Abstract
In this study, we propose a hybrid approach involving feature extraction, feature selection, and optimized learning for the diagnosis of multiple sclerosis (MS), which can detect the lesion caused by MS plaques in the brain using magnetic resonance imaging analysis. A major challenge associated with lesion diagnosis by neurologists is that it is a time-consuming process and demands high expertise; therefore, researchers have been stimulated to find an auto-diagnose method of the disease. Given the high resemblance of MS plaque-induced lesions and other lesions such as Alzheimer’s or dementia, scant research has explored the diagnosis of MS-induced lesions, most of which suffering from the lack of an efficient and accurate method. Informed by the need for a precise hybrid model for the classification of MS plaques and other comparable lesions, a solution is proposed that utilizes an efficient model. In this method, after image preprocessing, the feature vector is created by applying fractal and Pseudo-Zernike Moments descriptors. Feature selection using the Difference Evolution) algorithm to select the minimum subset of features will reduce the number of Extreme Learning Machine (ELM) inputs for classification. To improve the classification effect, the ELM wavelet kernel parameters are also regulated by the Shuffled Frog-Leaping Algorithm. By applying the proposed model to a set of brain MR images obtained from healthy subjects and MS patients during different experimental iterations, an average accuracy of 97% was obtained. The results of the method were estimated under specific conditions, and finally the proposed model yielded desirable outputs compared to similar methods.
Similar content being viewed by others
1 Introduction
Multiple Sclerosis is an inflammatory disease in which myelin sheaths of the nerve cells in the brain and spinal cord are damaged [1,2,3,4,5,6]. This damage can disrupt the ability of parts of the nervous system which are responsible for communication and causes many signs and symptoms such as physical problems [7]. Regional estimates suggest that this disease has a moderate prevalence in countries and is in prevalence range of the European and Far East countries [8]. Symptoms of MS disease appear in several forms and its new symptoms occur either as step recurrence -multiple disease reversal or alternately over time [9,10,11]. Initially, recovery from attacks is almost complete, but slowly, neuropsychiatric disabilities with different degrees will remain from each attack [12]. MRI is the most practical method for detecting the masses left in the brain, which can greatly help the specialist. Sometimes there are different interpretations of MR brain images and so it is necessary to have an accurate statistical analysis of alternating signals. Sometimes due to the inherent nature of MRI, it is not possible to distinguish lesions caused by diseases such as Alzheimer’s, MS, and other common diseases in the brain. If it is possible to detect lesions from this disease at early stages, then it will be possible to treat the disease.
The discovery of lesions by the neurologist requires expedition, time and high accuracy and typically, with increasing number of MR images of the brain, the efficiency required for the diagnosis will be significantly reduced. The inadequacies of contrast and image clarity, as well as the similarity of lesions caused by the disease to other brain tissues, lead to different interpretations of brain MRI images; therefore, a precise analysis of the resulting images is required. Thus, employing a robust automated approach can provide satisfactory outcomes, whereas additionally dominant the time needed for diagnosis. The major contributions of MR processing work lie in applying the feature extraction and classification to detect the lesions and non-lesions as shown in Fig. 1.
Several strategies have been proposed to date in order to automate the intelligent brain disease diagnosis, especially MS. Among the studies in the field of diagnosis, separation and classification of lesions caused by MS disease, Ballin et al. [13] mapped a three-dimensional lesion of the MS onto the MR image in addition to conduct classification by combination methods. Zhang et al. [14] also used tissue analysis in order to diagnose MS by extracting the features and then selecting the best ones in brain MR images. The best feature analysis had been used by linear methods and statistical analysis and the diagnostic accuracy of this disease in MR images can reach up to 88% by choosing the intelligent tissue analysis method.
Roy et al. [15] provided an automated method using tissue features and support vector machines, in which the brain lesion separation process was performed and then the classification occurred. The strong point of their work was to use the cumulative distribution function and also normalization of the area of interest to improve the image quality. The texture features, local brightness, and initial spatial information were used as the main features and in fact, the classification process occurred for image pixels, not for the image itself.
Elliott et al. [16] proposed the consistent probabilistic detection for new MS lesions in MR images. They used a series of sequential MR scans. Their work was a two-step classification; first, they used a classifier to find out the possibility of a voxel for being lesion. Cabezas et al. [17] suggested conducting the classification mechanism through the boosting classification technique in order to classify the classification for a set of features. They prepared MR images from 45 subjects during the three recording periods. This set of techniques has led to a typical, precise separation and as a result, parts of the MR image with the potential of disease are revealed.
Sweeney et al. [18] compared different methods of machine learning to build the best feature vector and they did this using a multi-quality structure in MR images. This resulted in a fairly precise separation of the images and the separation of lesions caused by MS disease. Ardakani et al. [19] investigated MS diagnosis through tissue analysis. They extracted features from the images for 50 patients through the analysis of MR images, initially by analyzing the main components. Then, they perform classification by relying on the linear separation method and obtained the sensitivity of 100% and the surface below the ROC curve had the value of 1 in their work. Separating a favorite area and classifying the three classes was one of the innovations of their work.
Liu et al. [20] diagnosed MS by removing confounding factors that did not include information on the lesion area and they did this using limited clustering. Weygandt et al. [21] relied on the pursuit of biomarkers based on segmentations such as thresholding and combining atlas images and brain MR to find the location of brain lesions based on which, the early stages of the formation of these lesions were possible from the images. Karimaghaloo et al. [22] proposed a system based on randomized conditional stratification and classification to separate the areas suspected to MS-related lesions, which distinguished small pieces which were considered as lesion.
Brosch [23] used three-dimensional encoded deep convolutional neural networks to detect lesions caused by MS disease. Zhang et al. [24] relied on methods such as static wavelet and entropy, and statistical characteristics. They suggested using the decision tree, k-NN classifier, and supporting vector machines for the classification process. They then achieved the accuracy of 97.9% for their data sets, and sensitivity and specificity values were higher than 95%. The use of two-dimensional separation and 3D mapping and final detection has been proposed through deep convolutional neural networks [25].
Recently, Gheshlaghi et al. [26] proposed a super-pixel segmentation based technique for multiple sclerosis lesion detection. Their study includes SVM as an effective classifier with polynomial kernels that is applied to have better performance in distinguishing specified decision classes. Also, they use discrete wavelet decomposition (DWT) which extracts local features from analyzing MR images.
Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach have been conducted by Valverde et al. [27]. They propose an automated White Matter (WM) lesion segmentation method for MS patient images and their approach relies on a cascade of two 7-layer convolutional neural networks. The comparison of the previous approaches over the last few years has been shown in Table 1.
In this study, different lesions of the MR images are segmented by using the clustering method, and then, features of segmented areas will be extracted. There are some objectives in the research, including the following: (1) Detection of MS lesions from magnetic resonance images in two classes of no disease and disease incidence; (2) Promoting the feature vector structure by aggregating results from descriptors such as Fractal and PZM. Additionally, we focus to demonstrate the following hypotheses: (1) Compatible extracted features based on efficient descriptors decrease the time and space complexity for building the prediction model. Also, feature subset selection by an evolutionary-differential algorithm improves the prediction accuracy of the ELM classifier and reduces the false prediction ratio. (2) Appropriate ELM configuration parameters, such as the number of neurons in hidden layer and the training error optimized by the memetic meta-heuristic SFLA algorithm. The performance of proposed methods is analyzed with the focus on dimensionality reduction of extracted features and improvement of classification accuracy using MR images.
The rest of this paper is organized as follows: Sect. 2 describes the proposed method with the different sections of the algorithm. Section 3, considers the implementation of the algorithm and the experimental results. Finally, the conclusion of the paper will be presented in Sect. 4.
2 Materials and methods
Separation in image processing is an approach which is conceptually used in the image segmentation process. We first described the MRI data used for the evaluation of the proposed segmentation and classification approach. Then, the different steps of the proposed classification process were detailed. The trend of the proposed stages could be summarized according to the following Fig. 2.
2.1 Pre-processing
In the preprocessing step, two steps are implemented on the basis of slices received from a person, which are Histogram equalization and segmentation by csFCM.
2.1.1 Histogram equalization
Retinex multi-scale transformation method was used for histogram equalization, which is one of the optimization methods, which also has acceptable results in the preprocessing. In the desired transform, first, the image brightness pattern is calculated based on the weighted neighboring mean in the specified radius for each pixel. The Retinex multi-scale transformation is expressed according to Eq. (1) [37]:
where I is the input image received from MRI devices and W is the weight of the matrix. Neighborhood weights (NW) are calculated for each pixel (icent, jcent) based on the neighborhood of pixels and in accordance with Eq. (2) [37]:
The cent refers to the pixel coordinates where we want to calculate their brightness and the Radius term is the default radius for the neighborhood, where different values can be adopted. Finally, the final image is calculated by difference of the brightness pattern from the original image in the logarithmic range. In other words, R and L are respectively the components of the reflection and the component of the image brightness:
2.1.2 csFCM
The conditional spatial fuzzy C-means (csFCM) method is derived from the clustering technique followed by the segmentation of magnetic resonance images [38] which can be used for noise images and provides good results. The process of the csFCM algorithm has been displayed in the Region of Interest (ROI) segmentation in Algorithm 1.
In the set of equations of Algorithm 1, xk is the equivalent of the pixel k of the magnetic resonance image. The conditional spatial FCM algorithm with parameter p and q is denoted as csFCMp,q. Again, it may be noted that csFCM1,0 is identical to the conventional FCM algorithm.
2.2 Feature extraction and selection
In the process of extracting and selecting features from the pre-processing images, the feature vector is constructed from the combination of features extracted by the Pseudo-Zernike moments and fractal algorithms and, then the modified DE algorithm reduces the extracted vector dimension.
2.2.1 Pseudo-Zernike moments (PZm)
Pseudo-Zernike moments (PZMs) are a set of complex polynomials which form an orthogonal set within a unit circle (x2 + y2 = 1) which is represented by Vnm(x, y) and its structure is defined as PZMs Eq. (4) [39]:

Where j = (− 1)0.5, θ = tan−1(yx−1), |ρ| ≤ 1, n ≥ 0, m ≤ n, n − |m| = even, and R is a radial polynomial and it should also be noted that ρ is the source vector length to the point (x, y) and θ is the angle between the vector ρ and the axis x in the direction of counterclockwise. In the above relation n is a non-negative integer that shows the polynomial order. M is a negative or positive integer that shows the order of the horizontal arc repetition and its absolute value is always smaller or equal to n, and the difference m of n is always an even value. In the above relation, Rnm are radial polynomials calculated in accordance with relationships (5) and (6) [39]:
Zernike moments (ZMs) are a mapping of the image into a set of Zernike complex polynomials. One of the important features of Zernike moments is their orthogonally; for this reason, image features can be displayed without any redundancy in information or overlap between moments. If the rotation is made in the images, it will not have any effect on its Zernike moments. The complex Zernike moments of the order n and with the repetition of m are calculated using Eq. (7) [39]:
where, f (x, y) is the brightness intensity function of a digital image at the location of x and y and the * sign also denotes the complex conjugate.
2.2.2 Fractal
A fractal descriptor, known as one of the tissue descriptors of the image, is implemented in two stages based on segmentation [40,41,42]; first, the gray level image is divided into a group of images with binary values, and this process is performed by the bi-threshold binary decomposition algorithm. The bi-threshold binary decomposition algorithm considers the image I(x, y) as an input and retrieves binary images. By bi-threshold binary decomposition, a pair of thresholds is selected from T as a result; the segmentation is carried out in accordance Eq. (8) [40]:
where tl and tu respectively represent the minimum and maximum threshold values. By applying Eq. (8), the binary images are decomposed and then proceeded from T U {nl}. On the one hand, all threshold pairs of {t, nl}, t ∈ T can be achieved in which, nl represent the maximum possible gray level in I (x, y). So the number of binary images is 2nt and the value of nt is set to 8 value. After applying the bi-threshold binary decomposition, the fractal tissue analysis feature vector is constructed according to the segmentation based on the size of the binary images, the mean gray level and the fractal dimensions of the boundaries. The boundaries of the image regions Ib (x, y) are selected as marginal segments, which are represented by Δ(x, y) and calculated as Eq. (9) [40]:
where NB[(x, y))] represents a group of pixels with 8 connections to (x, y). Also, Δ(x, y) will have a value of 1 if the pixel is at the position (x, y), which refers to the binary image Ib and, if there is at least one neighborhood with a pixel with zero has the sum of 1. Figure 3 indicates the implementation stages of fractal algorithm. In this figure, area (A1, A2,…, An) and mean gray level (V1, V2,…, Vn) features are computed directly from the binary image. Fractal dimension (D1, D2,…, Dn) features is computed from the border or margin image. In these dimension, we select n equal 8 because this would result in a lower error rate.
2.2.3 Differential Evolution (DE) algorithm
We conduct the feature selection with a modified Differential Evolution (DE) algorithm. In the feature selection problem, the most effective subset features is chosen which has a better solution. The general evolutionary-differential algorithm has been shown in Fig. 4.
In order to increase the speed and at the same time maintain the heuristic nature of the algorithm and prevent early convergence, in both Cross Over and Mutation vectors, we have established a balance between local and global search, where random and uniform numerical definitions in the range of zero to one can be used. Additionally, the use of additional parameters in the control of the algorithm is eliminated and therefore, adjusting the DE algorithm in the feature selection is greatly optimized and finally we do not eliminate the automatic and random mode of the algorithm. In the proposed DE algorithm, the mutation vector controls the mutation effect in adaptive form using the feedback and thus, a kind of self-adjustment will increase the accuracy and speed of the algorithm. In other words, when the average fitness approaches the best fitness, it indicates that the algorithm reaches the best final value. At this time, the effect of the mutation should be reduced so that the algorithm does not go away from the final solution. Conversely, when the difference between the average fitness and the best fitness is high, it is necessary to map the problem space algorithm to increase the effect of the mutation.
An ELM based on wavelet kernel is used to calculate the feature selection error and in the fitness function, the initial train data is divided into new train data and validation data with the K-fold cross validation method, with K equal to 5 and, the optimal subset is selected after a limited number of repetitions. Therefore, the error resulting from the classification of the validation data along with the minimum number of features will be the output of the cost function.
2.3 Evolutionary extreme learning machine
The Extreme Learning Machine (ELM) is a configuration with a hidden layer where the weights between the first layer and the random hidden layer are updated and, the weights of its second layer are weighted normally similar to the performance of the nervous system. ELM outputs with m neurons and the activation function f can be displayed as follows:
The algorithm is fast but may offer a better overall performance [43,44,45]. Learning error and output weights should be minimized simultaneously to reduce learning errors in the ELM algorithm. Therefore, the overall performance of the neural networks will increase:
where, it can be written that:
where, E is the adjustment coefficient, A is the output matrix of the hidden layer, and C is the expected output matrix of the samples. Therefore, the output function of the ELM algorithm can be written as Eq. (13):
If the feature vector v(r) is unspecified, the ELM kernel matrix can be rewritten based on the Mercer conditions:
where, u(r) is the output function of the wavelet kernel for ELM, which can be represented as Eq. (15):
where, D = AAT and b(r, g) is the kernel function ELM. In this case, there are some kernel functions including linear kernel, Polynomial kernel, Gaussian kernel, and Exponential kernel. But the Wavelet kernel function is beneficial for simulations and performance [14]:
Determining these parameters will greatly affect the desired accuracy. To find the best structure of ELM, we suggest that the Shuffled Frog-Leaping Algorithm (SFLA) algorithm perform the optimization. The steps of SFLA to find best parameters of wavelet kernel in ELM classifier are as follows:
-
A.
Global search
In SFLA, the combination increases the quality of the memetics affected by different subgroups. Global and Local searches continue to meet the convergence condition. The balance between global transfer and local search allows the algorithm to leap easily from the local minimum and extend until reaching optimization. One of the features of the SFLA algorithm is fast convergence. Global search to find best parameters is conducted based on Algorithm 2.
Algorithm 2. Global search by SFLA
Step 1) | Initialization: select m and n, m is the number of memeplexes, and n is the number of frogs per memeplex, thus the total population in the pool is obtained through the relation F = m.n. |
Step 2) | Virtual population generation: F virtual frogs (U(1)), U(2),…, U(F)) are sampled from the possible space. The value of fitness f(i) of each frog U(i) is calculated for each U(i)=(U1i, U2i,…, Udi) in which d is the number of decision variables. |
Step 3) | Frog Ranking: Frogs are sorted according to their fitness in descending order and stored in the array X = {U (i), f (i), i = 1,…, F}. The best frog position of PX is selected in the entire population (Px = U(1)) |
Step 4) | Partitioning frogs into mempelexes: The array X is divided into m memeplex (Y1, Y2,…, Ym), each containing n frogs. |
Step 5) | Memetic evolution in each memeplex: each memeplex (Yk, k = 1,…, m) evolves by local search. |
Step 6) | Shuffling memeplexes: After a certain number of memetic evolutions are completed in each memeplex, memeplexes (Y1,…, Ym) are placed in X so that the relation X = (Yk, k = 1,…, m) is established. The best frog position in the PX population is updated. If convergence conditions are met, it stops. Otherwise it goes to the fourth stage of global search. |
-
B.
Local search
In the fifth step of global search, the evolution of each memeplex is performed for n times independently. After the memeplex evolved, the algorithm returns to the global search for the shuffling. The details of local search are described in each memeplex. Weights are assigned with triangular probability distribution according to Eq. (17).
where, j is the index of j-th member and n is the number of elements. To construct submemeplex array, q frog is randomly selected from each n frogs in each memeplex. The frogs in the submemeplex are arranged in descending order in terms of their fitness. The positions of the best frog and the worst frog in the submemeplex are identified by PB and Pw, respectively. The new position of the worst frog in the submemeplex that is the frog with the worst performance and then is calculated through Eq. (18). Also, S is the step size or mutation rate of the frog and is calculated as Eq. (19):
Also, we can conduct the local search based on Algorithm 3.
Algorithm 3. Local search by SFLA
Step 1) | Initialization: Parameters im and in get the initial value of zero. In this concept, im counts the number of memeplexs and in counts the number of evolution stages. |
Step 2) | Generating submemeplexes: The goal of the frogs is to move optimal positions by improving their memeplexes. The submemeplex selection method is to allocate higher weights to frogs with higher performance and lower weights to the frogs with lower performance. Weights are assigned with triangular probability distribution according to Eq. (17). |
Step 3) | Correcting the position of the worst frog based on Eq (18). If the new position is better than the previous position, then replace the new U(q) with the previous U(q) and goes to step 6 of the local search. Otherwise, go to step 4 of the local search. |
Step 4) | S or step size of the frog and is calculated by Eq. (19). If U(q) is within the possible space, the new performance value f(q) is calculated. If the new f(q) is better than the previous one, then U(q) replaces the previous U(q) and goes to step 6 of the local search. Otherwise, it goes to step 5 of the local search. |
Step 5) | Calculating step size by Px: If better result is not obtained in the step 3, then after calculating the step size of the frog, the new position U(q) is calculated by (19). |
Step 6) | Censorship: If the new position is not in the possible area or not better than the previous position, a new frog (r) will randomly be generated in a possible position and replaces by a frog whose new position is not suitable for progress. f(r) is calculated and U(q) is set to r and f(q) equal to f(r)). |
Step 7) | Upgrading memeplex: After changing the memetic of the worst frog in the submemeplex, the frogs in Z are placed in their original position on YIm. YIm is sorted by descending order in terms of performance. If In < n, it goes to step 3 of the local search. If Im < m, go to step 1 of the local search. Otherwise, it will return to global search for shuffling the memeplexes. |
3 Experimental results
3.1 Data
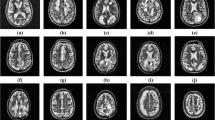
The main data, including slices selected by a specialist − 10 to 15 important slices have been separated from each person from a variety of people with different levels of MS disease which was collected during the course of a year and a half from the Vasei Hospital in Sabzevar, Iran from 2016 to 2018. The sample of MRI selected from MS patients has been shown in Fig. 5. Healthy individuals are also included in this research to be considered as control or study variables. Patients are divided into two groups with and without the history of pain inflammation in the head region and various magnetic resonance imaging centers provided reports on imaging. People with MS disease with different stages of the disease have long been monitored by a doctor and the definitive symptoms of the disease have been listed. Sampling was done randomly according to the opinion of the specialist. The personal characteristics of the patients after imaging were recorded in the questionnaires or along with two DICOMDIR and Radiant setup software and complete neurological and visual examination has been performed on patients. Data gathering research was a two-group cross-sectional analytical study.
The sample of the study including 125 subjects (at least 10 slices according to physician diagnosis) in patient subjects, definitive diagnosis of disease and age below 50 years old referring to the MS clinic, neurology ward and, patients referred to the magnetic resonance imaging center of Vasei Hospital in Sabzevar, in two groups 64 people and 61 people were examined. 64 patients were diagnosed with MS and 61 subjects were healthy without any complications. The two experts have definitely commented on the disease and even estimated its possible location. FLAIR images and often T1 and T2 modes have been studied.
3.1.1 Evaluations
Three factors of accuracy, sensitivity and specificity, which have been introduced to measure the diagnosis accuracy of the proposed system, are calculated in accordance with Eqs. (21) to (23) and used to evaluate the system:
where NTP is the number of brain MRI images containing MS-induced lesion, and the proposed algorithm has correctly diagnosed the disease. In addition, NTN indicates the number of brain MRI images not induced by MS lesions and the proposed algorithm has correctly detected the absence of disease. NFP is the number of brain MRI images that did not contain any MS-induced lesions, but the proposed algorithm had erroneously diagnosed the disease. Finally, NFN indicates the number of brain MRI images that contained MS-caused lesion, but the proposed algorithm had erroneously ruled out the disease.
3.2 Model initializing
Because the number of areas suspected to MS lesion may vary in the preprocessing stage that is, cases that resemble lesions resulting from Alzheimer’s disease, tumor or pathologic damage, or masses other than the target class, the experiment was repeated for 10 times. Thus, all images with an initial dimension after the pre-processing are imported to the optimized decision maker network as input.
The parameters of the DE algorithm in feature selection are based on the cost function due to the classification using the ELM (wavelet kernel) neural network. Table 2 represents the parameters of the DE algorithm in order to select the best features.
3.3 Results
The proposed algorithm was implemented in Matlab and the required system for data processing included Due CPU 2.6 GHz with a RAM of 4 GB and performance of the algorithm was between 10 and 20 s in short term. The change in accuracy is due to the resizing of the image that, in the current research it has been tried to consider three different models of image resizing. As mentioned, the first-stage benchmarks have been represented by the K-fold validation method in Tables 3, 4 and 5 for dimensional change. Each table consists of train and test stage, and accuracy varies between 90% and 98% due to the changes in dimensions. Also Figs. 6, 7, 8 and 9 indicate an error reduction and convergence due to the feature selection and kernel parameters regularization from all features as one-third of the total features of the model.
4 Discussion
Experiments were carried out in the feature selection phase, in which the appropriate subset selection was made based on which selection of subsets including 10%, 15%, 20%, 30%, 40%, 50%, 60%, 65%, 70%, 80%, 90%, and 100% of the total features, and finally all the features are included in the calculation and in Table 6, the accuracy level changes in 10 repetitions (the 5-fold final output) are observed. In the feature selection step, two 5-fold cross validation stages were used in the cost function for generating three train, validation, and test data.
Figure 10 shows a comparison between features selection approaches that output based on the lowest error rate with 5-fold cross validation and it performed with other methods such as Genetic algorithm (GA), Particle Swarm Optimization (PSO) and Ant Colony Optimization (ACO) and the most optimal features have been computed in comparison with the DE method of this study in four repetitions. DE performance is expressed based on Kappa coefficient by Eq. (23) in which was more appropriate than other solutions in feature selection level.
where, N is the total number of samples, r is the number of classes, xii is the main diagonal of adjacency matrix of error, xi+ is the marginal sum of the rows and x+i is the marginal sum of the columns.
Using other evolutionary algorithms such as PSO, ACO, GA, instead of the SFLA algorithm in optimizing the wavelet kernel of the ELM and its training did not result in less errors. The main reason for this is the ability of the SFLA to find the global and local optimum and persistence to achieve the optimal solution. Additionally, the processing time of the SFLA algorithm is more appropriate than other solutions and the advantage of not having much parameters and simplicity in implementation can be considered as the main reason for choosing an algorithm to improve ELM. Hence, in Fig. 10, SFLA optimizer performance has also been compared to improve ELM performance with other evolutionary optimizers such as PSO, ACO and GA and, the error in any type of data due to lower optimality has been reported.
The overall performance of the ELM algorithm has been shown after the improvement by the SFLA in the set of Fig. 11, in which the error converges to the minimum values. Figure 12 also compared ELM classifier with linear, RBF, polynomial, and wavelet kernels as decision makers for each class of sample slices, with and without MS and, the final result indicates the proper performance of this wavelet kernel in distinguishing healthy individuals and MS patients. The ELM classifier with different kernels is optimized by SFLA method.
The use of fractal and PZMs descriptors to extract feature yielded optimal responses. The primary reason for employing the proposed methods in feature extraction is its high accuracy in the analysis of MR images, in addition to its novelty and limited application in MR image analysis. Second, the set of images was analyzed by identical methods including GLCM, LBP, and HOG, which were compared with the proposed method in terms of feature extraction. Figure 13 shows that the calculation of performance accuracy at different test data by fractal and PZMs aggregation can produce responses that are more desirable than that of conventional methods such as GLCM, LBP and HOG. The third reason is that due to the nature of these descriptors, as explained in [39, 46,47,48], they are capable of extracting features even in small-scaled or rotated image or where the mass has been altered [47]. The fractal descriptor yielded high accuracy in Lahmiri’s research for the detection of Alzheimer’s lesions. It has been shown that fractal is effectively correlated with human perception of surface roughness.
In addition to the comparison of descriptors, the repeatability of the algorithm was tested in four random datasets by calculating Accuracy, Sensitivity, and Specificity factors. Figure 14 shows the algorithm’s repeatability with the minimum dispersion in the outcomes.
The algorithm performance was evaluated by calculating the evaluation factors and calculating their mean values, in such a way that:
-
(A)
Processing model: Information processing was performed on the basis of the comparison between the solutions of the magnetic resonance image analysis with the proper accuracy.
-
(B)
Repetition: The variance in responses is low and un-modeled uncertainty is solvable.
-
(C)
Processing time: The time reduction rate was lower during the design phase and evaluation. However, the data processing time is half the overall time, which can be important in saving time.
Consisting of descriptions of features based on fractal and PZM methods, feature selection with DE algorithm and wavelet kernel optimization by SFLA, the ELM performance is better than other similar classifiers. In overall, we achieve accuracy higher than 97% at train and test stage via dividing the data by K-Fold cross-validation.
5 Conclusion
Considering the nature of magnetic resonance image of brain, cancerous masses, brain lesions and their areas, in this paper, a method has been proposed on the basis of MS diagnosis based on the use of automatic processing technique. MS-related lesions have often abnormal position in MRI and because of compression in their slice, they exhibit a different brightness. Using csFCM, pixels containing probable MS lesions were separated from others. Feature extraction from the same separated pixels, the type and shape of the lesion by the two descriptors, makes it possible to create the proper feature vector of slices. The DE algorithm removed some of the data from the feature vector that, the DE algorithm function cost function was achieved by data segmentation through 5-fold cross validation on the basis of the calculation of ELM class error. The optimal ELM classifier by the SFLA has better capability for classification and the existing claims were proven. By repeating the test, there was a slight difference in the responses obtained which indicated un-modeled uncertainty. Undoubtedly, more improvements can be made in terms of learning and processing time and the error can be minimized. The suggestion is that a larger volume of slices of each subject will be introduced into the classifier by modeling methods such as deep learning.
References
Filippi M, Brück W, Chard D, Fazekas F, Geurts JJ, Enzinger C, Hametner S, Kuhlmann T, Preziosa P, Rovira À, Schmierer K (2019) Association between pathological and MRI findings in multiple sclerosis. Lancet Neurol 18(2):198–210
Eichinger P, Schön S, Pongratz V, Wiestler H, Zhang H, Bussas M, Hoshi MM, Kirschke J, Berthele A, Zimmer C, Hemmer B (2019) Accuracy of unenhanced MRI in the detection of new brain lesions in multiple sclerosis. Radiology 291(2):429–435
Schmierer K, McDowell A, Petrova N, Carassiti D, Thomas DL, Miquel ME (2018) Quantifying multiple sclerosis pathology in post mortem spinal cord using MRI. Neuroimage 15(182):251–258
Fechner A, Savatovsky J, El Methni J, Sadik JC, Gout O, Deschamps R, Gueguen A, Lecler A (2019) A 3T phase-sensitive inversion recovery MRI sequence improves detection of cervical spinal cord lesions and shows active lesions in patients with multiple sclerosis. Am J Neuroradiol 40(2):370–375
Silveira F, Sánchez F, Miguez J, Contartese L, Gómez A, Patrucco L, Cristiano E, Rojas JI (2020) New MRI lesions and topography at 6 months of treatment initiation and disease activity during follow up in relapsing remitting multiple sclerosis patients. Neurol Res 42(2):148–152
Rovira À, Wattjes MP, Tintoré M, Tur C, Yousry TA, Sormani MP, De Stefano N, Filippi M, Auger C, Rocca MA, Barkhof F (2015) Evidence-based guidelines: MAGNIMS consensus guidelines on the use of MRI in multiple sclerosis—clinical implementation in the diagnostic process. Nat Rev Neurol 11(8):471
Ysrraelit MC, Fiol MP, Gaitán MI, Correale J (2018) Quality of life assessment in multiple sclerosis: different perception between patients and neurologists. Front Neurol 11(8):729
Brundin L, Kobelt G, Berg J, Capsa D, Eriksson J (2017) European multiple sclerosis platform: new insights into the burden and costs of multiple sclerosis in Europe: Results for Sweden. Mult Scleros J 23(2):179–191
Khosravi M, Newberg A, Jahangiri P, Raynor W, Al-zaghal A, Werner T, Alavi A (2018) Innovative applications of combined PET/MRI modality in diagnosis and follow-up of Multiple Sclerosis. J Nucl Med 59(supplement 1):1220
Novakova L, Axelsson M, Khademi M, Zetterberg H, Blennow K, Malmeström C, Piehl F, Olsson T, Lycke J (2017) Cerebrospinal fluid biomarkers as a measure of disease activity and treatment efficacy in relapsing-remitting multiple sclerosis. J Neurochem 141(2):296–304
Thompson AJ, Banwell BL, Barkhof F, Carroll WM, Coetzee T, Comi G, Correale J, Fazekas F, Filippi M, Freedman MS, Fujihara K (2017) Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 1:1
Rezaee K, Azizi E, Ghezelbash MR, Madanian M, Haddania J (2012) A novel intelligent system to accurately segmentation of brain tumors in MR images by using image processing and discrete wavelet transform. Majlesi J Multimed Process 1(4):1
Akselrod-Ballin A, Galun M, Basri R, Brandt A, Gomori MJ, Filippi M, Valsasina P (2006) An integrated segmentation and classification approach applied to multiple sclerosis analysis. In: IEEE computer society conference on computer vision and pattern recognition, vol 1. IEEE, pp 1122–1129
Zhang J, Wang L, Tong L (2007) Feature reduction and texture classification in MRI-Texture analysis of multiple sclerosis. In: IEEE/ICME international conference on complex medical engineering, 2007, CME 2007. IEEE, pp 752–757
Roy PK, Bhuiyan A, Ramamohanarao K (2013) Automated segmentation of multiple sclerosis lesion in intensity enhanced flair MRI using texture features and support vector machine. In: IEEE international conference on image processing 2013. IEEE, pp 4277–4281
Elliott C, Arnold DL, Collins DL, Arbel T (2013) Temporally consistent probabilistic detection of new multiple sclerosis lesions in brain MRI. IEEE Trans Med Imaging 32(8):1490–1503
Cabezas M, Oliver A, Valverde S, Beltran B, Freixenet J, Vilanova JC, Ramió-Torrentà L, Rovira À, Lladó X (2014) BOOST: a supervised approach for multiple sclerosis lesion segmentation. J Neurosci Methods 30(237):108–117
Sweeney EM, Vogelstein JT, Cuzzocreo JL, Calabresi PA, Reich DS, Crainiceanu CM, Shinohara RT (2014) A comparison of supervised machine learning algorithms and feature vectors for MS lesion segmentation using multimodal structural MRI. PLoS One 9(4):e95753
Ardakani AA, Gharbali A, Saniei Y, Mosarrezaii A, Nazarbaghi S (2015) Application of texture analysis in diagnosis of multiple sclerosis by magnetic resonance imaging. Glob J Health Sci 7(6):68
Liu J, Brodley CE, Healy BC, Chitnis T (2015) Removing confounding factors via constraint-based clustering: an application to finding homogeneous groups of multiple sclerosis patients. Artif Intell Med 65(2):79–88
Weygandt M, Hummel HM, Schregel K, Ritter K, Allefeld C, Dommes E, Huppke P, Haynes J, Wuerfel J, Gärtner J (2015) MRI-based diagnostic biomarkers for early onset pediatric multiple sclerosis. NeuroImage Clin 7:400–408
Karimaghaloo Z, Rivaz H, Arnold DL, Collins DL, Arbel T (2015) Temporal hierarchical adaptive texture CRF for automatic detection of gadolinium-enhancing multiple sclerosis lesions in brain MRI. IEEE Trans Med Imaging 34(6):1227–1241
Brosch T, Tang LY, Yoo Y, Li DK, Traboulsee A, Tam R (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging 35(5):1229–1239
Zhang Y, Lu S, Zhou X, Yang M, Wu L, Liu B, Phillips P, Wang S (2016) Comparison of machine learning methods for stationary wavelet entropy-based multiple sclerosis detection: decision tree, k-nearest neighbors, and support vector machine. Simulation 92(9):861–871
Yoo Y, Tang LY, Brosch T, Li DK, Kolind S, Vavasour I, Rauscher A, MacKay AL, Traboulsee A, Tam RC (2018) Deep learning of joint myelin and T1w MRI features in normal-appearing brain tissue to distinguish between multiple sclerosis patients and healthy controls. NeuroImage Clin 17:169–178
Gheshlaghi SH, Ranjbar A, Suratgar AA, Menhaj MB, Faraji F (2019) A superpixel segmentation based technique for multiple sclerosis lesion detection. arXiv preprint arXiv:1907.03109
Valverde S, Cabezas M, Roura E, González-Villà S, Pareto D, Vilanova JC, Ramió-Torrentà L, Rovira À, Oliver A, Lladó X (2017) Improving automated multiple sclerosis lesion segmentation with a cascaded 3D convolutional neural network approach. NeuroImage 15(155):159–168
Souplet, JC, Lebrun C, Ayache N, Malandain G (2008) An automatic segmentation of T2-FLAIR multiple sclerosis lesions. In: Grand challenge work: multiple sclerosis lesion segmentation. Challenge. pp 1–11
Schmidt P, Gaser C, Arsic M, Buck D, Förschler A, Berthele A, Hoshi M, Ilg R, Schmid VJ, Zimmer C, Hemmer B (2012) An automated tool for detection of FLAIR-hyperintense white-matter lesions in multiple sclerosis. Neuroimage 59(4):3774–3783
Kwok PP, Ciccarelli O, Chard DT, Miller DH, Alexander DC (2012) Predicting clinically definite multiple sclerosis from onset using SVM. In: Machine learning and interpretation in neuroimaging 2012. Springer, Berlin, pp 116–123
Cerasa A, Bilotta E, Augimeri A, Cherubini A, Pantano P, Zito G, Lanza P, Valentino P, Gioia MC, Quattrone A (2012) A cellular neural network methodology for the automated segmentation of multiple sclerosis lesions. J Neurosci Methods 203(1):193–199
Cabezas M, Oliver A, Roura E, Freixenet J, Vilanova JC, Ramió-Torrentà L, Rovira À, Lladó X (2014) Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding. Comput Methods Programs Biomed 115(3):147–161
Kawahara J, McIntosh C, Tam R, Hamarneh G (2014) Novel morphological and appearance features for predicting physical disability from MR images in multiple sclerosis patients. In: Computational methods and clinical applications for spine imaging. Springer, Cham, pp 61–73
Siddiqui MF, Reza AW, Kanesan J (2015) An automated and intelligent medical decision support system for brain MRI scans classification. PLoS One 10(8):e0135875
Deshpande H, Maurel P, Barillot C (2015) Classification of multiple sclerosis lesions using adaptive dictionary learning. Comput Med Imaging Graph 1(46):2–10
Alshayeji MH, Al-Rousan MA, Ellethy H (2018) An efficient multiple sclerosis segmentation and detection system using neural networks. Comput Electr Eng 1(71):191–205
Jobson DJ, Rahman ZU, Woodell GA (1997) Properties and performance of a center/surround retinex. IEEE Trans Image Process 6(3):451–462
Adhikari SK, Sing JK, Basu DK, Nasipuri M (2015) Conditional spatial fuzzy C-means clustering algorithm for segmentation of MRI images. Appl Soft Comput 1(34):758–769
Gorji HT, Haddadnia J (2015) A novel method for early diagnosis of Alzheimer’s disease based on pseudo Zernike moment from structural MRI. Neuroscience 1(305):361–371
Costa AF, Humpire-Mamani G, Traina AJ (2012) An efficient algorithm for fractal analysis of textures. In: 25th SIBGRAPI conference on graphics, patterns and images. IEEE, pp 39–46
Khawaled S, Zibulevsky M, Zeevi YY (2019) Texture and Structure Two-view Classification of Images. arXiv preprint arXiv:1908.09264
Krohn S, Froeling M, Leemans A, Ostwald D, Villoslada P, Finke C, Esteban FJ (2019) Evaluation of the 3D fractal dimension as a marker of structural brain complexity in multiple-acquisition MRI. Human brain mapping
Ahmadi A, Davoudi S, Daliri MR (2019) Computer aided diagnosis system for multiple sclerosis disease based on phase to amplitude coupling in covert visual attention. Comput Methods Programs Biomed 1(169):9–18
Zhang YD, Zhao G, Sun J, Wu X, Wang ZH, Liu HM, Govindaraj VV, Zhan T, Li J (2018) Smart pathological brain detection by synthetic minority oversampling technique, extreme learning machine, and Jaya algorithm. Multimed Tools Appl 77(17):22629–22648
Nayak DR, Dash R, Majhi B (2017) An improved extreme learning machine for pathological brain detection. In: TENCON 2017—2017 IEEE region 10 conference. IEEE, pp 13–18
Zhang YD, Jiang Y, Zhu W, Lu S, Zhao G (2018) Exploring a smart pathological brain detection method on pseudo Zernike moment. Multim Tools Appl 77(17):22589–22604
Lahmiri S (2016) Image characterization by fractal descriptors in variational mode decomposition domain: application to brain magnetic resonance. Physica A 15(456):235–243
Backes AR, Casanova D, Bruno OM (2012) Color texture analysis based on fractal descriptors. Pattern Recogn 45(5):1984–1992
Acknowledgements
The authors sincerely thank the anonymous reviewers for their valuable comments. The authors wish to thank Dr. Mohammad R. Khosravi for his suggestions on preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We have no conflict of interest to declare.
Ethical approval
We have all permissions for working on the real medial data from the ethical committee of Vasei hospital in Sabzevar, Iran, regarding the healthy and patient individuals. Also, the results of our research can be reported based on the same permission received.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rezaee, A., Rezaee, K., Haddadnia, J. et al. Supervised meta-heuristic extreme learning machine for multiple sclerosis detection based on multiple feature descriptors in MR images. SN Appl. Sci. 2, 866 (2020). https://doi.org/10.1007/s42452-020-2699-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2699-y