed antimicrobial resistance. It is possible that this resistance is acquired by the contact of the animals with waterways contaminated with waste from sewage effluent. Sewage effluent is known to harbor several resistant microorganisms and consequently provides a route for horizontal transfer of resistance genes, which is the main hypothesis for the higher rate of AMR in rodents in park 1 than in park 2 [52, 56,57,58].
The high incidence of potentially pathogenic staphylococci to humans, as well as the high rate of AMR, including methicillin-resistant and other MDR staphylococci, is of concern mainly for park 1. Methicillin-resistant staphylococci confer resistance to at least all beta-lactam antimicrobials, which excludes most of the first-choice treatment options for both animals and humans, substantially reducing therapeutic alternatives [57]. There are several reports on the colonization and infection of companion animals that transmit methicillin-resistant staphylococci and MDR [59,60,61], showing the relevance of studies monitoring the occurrence of these resistant bacteria in animals.
The isolation rate of C. difficile in the present study (1.5%) was lower than that reported previously with urban rodents, which varied between 4.3 and 35% [13, 62, 63] and with rodents trapped in or around farms, which returned between 24 and 39.2% [63, 64]. A previous study from 2014 with South American coati (Nasua nasua) in park 2 also reported a low isolation rate (6.5%) of C. difficile [16], whereas all animals sampled in park 2 were negative for C. difficile in the present study. It is believed that pests reflect environmental contamination with C. difficile spores [65], and therefore this difference among findings is expected, suggesting that both parks have low C. difficile contamination.
One of the C. difficile strains was toxigenic and classified as ribotype 064. C. difficile ribotypes associated with CDI in Brazil are still largely unknown because of the lack of large-scale studies [66]. RT064 currently has not been reported in either humans or animals in Brazil, including studies specifically in Belo Horizonte, the same city where the two parks are located [66,67,68]. RT064 has previously been reported in animals elsewhere [69] and has also been shown to infect humans [70, 71].
Resistance to rifampicin was detected in a non-toxigenic C. difficile strain, but both isolates were susceptible to all other antimicrobials tested. This result contrasts with other studies with rodents, which showed high rates of MDR C. difficile strains isolated from rodents [65]. At the same time, the detection of non-toxigenic strains resistant to rifampicin contributes to the growing concern regarding the role of non-toxigenic strains, including isolates from rodents, in the spread of resistance patterns of C. difficile, which was previously only focused on toxigenic isolates [65, 72].
Conclusion
In conclusion, the present work suggests that free-living rodents in Belo Horizonte (Brazil) are commonly colonized by S. saprophyticus and can harbor MDR and methicillin-resistant Staphylococcus strains. C. difficile strains with antimicrobial resistance and those from a ribotype previously reported in humans were also recovered from these animals. With regard to marsupials, this is the first study to evaluate the colonization and antimicrobial resistance profile of staphylococci isolated from the feces of free-living opossums, and despite the small sample size, the results suggest low fecal elimination of staphylococci by these animals.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J (2010) Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol 8:251–259. https://doi.org/10.1038/nrmicro2312
Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. https://doi.org/10.1038/nrmicro1234
Otto M (2013) Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: Staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 35, 4–11. https://doi.org/10.1002/bies.201200112.
Becker K, Heilmann C, Peters G (2014) Coagulase-negative staphylococci. Clin Microbiol Rev 27:870–926. https://doi.org/10.1128/CMR.00109-13
Wieler LH, Ewers C, Guenther S, Walther B, Lübke-Becker A (2011) Methicillin-resistant staphylococci (MRS) and extended-spectrum beta-lactamases (ESBL)-producing Enterobacteriaceae in companion animals: nosocomial infections as one reason for the rising prevalence of these potential zoonotic pathogens in clinical samples. International Journal of Medical Microbiology: IJMM 301:635–641. https://doi.org/10.1016/j.ijmm.2011.09.009
Walther B, Tedin K, Lübke-Becker A (2017) Multidrug-resistant opportunistic pathogens challenging veterinary infection control. Vet Microbiol 200:71–78. https://doi.org/10.1016/j.vetmic.2016.05.017
Heilmann C, Ziebuhr W, Becker K (2019) Are coagulase-negative staphylococci virulent? Clinical Microbiology and Infection: The Official Publication of the European Society of Clinical Microbiology and Infectious Diseases, 25, 1071–1080, https://doi.org/10.1016/j.cmi.2018.11.012.
Himsworth CG, Miller RR, Montoya V, Hoang L, Romney MG, Al-Rawahi GN, Kerr T, Jardine CM, Patrick DM, Tang P, Weese JS (2014) Carriage of methicillin-resistant Staphylococcus aureus by wild urban Norway rats (Rattus norvegicus). PLoS ONE 9:e87983. https://doi.org/10.1371/journal.pone.0087983
Ge J, Zhong XS, Xiong YQ, Qiu M, Huo ST, Chen XJ, Mo Y, Cheng MJ, Chen Q (2019) Methicillin-resistant Staphylococcus aureus among urban rodents, house shrews, and patients in Guangzhou. Southern China BMC Veterinary Research 15:260. https://doi.org/10.1186/s12917-019-2012-8
Jahan NA, Lindsey LL, Larsen PA (2021) The role of peridomestic rodents as reservoirs for zoonotic foodborne pathogens. Vector Borne Zoonotic Dis 21:133–148. https://doi.org/10.1089/vbz.2020.2640
Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ (2016) Clostridium difficile infection. Nat Rev Dis Primers 2:16020. https://doi.org/10.1038/nrdp.2016.20
Burt SA, Siemeling L, Kuijper EJ, Lipman LJ (2012) Vermin on pig farms are vectors for Clostridium difficile PCR ribotypes 078 and 045. Vet Microbiol 160:256–258. https://doi.org/10.1016/j.vetmic.2012.05.014
Himsworth CG, Patrick DM, Mak S, Jardine CM, Tang P, Weese JS (2014) Carriage of Clostridium difficile by wild urban Norway rats (Rattus norvegicus) and black rats (Rattus rattus). Appl Environ Microbiol 80:1299–1305. https://doi.org/10.1128/AEM.03609-13
Jardine CM, Reid-Smith RJ, Rousseau J, Weese JS (2013) Detection of Clostridium difficile in small and medium-sized wild Mammals in Southern Ontario Canada. Journal of Wildlife Diseases 49:418–421. https://doi.org/10.7589/2012-04-120
Krijger IM, Meerburg BG, Harmanus C, Burt SA (2019) Clostridium difficile in wild rodents and insectivores in the Netherlands. Lett Appl Microbiol 69:35–40. https://doi.org/10.1111/lam.13159
Silva ROS, Almeida LR, Oliveira CA Jr, Soares DFM, Pereira PLL, Rupnik M, Lobato FCF (2014) Carriage of Clostridium difficile in free-living South American coati (Nasua nasua) in Brazil. Anaerobe 30:99–101. https://doi.org/10.1016/j.anaerobe.2014.09.012
Costa GB, Ribeiro de Almeida L, Cerqueira AGR, Mesquita WU, Silva de Oliveira J, Miranda JB, Saraiva-Silva AT, Abrahão JS, Drumond BP, Kroon EG, Pereira PLL, Soares DFM, Trindade GS (2018) Vaccinia virus among domestic dogs and wild coatis, Brazil, 2013–2015. Emerging Infectious Diseases, 24, 2338–2342, https://doi.org/10.3201/eid2412.171584.
Almeida LR, Souza JGR, Santos HA, Torres EJL, Vilela RV, Cruz OMS, Rodrigues L, Pereira CAJ, Maldonado A Jr, Lima WS (2020) Angiostrongylus minasensis n. sp.: new species found parasitizing coatis (Nasua nasua) in an urban protected area in Brazil. Revista Brasileira de Parasitologia Veterinária 29:e018119. https://doi.org/10.1590/S1984-29612019103
Estevam LGTM, Fonseca Junior AA, Silvestre BT, Hemetrio NS, Almeida LR, Oliveira MM, Silva SM, Ribeiro MFB, Silveira JAG (2020) Seven years of evaluation of ectoparasites and vector-borne pathogens among ring-tailed coatis in an urban park in southeastern Brazil. Veterinary Parasitology, Regional Studies and Reports, 21, 100442, https://doi.org/10.1016/j.vprsr.2020.100442.
Torres PF (2012) Uso de ambientes por anfíbios anuros em seis parques urbanos de Belo Horizonte, Minas Gerais. Dissertação (Dissertação em Ciências Biológicas) – UFMG. Minas Gerais, p. 113
Parque Municipal Jacques Cousteau (2019) Disponível em: https://prefeitura.pbh.gov.br/fundacao-de-parques-e-zoobotanica/informacoes/parques/parque-jacques-cousteau . Acesso em: 2 abr 2021.
Parque Municipal das Mangabeiras (2020) Disponível em: https://prefeitura.pbh.gov.br/fundacao-de-parques-e-zoobotanica/informacoes/parques/parque-das-mangabeiras . Acesso em: 2 abr 2021.
Gómez P, González-Barrio D, Benito D, García JT, Viñuela J, Zarazaga M, Ruiz-Fons F, Torres C (2014) Detection of methicillin-resistant Staphylococcus aureus (MRSA) carrying the mecC gene in wild small mammals in Spain. J Antimicrob Chemother 69:2061–2064. https://doi.org/10.1093/jac/dku100
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156. https://doi.org/10.1111/j.1472-765X.1989.tb00262.x
Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S (1991) Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol 29:2240–2244. https://doi.org/10.1128/JCM.29.10.2240-2244.1991
Clinical and Laboratory Standards Institute (CLSI) (2020) Wayne, PA, USA. Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI supplement M100, 19087
Clinical and Laboratory Standards Institute (CLSI) (2018) Wayne, PA, USA. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 5th ed. CLSI standard VET01
Sweeney MT, Lubbers BV, Schwarz S, Watts JL (2018) Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J Antimicrob Chemother 73:1460–1463. https://doi.org/10.1093/jac/dky043
Silva ROS, Ribeiro MG, Palhares MS, Borges AS, Maranhão RPA, Silva MX, Lucas TM, Olivo G, Lobato FCF (2013) Detection of A/B toxin and isolation of Clostridium difficile and Clostridium perfringens from foals. Equine Vet J 45:671–767. https://doi.org/10.1111/evj.12046
Silva ROS, Salvarani FM, Cruz Jr, ECC, Pires PS, Santos RL, Assis RA, Guedes RMC, Lobato FCF (2011) Detection of enterotoxin A and cytotoxin B, and isolation of Clostridium difficile in piglets in Minas Gerais, Brazil. Ciência Rural, 41, 1430–1435. https://doi.org/10.1590/S0103-84782011005000100.
Janezic S, Rupnik M (2010) Molecular typing methods for Clostridium difficile: pulsed-field gel electrophoresis and PCR ribotyping. Methods Mol Biol 646:55–65. https://doi.org/10.1007/978-1-60327-365-7_4
Pirš T, Avbersek J, Zdovc I, Krt B, Andlovic A, Lejko-Zupanc T, Rupnik M, Ocepek M (2013) Antimicrobial susceptibility of animal and human isolates of Clostridium difficile by broth microdilution. J Med Microbiol 62:1478–1485. https://doi.org/10.1099/jmm.0.058875-0
Clinical and Laboratory Standards Institute (CLSI) (2015) Wayne, PA, USA. Performance standards for antimicrobial susceptibility testing; Twenty-Fifth Informational Supplement
The European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2019) Breakpoint tables for interpretation of MICs and zone diameters. In: European Society of Clinical Microbiology and Infectious Diseases Basel
Himsworth CG, Parsons KL, Jardine C, Patrick DM, Rats C (2013) Rats, cities, People, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector Borne Zoonotic Dis 13:349–359. https://doi.org/10.1089/vbz.2012.1195
Feng AYT, Himsworth CG (2014) The secret life of the city rat: a review of the ecology of urban Norway and black rats (Rattus norvegicus and Rattus rattus). Urban Ecosystems 17:149–162. https://doi.org/10.1007/s11252-013-0305-4
Cáceres NC, Moraes MM, Melo GL, Meloro C, Sponchiado J, Carvalho RS, Bubadué JM (2016) Which factors determine spatial segregation in the South American Opossums (Didelphis aurita and D. albiventris)? An ecological niche modelling and geometric morphometrics approach. PLoS ONE, 11, https://doi.org/10.1371/journal.pone.0157723.
Byers KA, Lee MJ, Patrick DM, Himsworth CG (2019) Rats about town: a systematic review of rat movement in urban ecosystems. Frontiers in Ecology and Evolution, 7, https://doi.org/10.3389/fevo.2019.00013
Nagase N, Sasaki A, Yamashita K, Shimizu A, Wakita Y, Kitai S, Kawano J (2002) Isolation and species distribution of staphylococci from animal and human skin. J Vet Med Sci 64:245–250. https://doi.org/10.1292/jvms.64.245
Hauschild T, Kehrenberg C, Schwarz S (2003) Tetracycline resistance in staphylococci from free-living rodents and insectivores. Journal of Veterinary Medicine B, Infectious Diseases and Veterinary Public Health 50:443–446. https://doi.org/10.1046/j.0931-1793.2003.00706.x
Kmeť V, Čuvalová A, Stanko M (2018) Small mammals as sentinels of antimicrobial-resistant staphylococci. Folia Microbiol 63:665–668. https://doi.org/10.1007/s12223-018-0594-3
Latham RH, Running K, Stamm WE (1983) Urinary tract infections in young adult women caused by Staphylococcus saprophyticus. JAMA 250:3063–3066
Von Eiff C, Peters G, Heilmann C (2002) Pathogenesis of infections due to coagulase negative staphylococci. Lancet Infect Dis 2:677–685. https://doi.org/10.1016/S1473-3099(02)00438-3
Raz R, Colodner R, Kunin CM (2005) Who are you – Staphylococcus saprophyticus? Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 40:896–898. https://doi.org/10.1086/428353
Fariña N, Carpinelli L, Samudio M, Guillén R, Laspina F, Sanabria R, Abente S, Rodas L, González P, de Kaspar HM (2013) Clinically significant coagulase-negative staphylococci: most frequent species and virulence factors. Rev Chilena Infectol 30:480–488. https://doi.org/10.4067/S0716-10182013000500003
Sousa VS, Rabello RF, Dias RCS, Martins IS, Santos LBGS, Alves EM, Riley LW, Moreira BM (2013) Time-based distribution of Staphylococcus saprophyticus pulsed field gel-electrophoresis clusters in community-acquired urinary tract infections. Memorias Do Instituto Oswaldo Cruz 108:73–76. https://doi.org/10.1590/s0074-02762013000100012
Lo DS, Shieh HH, Barreira ER, Ragazzi SLB, Gilio AE (2015) High frequency of Staphylococcus saprophyticus urinary tract infections among female adolescents. Pediatr Infect Dis J 34:1023–1025. https://doi.org/10.1097/INF.0000000000000780
Weese JS, Van Duijkeren E (2010) Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Vet Microbiol 140:418–429. https://doi.org/10.1016/j.vetmic.2009.01.039
Edmiston CE, Mcbain AJ, Kiernan M, Leaper DJ (2016) A narrative review of microbial biofilm in postoperative surgical site infections: clinical presentation and treatment. J Wound Care 25:693–702. https://doi.org/10.12968/jowc.2016.25.12.693
Chalmers SJ, Wylam ME (2020) Methicillin-resistant Staphylococcus aureus infection and treatment options. In: Methods in Molecular Biology Ji, Y. (Ed.). Methicillin-Resistant Staphylococcus Aureus (MRSA) Protocols: Cutting-Edge Technologies and Advancements. New York, NY: Springer, 2069, 229–251, https://doi.org/10.1007/978-1-4939-9849-4_16.
Yalden DW, Harris S (2008) Mammals of the British Isles: Handbook. 4th ed,
Furness LE, Campbell A, Zhang L, Gaze WH, Mcdonald RA (2017) Wild small mammals as sentinels for the environmental transmission of antimicrobial resistance. Environ Res 154:28–34. https://doi.org/10.1016/j.envres.2016.12.014
Chen MMS, Boardman WSJ, Smith I, Egoodman AE, Brown MH (2014) Nasal colonization of Staphylococcus spp. among captive and free-ranging wallabies in South Australia. Journal of Veterinary Science and Medical Diagnosis, 03. https://doi.org/10.4172/2325-9590.1000136.
Beims H, Overmann A, Fulde M, Steinert M, Bergmann S (2016) Isolation of Staphylococcus sciuri from horse skin infection. Open Veterinary Journal 6:242–246. https://doi.org/10.4314/ovj.v6i3.14
Schoenfelder SMK, Dong Y, Feßler AT, Schwarz S, Schoen C, Köck R, Ziebuhr W (2017) Antibiotic resistance profiles of coagulase-negative staphylococci in livestock environments. Veterinary Microbiology Resistance 200:79–87. https://doi.org/10.1016/j.vetmic.2016.04.019
Baquero F, Martínez JL, Cantón R (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19:260–265. https://doi.org/10.1016/j.copbio.2008.05.006
Gaze W, O’Neill C, Wellington E, Hawkey P (2008) Antibiotic resistance in the environment, with particular reference to MRSA. Adv Appl Microbiol 63:249–280. https://doi.org/10.1016/S0065-2164(07)00007-X
Jobbins SE, Alexander KA (2015) From whence they came – antibiotic-resistant Escherichia coli in African wildlife. J Wildl Dis 51:811–820. https://doi.org/10.7589/2014-11-257
Van Duijkeren E, Kamphuis M, Van der Mije IC, Laarhoven LM, Duim B, Wagenaar JA, Houwers DJ (2011) Transmission of methicillin-resistant Staphylococcus pseudintermedius between infected dogs and cats and contact pets, humans and the environment in households and veterinary clinics. Vet Microbiol 150:338–343. https://doi.org/10.1016/j.vetmic.2011.02.012
Paul NC, Moodley A, Ghibaudo G, Guardabassi L (2011) Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission. Zoonoses Public Health 58:533–539. https://doi.org/10.1111/j.1863-2378.2011.01398.x
Pomba C, Rantala M, Greko C, Baptiste KE, Catry B, Van Duijkeren E, Mateus A, Moreno MA, Pyörälä S, Ružauskas M, Sanders P, Teale C, Threlfall EJ, Kunsagi Z, Torren-Edo J, Jukes H, Törneke K (2017) Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother 72:957–968. https://doi.org/10.1093/jac/dkw481
Williams SH, Che X, Paulick A, Guo C, Lee B, Muller D, Uhlemann AC, Lowy FD, Corrigan RM, Lipkin WI (2018) New York City house mice (Mus musculus) as potential reservoirs for pathogenic bacteria and antimicrobial resistance determinants. mBio, 9, e00624–18
Burt SA, Meijer K, Burggraaff P, Kamerich WS, Harmanus C (2018) Wild mice in and around the city of Utrecht, the Netherlands, are carriers of Clostridium difficile but not ESBL-producing Enterobacteriaceae, Salmonella spp. or MRSA. Letters in Applied Microbiology 67:513–519
Krijger IM, Meerburg BG, Harmanus CC, Burt SA (2019) Clostridium difficile in wild rodents and insectivores in the Netherlands. Letters in Applied Microbiology 69:35–40
Andres-Lasheras S, Bolea R, Mainar-Jaime RC, Kuijper E, Sevilla E, Martín-Burriel I, Chirio-Trejo M (2016) Presence of Clostridium difficile in pig faecal samples and wild animal species associated with pig farms. J Appl Microbiol 122:462–472. https://doi.org/10.1111/jam.13343
Trindade CNR, Domingues RMCP, Ferreira EO (2019) The epidemiology of Clostridioides difficile infection in Brazil: a systematic review covering thirty years. Anaerobe 58:13–21. https://doi.org/10.1016/j.anaerobe.2019.03.002
Silva ROS, Rupnik M, Diniz AN, Vilela EG, Lobato FCF (2015) Clostridium difficile ribotypes in humans and animals in Brazil. Mem Inst Oswaldo Cruz 110:1062–1065. https://doi.org/10.1590/0074-02760150294
Diniz AN, Oliveira CA Jr, Vilela EG, Figueiredo HCP, Rupnik M, Wilcox MH, Fawley WN, Blanc D, Lobato FCF, Silva ROS (2019) Molecular epidemiology of Clostridioides (previously Clostridium) difficile isolates from a university hospital in Minas Gerais. Brazil Anaerobe 56:34–39. https://doi.org/10.1016/j.anaerobe.2019.01.010
Knight DR, Thean S, Putsathit P, Fenwick S, Riley TV (2013) Cross-sectional study reveals high prevalence of Clostridium difficile non PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol 79:2630–2635. https://doi.org/10.1128/AEM.03951-12
Knight DR, Thean S, Putsathit P, Fenwick S, Riley TV (2013) Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol 79:2630–2635. https://doi.org/10.1128/AEM.03951-12
Alfayyadh M, Collins DA, Tempone S, McCann R, Armstrong PK, Riley TV, Cook A (2019) Recurrence of Clostridium difficile infection in the Western Australian population. Epidemiol Infect 147:e153. https://doi.org/10.1017/S0950268819000499
Álvarez-Pérez S, Blanco JL, Peláez T, Lanzarot MP, Harmanus C, Kuijper E, García ME (2015) Faecal shedding of antimicrobial-resistant Clostridium difficile strains by dogs. J Small Anim Pract 56:190–195. https://doi.org/10.1111/jsap.12311
Acknowledgements
We thank Fundação de Parques Municipais da prefeitura de belo Horizonte, including Jacques Cousteau Municipal Park and Mangabeiras Municipal Park, for the support and animal samples. We thank Gustavo Canesso Bicalho for his valuable contribution in the making of the map of the evaluated parks. We also thank CAPES, CNPq, FAPEMIG, and PRPq/UFMG for all the financial support.
Funding
This work was supported by funds from the Coordination for the Improvement of Higher Education Personnel (CAPES–Prêmio CAPES 2015–0774/2017), the National Council for Scientific and Technological Development (CNPq-406402/2018–3), Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG- PQ-00524–17), and Pró-Reitoria de Pesquisa da Universidade Federal de Minas Gerais (PRPq/UFMG) and the MCTIC/FNDCT-CNPq/MEC-CAPES/Grant 440593/2016–6.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and samples collection were performed by Salene Angelini Colombo, Lara Ribeiro de Almeida, and Brendhal Almeida Silva. Laboratory analysis were performed by Jordana Almeida Santana, Salene Angelini Colombo, Brendhal Almeida Silva, Amanda Nádia Diniz, and Carlos Augusto Oliveira Junior. The first draft of the manuscript was written by Jordana Almeida Santana, Rodrigo Otávio Silveira Silva, Giliane de Souza Trindade, Adriano Pereira Paglia, and Francisco Carlos Faria Lobato. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by the Ethical Committee on Animal Use (CEUA) of the Federal University of Minas Gerais under protocol 306/2017 and by the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) under protocol 12989–2.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Santana, J.A., Colombo, S.A., Silva, B.A. et al. Clostridioides difficile and multi-drug-resistant staphylococci in free-living rodents and marsupials in parks of Belo Horizonte, Brazil. Braz J Microbiol 53, 401–410 (2022). https://doi.org/10.1007/s42770-021-00640-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00640-x