Abstract
DNA storage is a new digital data storage technology based on specific encoding and decoding methods between 0 and 1 binary codes of digital data and A-T-C-G quaternary codes of DNAs, which and is expected to develop into a major data storage form in the future due to its advantages (such as high data density, long storage time, low energy consumption, convenience for carrying, concealed transportation and multiple encryptions). In this review, we mainly summarize the recent research advances of four main encoding and decoding methods of DNA storage technology: direct mapping method between 0 and 1 binary and A-T-C-G quaternary codes in early-stage, fountain code for higher logical storage density, inner and outer codes for random access DNA storage data, and CRISPR mediated in vivo DNA storage method. The first three encoding/decoding methods belong to in vitro DNA storage, representing the mainstream research and application in DNA storage. Their advantages and disadvantages are also reviewed: direct mapping method is easy and efficient, but has high error rate and low logical density; fountain code can achieve higher storage density without random access; inner and outer code has error-correction design to realize random access at the expense of logic density. This review provides important references and improved understanding of DNA storage methods. Development of efficient and accurate DNA storage encoding and decoding methods will play a very important and even decisive role in the transition of DNA storage from the laboratory to practical application, which may fundamentally change the information industry in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
DNA storage technology is a new data storage technology through DNA storage medium, which can achieve digital data storage (text, image, audio, video, etc.) by encoding and decoding for the synthesized DNAs with specific sequences based on certain encoding/decoding methods. Specially, according to certain encoding methods/rules of DNA storage, the 0–1 binary codes encoding for various digital data (text, image, audio, video, etc.) can be converted to corresponding DNA quaternary codes (i.e., combinations of A, T, C, and G), and the corresponding DNAs were then synthesized to store the digital data information into the DNAs with specific sequences. Conversely, based on corresponding decoding methods/rules, the stored DNAs can be sequenced to obtain DNA quaternary codes, further restoring to the digital data with 0–1 binary codes. Here the encoding and decoding methods follow the same “codebook”, and the encoding is the reverse process of the decoding (Fig. 1).
Schematic showing DNA storage. DNA storage system generally consists of encoding process, DNA synthesis and storage, DNA sequencing, and decoding process. In encoding process, the 0–1 binary codes can be converted to 0–1 binary codes according to the same codebook. The corresponding DNAs are then synthesized and stored. In decoding process, the synthesized DNAs are sequenced through high throughput sequencing platforms and then recovered to the digital data with 0–1 binary codes
DNA storage breaks through the limitation of existing storage medium of silica-based materials (such as hard disk, optical disk, and removable magnetic disk): compared with the existing digital data storage technologies, DNA storage technology has the advantages of high data density, long storage time, low energy consumption, convenience for carrying, concealed transportation, and multiple encryptions (De Silva and Ganegoda 2016). Furthermore, with rapid development of biotechnology and information technology (BT&IT), DNA storage is expected to fundamentally change the pattern of data storage and transmission, further leading to revolutionary changes in various important areas of the national economy such as manufacturing, internet industry, and national security (The DNA data storage alliance 2021).
In this review, we mainly introduce some mainstream encoding and decoding methods of DNA storage technology. As mentioned above, based on the same “codebook”, the encoding method of DNA storage is to convert 0–1 binary codes to A-T-C-G DNA quaternary codes, while the decoding method is to convert quaternary sequencing data to 0–1 binary files. Herein, the sequence structure of encoding and decoding DNAs generally includes three parts: address information, data payload and error-correcting code (Fig. 2), which is commonly less than 250 nt due to the limitations of current synthesis and sequencing technologies (Kumar et al. 2019; Liu et al. 2012).
2 Results
2.1 Codebook (mapping rule)
The codebook reflects the encoding and decoding mapping rule between 0–1 binary codes and DNA A-T-C-G quaternary codes, which guides the conversion between digital bitstream and DNA storage file (Clelland et al. 1999). The codebook should follow certain rules to avoid systematic errors during DNA data storage, such as DNA secondary structure, high GC content, and homopolymer (Organick et al. 2018). To date, there are two types of codebooks: “bit-base” (Goldman et al. 2013) and “symbol-codon” (Fig. 3) (Clelland et al. 1999). “Bit-base” indicates the mapping rule between bits and bases. For example, Fig. 3a showed the mapping relationship between “0” and “(A)C/(C)G/(G)T/(T)A”. “Symbol-codon” indicates the mapping relationship between a symbol and a codon (Fig. 3b). Before use, the codons in the DNA storage file should be screened by DNA sequence characteristics to avoid systematic errors. Although the former is simpler than the latter, the latter is able to avoid systematic errors to some extent during DNA data storage.
2.2 Mainstream encoding and decoding methods: Church encoding and decoding method
In August 2012, the George Church group of Harvard University published a landmark paper in the research field of DNA storage and achieved the DNA storage for a 5.2 MB data of HTML files, JPG images, and JavaScript programs (Church et al. 2012). They first proposed the “bit-base” mapping rule/codebook: one bit per base. Specially, each bit corresponds to a nucleotide, in which A or C represents 0, and T or G represents 1 (Fig. 3a). In the codebook, the generated DNA sequences with more than 3 nt homopolymer were excluded. Here, the sequence structure of encoding and decoding DNAs (159 nt) includes a 96 nt DNA fragment for data payload, a 19 nt fragment for address information, and two 22 nt fragments as PCR amplification primers on both ends (Fig. 4). For decoding, we constructed a paired-end library for each 159 nt DNA fragment and then sequenced it on an Illumina HiSeq 2000 platform in 100-bp paired-end mode. Further, the sequenced pair-end reads were combined into a single contig by overlapping to reduce the systematic sequencing errors. However, due to systematic errors and without using error-correcting code during DNA storage, the data could not be recovered completely in spite of higher sequencing depth (average coverage: 3000 ×).
In 2016, Blawat improved Church's method using triple error-detecting/correcting codes to achieve the DNA storage and completely accurate recovery for a 22 MB data of a MPEG compressed movie (Blawat et al. 2016): the address information was horizontally protected by Bose-Chaudhuri-Hocquenghem codes (BCH) (63, 39) with the minimum Hamming distance of 9 (occupying 39 bits); the consecutive data blocks (223 bits for data payload) were vertically protected by Reed–Solomon codes (RS) (255, 223, 33); 16-bit Cyclic Redundancy Check (CRC) was used for error detection of address information and data payload (Fig. 5).
Blawat’s error-detecting/correcting codes (Blawat et al. 2016)
2.3 Mainstream encoding and decoding methods: Goldman encoding and decoding method
In 2013, Goldman’s group achieved the DNA storage and completely accurate recovery for a 739 KB data file (including text, JPG images, and JavaScript program) only using lower sequencing depth (average coverage: ~ 51 ×) (Goldman et al. 2013). Goldman’s encoding and decoding method belong to an improved “bit-base” mapping rule of DNA storage. Compared with the Church method, the 0–1 binary codes of digital data were first subjected to data compression by 3-base Huffman code and then converted to A-T-C-G quaternary codes. Besides, a parity base and an overlap of 75 bases were used in the sequence structure to increase the robustness against systematic errors during DNA storage. Here, the sequence structure of encoding and decoding DNAs (117 nt) includes a 100 nt DNA fragment for data payload, a 2 + 12 nt fragment for address information, one parity nucleotide, and one nucleotide at each end for the alternate segments of reverse complemented DNA sequences (Fig. 6).
Goldman’s encoding and decoding method (Goldman et al. 2013). a The mapping rule from 0–1 binary code to ternary Huffman code then to A-T-C-G quaternary DNA code in Goldman’s encoding/decoding method. b Sequence structure of Goldman’s encoding/decoding method
Based on Goldman’s work, Bornholt et al. successfully introduced RAID (Redundant Array of Independent Disks) to lower the systematic errors of DNA storage (Bornholt et al. 2016), and achieved the DNA storage and completely accurate recovery for a 151 KB data file using lower sequencing depth (average coverage: ~ 40 ×). RAID5 is a main data storage system for error correction in computer technology (Microsemi Corporate Headquarters 2017). Here the data information and its verification information are stored on different disks. The data information can be rebuilt through the verification information when they are damaged only on one disk. Similarly, Bornholt et al. produced a new exclusive-or A ⊕ B DNA strand as the verification information of A + B DNA strands (data information). The data information of A + B DNA strands can be recovered through the new exclusive-or A ⊕ B DNA strand if we cannot obtain the completely sequencing data on one strand (A or B) (Fig. 7). Compared with Goldman’s method, another advantage of this improved method is to decrease the redundancy (2–3 times) by avoiding the overlaps.
2.4 Mainstream encoding and decoding methods: Grass encoding and decoding method
In 2015, Grass’ group put forward a new “Symbol-codon” encoding and decoding method with inner and outer codes, in which a redundant DNA sequence was added to each row and column separately based on Reed–Solomon code to do error corrections in two dimensions (Grass et al. 2015). Specially, the 0–1 binary data are first arranged in the matrix blocks. For each matrix, the outer code, redundancy A is generated through the Reed–Solomon code for each row; the inner code, redundancy B is generated by the Reed–Solomon code for each column. Finally, each column of 0–1 binary data can be converted to A-T-C-G quaternary codes based on the corresponding “Symbol-codon” codebook (Fig. 8), and the related DNAs are synthesized and stored. Through this method, Grass’ group achieved the DNA storage and completely accurate recovery for an 83 KB data file (text) using lower sequencing depth (average coverage: ~ 372 ×). Here, the DNA sequence structure (158 nt) includes a 117 nt DNA fragment for data payload and one adapter at each end for sequencing.
Grass encoding–decoding method (Grass et al. 2015). a The inner and outer code. b The sequence structure for Grass’s encoding/decoding method. c The codebook of Grass encoding/decoding method
In 2018, based on the Grass encoding and decoding method, Microsoft and the University of Washington proposed a random-access encoding method. They successfully stored and individually recovered 35 distinct files (average coverage: ~ 5 ×) through extra addressing primers (20 nt) at both ends, in which the addressing primers should be satisfied with design criteria, such as GC content, homopolymers, hamming distance. Furthermore, to avoid the collision of addressing primers and payloads, the mapping rules of the codebook can be dynamically changed to reduce the high similarity probability between addressing primers and payloads (Fig. 9).
Microsoft random access encoding–decoding method (Organick et al. 2018)
2.5 Mainstream encoding and decoding methods: Fountain code
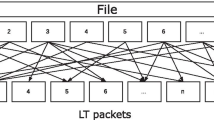
In 2017, Yaniv Erlich's research team at Columbia University reported a new DNA storage method based on fountain code (LT code) and achieved the DNA storage and completely accurate recovery for a 2.15 MB data file (including text, operating system, image, PDF, movie, and malware) using lower sequencing depth (average coverage: ~ 10.5 ×) (Erlich and Zielinski 2017). Fountain code is known as many sending water droplets with data information at the sending ends like fountain. Here, the data can be completely recovered when the water droplets in a whole bucket are collected at the receiving ends. Compared with previous encoding–decoding methods, this method reduces the redundancy significantly since it does not need overlaps for sequencing assembly. This method can achieve a higher storage capacity of 1.57 bit/nt: the 0–1 binary data are first divided into many data packets like water droplets in a fountain, followed by randomly choosing a pseudo-random seed as the packet degree from the robust soliton distribution. The selected packets are then put together by exclusive-or operation. In the end, a 4-byte seed is attached to a 32-byte bitwise addition to generate a droplet. Here the DNAs with homopolymers were avoided to lower systematic errors during DNA storage. The DNA sequence structure (200 nt) includes a 128 nt DNA fragment for data payload, a 16 nt DNA fragment for pseudo-random seed, 8 nt DNA fragment for Reed–Solomon code, and one adapter (24 nt) at each end for sequencing (Fig. 10).
DNA fountain code (Erlich and Zielinski 2017). a DNA Fountain encoding method used in the DNA storage of a file of 32 bits. b The sequence structure of fountain code encoding method
Leon Anavy et al. improved DNA fountain encoding method to achieve higher logical density by using compound DNA alphabet, which could convert binary code to q-ary rather than quaternary (Anavy et al. 2019). The compound DNA alphabet consists of different proportions of A, C, G, T letters. For example, a six-letter alphabet can be denoted as {A, C, G, T, M, K}, in which M is a mixture of A and C (1:1), and K is a mixture of G and T (1:1) (Fig. 11). Through this method, they achieved 2.15 MB data storage by 152 nt oligos with 58,000 six-letter composites. Compared with Yaniv Erlich's fountain encoding method using 72,000 152 nt oligos, a ~ 24% increase in logical density was achieved by this improved method. However, considering synthesizing and sequencing costs, the length of the DNA compound alphabet is not unlimited since the larger size of the alphabet needs deeper sequencing depth to recover the data information. Through the simulation calculation of the resolution k, this system with 56 letters can achieve about 52% overall cost reduction compared with the quaternary system using k = 5.
Improved DNA fountain code using composite DNA letters (Anavy et al. 2019)
2.6 CRISPR mediated in vivo DNA storage technology
CRISPR is widely used for gene editing (Adli 2018). A Harvard team and a Columbia University team separately adapted this method for information storage in living bacteria. In 2016, the Church research group built the first CRISPR-based molecular recorder on the world (Shipman et al. 2016). Next year, they successfully achieved digital information encoding and storage of a short GIF movie (2.6 kb) with more than 90% accuracy in living bacterial genome according to chronological order, through capturing ability for foreign DNAs of CRISPR system (Figs. 12, 13) (Shipman et al. 2017). They first converted the digital information of a GIF movie into DNA codes and integrated the DNA codes with PAM sequences into the interval sequences. Then they provided a group of bacteria with a set of interval sequences, which were designed for sequential frames arranged in chronological order. Through overexpressing Cas1/Cas2, the set of interval sequences were added to the CRISPR array in their genomes. In the end, the data information of the GIF movie could be recovered by DNA sequencing data.
Flowchart showing CRISPR-based DNA storage technology (Shipman et al. 2017)
Based on Church’s CRISPR technology, in 2021, Wang’s team at Columbia University described a new electrical generation framework for directly storing digital data (72 bits) in living bacteria (Fig. 14) (Yim et al. 2021). Through electrical stimulation, a 3-bit binary code was encoded into the CRISPR-harboring bacterial arrays with multiplexed barcodes by electrical stimulation in a CRISPR-based DNA recorder. Here, the stored information in bacteria populations can be retained for many generations in a natural open environment. This work provides the potential for DNA data storage in vivo through direct communication in living cells, further establishing the data storage framework of digital biology of exchanged information between silicon and carbon-based entities.
A CRISPR-based DNA recorder using electronic signals (Yim et al. 2021)
3 Conclusion
This paper reviews the research advances of encoding and decoding methods in the field of DNA storage, covering several primary encoding and decoding methods (Table 1): direct mapping between 0–1 binary digital data and A-T-C-G quaternary DNA storage data in the early stages (Church, Blawat, Goldman and Bornholt labs), fountain code (Erlich & Zielinski and Anavy labs), inner and outer codes for random access DNA storage data (Grass and Organick labs), and CRISPR mediated in vivo DNA storage technology (developed by Church et al. 2012).
Each encoding and decoding method has its advantages and disadvantages. Here, the direct mapping method is easy and efficient but has a high error rate and low logical density. Fountain code made a breakthrough for logical density, reaching up to 80% of the upper limit of theoretical estimation of DNA storage, but it could not achieve random access. Grass's inner and outer code has an error-correction design (at the chain and data block levels) to realize random access at the expense of logic density. In all, the above three encoding/decoding methods belong to in vitro DNA storage, with the advantage of high-throughput storage and lower cost. CRISPR mediated DNA storage technology belongs to in vivo DNA storage, with the advantage of long-term stable storage and cheaper/random amplification of DNA storage data copies, but it is nowadays in the early exploration stage only for a small amount of data storage due to some insurmountable technical bottlenecks (such as biochemical restrictions in vivo).
At present, the existing data storage technologies are increasingly unable to meet the requirements of the explosive growth of data (Léquepeys et al. 2021). Due to many abovementioned advantages and achievements, DNA storage is expected to develop into a major data storage form in the future. Among them, the development of efficient and accurate DNA storage encoding and decoding methods and related error-correction algorithms will play a very important and even decisive role in the transition from the laboratory to practical application, which may fundamentally change the information industry in the future.
References
Adli, M.: The CRISPR tool kit for genome editing and beyond. Nat. Commun. 9(1), 1911 (2018). https://doi.org/10.1038/s41467-018-04252-2
Anavy, L., Vaknin, I., Atar, O., Amit, R., Yakhini, Z.: Data storage in DNA with fewer synthesis cycles using composite DNA letters. Nat. Biotechnol. 37(10), 1229–1236 (2019). https://doi.org/10.1038/s41587-019-0240-x
Blawat, M., Gaedke, K., Hütter, I., Chen, X.-M., Turczyk, B., Inverso, S., Pruitt, B.W., Church, G.M.: Forward error correction for DNA data storage. Proc Comput Sci 80, 1011–1022 (2016). https://doi.org/10.1016/j.procs.2016.05.398
Bornholt, J., Lopez, R., Carmean, D.M., Ceze, L., Seelig, G., Strauss, K.: A DNA-based archival storage system. SIGARCH ComputArch News 44, 637–649 (2016). https://doi.org/10.1145/2872362.2872397
Church, G.M., Gao, Y., Kosuri, S.: Next-generation digital information storage in DNA. Science 337(6102), 1628 (2012). https://doi.org/10.1126/science.1226355
Clelland, C.T., Risca, V., Bancroft, C.: Hiding messages in DNA microdots. Nature 399(6736), 533–534 (1999). https://doi.org/10.1038/21092
De Silva, P.Y., Ganegoda, G.U.: New trends of digital data storage in DNA. Biomed. Res. Int. (2016). https://doi.org/10.1155/2016/8072463
Erlich, Y., Zielinski, D.: DNA Fountain enables a robust and efficient storage architecture. Science 355(6328), 950–954 (2017). https://doi.org/10.1126/science.aaj2038
Goldman, N., Bertone, P., Chen, S., Dessimoz, C., LeProust, E.M., Sipos, B., Birney, E.: Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494(7435), 77–80 (2013). https://doi.org/10.1038/nature11875
Grass, R.N., Heckel, R., Puddu, M., Paunescu, D., Stark, W.J.: Robust chemical preservation of digital information on DNA in silica with error-correcting codes. Angew. Chem. Int. Ed. Engl. 54(8), 2552–2555 (2015). https://doi.org/10.1002/anie.201411378
Kumar, K.R., Cowley, M.J., Davis, R.L.: Next-generation sequencing and emerging technologies. Semin. Thromb. Hemost. 45(7), 661–673 (2019). https://doi.org/10.1055/s-0039-1688446
Léquepeys J-R, Duranton M, Bonnetier S, Catrou S, Fournel R, Ernst T, Hérault L, Louis D, Jerraya A, Valentian A (2021) Overcoming the data deluge challenges with greener electronics. ESSCIRC 2021-IEEE 47th European Solid State Circuits Conference (ESSCIRC), pp 7–14
Liu, L., Li, Y., Li, S., Hu, N., He, Y., Pong, R., Lin, D., Lu, L., Law, M.: Comparison of next-generation sequencing systems. J. Biomed. Biotechnol. (2012). https://doi.org/10.1155/2012/251364
Microsemi Corporate Headquarters: White paper: hardware RAID vs. software RAID: Which implementation is best for my application? MRCIOCHIP. http://ww1.microchip.com/downloads/en/DeviceDoc/Hardware_RAID_vs_Software_RAID__Which_Implementation_is_Best_for_my_Application_Whitepaper.pdf. Accessed Oct 2017
Organick, L., Ang, S.D., Chen, Y.-J., Lopez, R., Yekhanin, S., Makarychev, K., Racz, M.Z., Kamath, G., Gopalan, P., Nguyen, B., Takahashi, C.N., Newman, S., Parker, H.-Y., Rashtchian, C., Stewart, K., Gupta, G., Carlson, R., Mulligan, J., Carmean, D., Seelig, G., Ceze, L., Strauss, K.: Random access in large-scale DNA data storage. Nat. Biotechnol. 36(3), 242–248 (2018). https://doi.org/10.1038/nbt.4079
Shipman, S.L., Nivala, J., Macklis, J.D., Church, G.M.: Molecular recordings by directed CRISPR spacer acquisition. Science 353(6298), aaf1175 (2016). https://doi.org/10.1126/science.aaf1175
Shipman, S.L., Nivala, J., Macklis, J.D., Church, G.M.: CRISPR-Cas encoding of a digital movie into the genomes of a population of living bacteria. Nature 547(7663), 345–349 (2017). https://doi.org/10.1038/nature23017
The DNA Data Storage Alliance: Preserving our digital legacy: an introduction to DNA data storage. DNA DATA ALLIANCE (2017). https://dnastoragealliance.org. Accessed June 2021
Yim, S.S., McBee, R.M., Song, A.M., Huang, Y., Sheth, R.U., Wang, H.H.: Robust direct digital-to-biological data storage in living cells. Nat. Chem. Biol. 17(3), 246–253 (2021). https://doi.org/10.1038/s41589-020-00711-4
Acknowledgements
This work was supported by the National key research and development program (Grant No. 2020YFA0907000), and the National Natural Science Foundation of China (Grant No. 31971389).
Author information
Authors and Affiliations
Author notes
Chenyang Wang and Guannan Ma contributed equally to this work.
Contributions
CW: searching the literature, writing—original draft. GM: writing—original draft, searching the literature. DW: searching the literature. XZ: searching the literature. PW: writing—original draft. CL: writing—original draft. JX: critically revising. ZW: critically revising. BD: CRITICALLY revising. DY: critically revising. PW: searching the literature. DB: critically revising, Writing—review & editing. FC: conceptualization, critically revising, Writing—review & editing. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, C., Ma, G., Wei, D. et al. Mainstream encoding–decoding methods of DNA data storage. CCF Trans. HPC 4, 23–33 (2022). https://doi.org/10.1007/s42514-022-00094-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42514-022-00094-z