Abstract
Segmentation divides an image into discrete provinces containing pieces of pixels with analogous attributes. To be expressive and useful for image analysis and interpretation, the regions should strongly relate to depicted objects or features of interest. Different soft computing and hard computing methods are used for medical image segmentation for efficient accuracy. These are computing methods where hard computing is the conventional methodology, which relies on the principles of accuracy, certainty, and inflexibility. Conversely, soft computing is a modern approach premised on the idea of the approximation, uncertainty, and flexibility. Accurate segmentation is very necessary for medical images for better treatment planning. This article provides an efficient analysis of medical images using hard and soft computing. Further, it will explain data used, results obtained, and observation of the current literatures available for medical image segmentation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Image segmentation is the procedure of subdividing image into subsequent parts or objects. These objects are further utilized in image processing and image analysis. Each of these parts is made up of set of pixels. Image segmentation is highly application-dependent. During automatic image recognition and analysis problems, segmentation should stop when object or part of interest is encountered [1]. An efficient image segmentation process is a major factor for a successful image analysis. Researchers have been working to develop an automatic state-of-the-art process of image segmentation to coordinate with high-level machine-based image analysis. Different image segmentation processes are employed for different kind of image analysis [2]. Many a times, manual annotations to carry out image segmentations are used, but they are time-consuming, less efficient, and labor-intensive. Hence, the field of automating the image segmentation process is attracting a huge number of researchers, and proportional to that, there has been some major breakthrough in the same [3]. However, there are several researches happening based on medical image segmentation. Still, there is no exact way to implement segmentation on various datasets. This paper provides the brief review of several soft and hard computing approaches for medical image segmentation nowadays. Further, this paper is organized in such a way that Sect. 3 gives the overview of soft and hard computing techniques, and Sect. 4 gives the brief introduction of the several current approaches used for medical image segmentation based on soft and hard computing approaches. Conclusion is given in Sect. 5. Finally, the references are given in Sect. 6.
2 Study of important topics
2.1 Image segmentation
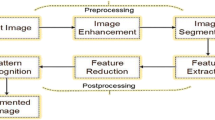
As discussed, image segmentation is the process of segregating images into numerous objects based on some homogenous features like intensity or color. This provides ease in analysis of the image by showcasing only the features of the complete image, which are needed for analysis. The extent of homogeneity of these features can be measured using image properties like pixels [4]. Segmentation is the most crucial and critical part of both image processing and image analysis on all levels. Fig. 1 gives the example of image segmentation. The subtask image analysis also uses image segmentation as its sub-process. Segmentation of image is a high benefactor in case of medical image process like brain tumor locators and breast cancer detectors [5]. It is responsible for lowering the load of work that needs to be done by the image processor. Hence, a great amount of research is going on to define and dedicate new methods for faster, user interaction efficient, and accurate algorithms to be employed for image segmentation. Presently, there is no standard method that could be applied for segmentation, and several methods have their own merits and demerits and are employed in an image processor accordingly.
Moreover, all the proposed methods for image segmentation can be classified into majorly two categories—hard computing methods or traditional methods and soft computing methods. Some of the hard computing techniques are mean shift method, edge-based segmentation (boundary localization), level-set method, ratio contour method, etc, while examples of some of the soft computing segmentation techniques are genetic algorithms, artificial neural network (ANN), convolutional neural network, recurrent neural network (RNN), swarm optimization, etc. [6]. Soft computing techniques are gaining more attention of the researchers due to their immense flexibility of working with ownership functions [7]. These techniques are highly adaptive in nature without any compromise in accuracy level, hence are highly preferred by researchers and implementors. They have several advantages over hard computing techniques some of which being their tolerance toward imprecision, approximations, and uncertainty. The principal methodology of soft computing techniques is genetic algorithms, several neural networks, and fuzzy logics. Soft computing techniques are usually employed along with some hard computing techniques, which finally results in cost-effective and high-performance results to a certain problem.
2.2 Soft and hard computing techniques
Soft computing offers us the opportunity to solve a problem being closer to the real world; with the use of neural networks and fuzzy logics, we can imitate a human mind working and generate outputs. It can be understood as a combination of biological structures and computing techniques; this combination provides the capacity to produce more competent and dependable solutions. It is believed that together genetic algorithms, fuzzy logics, and artificial neural networks have the capability of predicting uncertain events and work on incomplete truth [8]. Fuzzy logic is classified into two that is propositional logic and predictive logic, while the neural network is classified into feedforward neural network, feedback neural network, Hopfield neural network (HNN), recurrent neural network (RNN), and radial basis network. Figure 2 represents a flowchart depicting the major techniques used for image segmentation that has been covered in this paper.
Hard computing techniques in contrary to soft computing techniques provide us an opportunity to extract exact results with the use of binary and Boolean logics. They are based on analytical methods rather being depending on approximate modeling. They always provide us with accurate results and are consistent in nature. They tend to follow sequential computation [9]. Fig. 2 explains a simple FCN (fully convolutional neural network) used for brain data image segmentation (Fig. 3).
3 Description of different hard computing methods
3.1 Thresholding
Many a times, gray level of the pixels of the subject that is to be segmented out is completely different to the background, and thresholding is a frequently used segmentation tool in such conditions. Threshold techniques are classified into two based on their state and vicinity of action; they are namely local and global techniques. In global techniques, the same threshold is applied on the complete image, while in local technique, a series of thresholds are applied locally of several parts of the image. Different types of thresholding are global threshold, adaptive threshold, optimal threshold, and local threshold.
3.2 Edge-based method
Edge detection method is a critical and widely used techniques employed for image segmentation. It is mostly used for object detection, which is a key part of medical image processing. In this method, the original image is transformed into edge images based on the changes of the gray tones in the image. It is highly preferred for localization of certain geometric and physical traits of the image. It outlines the object and segments it out from the background for further analysis.
3.3 K-means clustering
In this method, clusters of similar elements are tried to be created; pixels of images are clustered on three different axes based on RGB. The clustering is implemented by a function, which minimizes the distance between the data points and the centroid or any central tendency of the cluster.
3.4 Markov random field
In most of the cases, images are homogenous in nature, that is, they have similar properties in all their neighboring areas. Markov random field works quite efficiently in these cases by differentiating between the similar properties like texture, intensity, and color. It is a probabilistic model to capture contextual constraints. Some of the major components of this model are labeling fields, neighboring pixels, Gibbs distribution, energy function, and cliques.
4 Description of different soft computing methods
4.1 Fuzzy-based image segmentation
It is based on the principle of degree of truth of false. It is a probabilistic method of segmentation based on a system of fuzzy logics. The input image is firstly passed through a process of image fuzzification, after which it is passed on for membership modification based on expert’s knowledge and a set of fuzzy logics and set theory. The modified image is passed through image defuzzification, and final result is obtained. Some of the fuzzy-based techniques are as follows: fuzzy thresholding, fuzzy integral-based decision making, and fuzzy c-means clustering.
4.2 Artificial neural network (ANN)-based image segmentation
Neural network architecture is completely based on human nervous system. Perceptrons are the building and learning block of the neural networks. They are self-trainable models, which learn with repetitive iterations and changes in weights of different features. Some of the neural networks are as follows: Hopfield neural network (HNN), constraint satisfaction neural network (CSNN), back-propagation neural networks (BPNN), feedforward neural network (FFNN), and pulse-coupled neural network (PCNN).
4.3 Deep learning-based image segmentation
Deep learning models imitate human cerebrum while making decisions. It is mostly employed for unstructured data and uses unsupervised learning. Most of the segmentation problems are being handled by deep learning. Most widely used is convolutional neural network (CNN). Between the first (input layer) and the last (output layer), a number of hidden layers are constructed. More and more features are extracted as the hidden layer increases, making the model capable to learn from data. There are several successful architectures based on CNN, most important of them is U-Net architecture, which comprises of equal number of upsampling and downsampling. Some variants of U-Net are also successful in segmentation of 3-D images.
5 Current approaches for medical image segmentation: soft and hard computing approaches
5.1 Deep learning with image-specific fine-tuning [10]
Dataset used (A) (2-D segmentation of multiple organs) Single-shot fast spin echo (SSFSE) is used to acquire stacks of T2-weighted MRIs.
(B) (3-D segmentation of brain tumors from T1c and FLAIR)—2015 Brain Tumor Segmentation Challenge (BRATS) training set is used.
Results obtained (A) proposed model is more successful to segment previously unseen objects than the conventional CNNs.
(B) Image-specific fine-tuning using the proposed weighted loss function gives more accurate results.
(C) User interaction efficiency is more as compared to CNNs.
Observations A deep learning-based framework that uses a bounding box-based CNN is used for interactive 2-D/3-D image segmentation. They are successful in segmentation of previously unseen objects. Image-specific fine-tuning based on a weighted loss function for both supervised and unsupervised refinements of initial segmentations is proposed.
5.2 DeepIGeoS: a deep interactive geodesic for medical image segmentation [11]
Dataset used Placenta dataset from 2-D fetal MRI and brain tumors dataset from 3-D FLAIR images.
Results obtained Better results as compared to automatic CNNs were achieved. Higher accuracy for 3-D brain tumor segmentation was seen. It resulted in lesser user interface time as compared to typical CNNs.
Observations An interactive framework using deep learning approach is proposed. The framework consists of two stages, first one is P-Net used to obtain an initial automatic segmentation. The second stage consists of a R-Net to further process the result based on user interaction that is integrated into the input of R-net after transformation into geodesic distance maps.
5.3 Deep convolutional network for semantic segmentation [12]
Dataset used For all experiments, two road scene datasets are used namely CamVid dataset from Cambridge University and the CityScape dataset. All the images in both the datasets were taken under good or moderate weather conditions.
Results obtained The network performed well in segmentation accuracy as well as class extension.
Observations A deep convolutional neural network with residual shortcuts, which act as an end-to-end structure providing pixel-wise segmentation, is introduced.
5.4 Deep conditional neural network [13]
Dataset used PASCAL VOC 2012 dataset. Berkley segmentation dataset (BSDS) and NYU-Depth V2 dataset. COCO dataset for training based on baseline model.
Results obtained The model yields competitive result as compared to the combination of convolutional neural networks (CNNs) and conditional random fields (CRFs). Conditional Boltzmann machine (CBM) has excellent representation capability in structured learnings used with structured random forest (SRF) shows better results as compared to other CBMs.
Observations Baseline model (CBM_VOC), which contains only CBMs and convolutional layers and trained on VOC dataset, reaches 63.9% IoU in segmentation. CBM using additional COCO dataset for training based on baseline model enhances the performance and manages to achieve 68.7% segmentation accuracy. The proposed model uses SRFs, which are used for objects border and its ownership detection along with CBMs and achieves 72.3% IoU.
5.5 Brain tumor segmentation using fuzzy c-means-based particle swarm optimization initialization and outlier rejection with level-set methods [14]
Dataset used Brain MRI images are used. Two synthetic gray images with different level of noise are used.
Results obtained Improved kernel possibilistic c-means (IKPCM), fuzzy c-means (FCM), robust FCM, and kernel possibilistic c-means (KPCM) were tested on the same image dataset, and IKPCM showed better result than others.
-
Partition coefficient for IKPCM model is 0.9835, 0.9723, and 0.9751 for 1%, 5%, and 9% Gaussian noise, respectively, in comparison with 0.9326, 0.9225, and 0.9128 partition coefficient for KPCM model for 1%, 5%, and 9% Gaussian noise, respectively.
-
The values of partition entropy experimentally determined are 0.0369, 0.0741, and 0.0826 for Gaussian noise of 1%, 5%, and 9%, respectively, in comparison with 0.1605, 0.1835, and 0.1943 partition entropy values for KPMC model for 1%, 5%, and 9% Gaussian noise, respectively.
Observations The proposed algorithm is the modified version of KPCM method, which considers fuzzy partition matrix and pixel’s spatial information. The following steps describe the proposed model:
(A) PSO algorithms used for initialization of cluster centers and memberships.
(B) Outlier rejection is considered for modifying membership function of the KPCM.
5.6 Intensity and contextual information by Markov random field [15]
Dataset used Experiments are carried out on 3 different noised image datasets: simulated MRI images, real MRI images, and synthetic images.
Results obtained Proposed method has satisfying performance and strong robustness. In comparison with some other methods and algorithms like fuzzy c-means (FCM), fast generalized FCM (FGFCM), and fuzzy local information c-means clustering algorithm (FLICM), the proposed method delivers best results both statistically and visually. The proposed method has average similarity index of 36.8%, 33.7%, and 2.75% increase against the other 3 methods.
Observations The proposed segmentation method, which combines fuzzy clustering and Markov’s random field, yields better results as compared to other state-of-the-art techniques. Hence, it is a robust and precise segmentation technique.
5.7 Fuzzy clustering [16]
Dataset used Dataset from brain web images is used with classical acquisition parameters using high-resolution T1-weighted phantom with slice thickness of 1 mm resolution.
Results obtained The proposed technique is employed in segmentation of medical images, and the efficiency with salt pepper noise is recorded as 99.86%.
Observations Efficiency of reformulated fuzzy logical information c-means clustering algorithm (RFLICM) is remarkably higher as compared to FCM and fuzzy local information c-means clustering algorithm (FLICM) segmentation methods for classifying tissues in brain MR images.
5.8 Posteriori probability for partial volume segmentation of brain MRI [17]
Dataset used The proposed MV-MAP method was tested using two types of datasets—digital phantom and clinical MRI images. Digital brain phantoms were obtained from McConnell Brain Imaging Center and Montreal Neurological Institute.
Results obtained The proposed model (PV-MAP) was compared to fuzzy c-means (FCM) model and adaptive fuzzy c-mean (AFCM) model. For comparison purpose, the measures used were true-positive fraction (TPF), false-positive fraction (FPS), and accurate segmentation ratio (ASR). For instance, white matter (WM) accuracy using TPF measure was recorded as 74.57, 76.58, and 91.96 for FCM, AFCM, and PV-MAP, respectively.
Observations A unified framework to improve brain MR image segmentation by considering all effects simultaneously using MAP probability principle is proposed.
5.9 3-D fast marching algorithm and single hidden layer feedforward neural network [18]
Dataset used Liver tumor images were obtained using 1.5 T magnetic resonance imaging (MRI). Pro-contrast images were used that were obtained by using T1-weighted volumetric interpolated breath-hold examination (VIBE).
Results obtained True-positive, false-negative, and false-positive criteria were used to compare the results with ground truth. Single-layer feedforward neural network (SLFN) combined with fast marching algorithm produces the classification results, which in comparison with manual method are closer to ground truth. The average tumor volume of the proposed model was 13.88 ± 33.21 cc while that of the manual method was 15.36 ± 36.77 cc.
Observations A model by combining neural network with fast marching algorithm is proposed for liver image segmentation. The result in comparison with ground truth is more efficient and accurate.
5.10 Multi-scale 3-D convolutional neural network to perform fast fully automatic segmentation of the abnormal human right ventricle from cardiovascular MRI [19]
Dataset used CMR images from Royal Brompton Hospital were used as dataset.
Results obtained Dice similarity coefficient and absolute volume difference were used as accuracy parameter for the proposed method. Dice similarity coefficient was found to be 0.8281 ± 0.1010, and absolute volume difference was found to be 12.6864 ± 12.9872%.
Observations 3-D multi-scale deep CNN is used to implement automatic image segmentation.
5.11 Hough CNN [20]
Dataset used Dataset consists of MRI of 55 subjects and was acquired using 3-D gradient-echo imaging, while the US dataset of 34 subjects was recorded with freehand 3-D sweeps through left and right temporal bone window.
Results obtained The proposed model yields better dice coefficient when bigger regions and high contrast area are segmented. It performed significantly well as compared to the voxel-wise semantic segmentation of CNNs.
Observations A patch-wise multi-atlas method namely Hough CNN was proposed. It implicitly encodes priors on anatomic shapes. The proposed method outperformed the usual CNNs even with lesser number of data and on all selection of parameters.
5.12 Deep neural networks segment neuronal membranes in electron microscopy images [21]
Dataset used Serial-section transmitted electron microscopy (ssTEM) data images are used as dataset.
Results obtained Three error metrics were used to measure results, namely rand error, warping and pixel error, which were found to be 48, 434, and 60, respectively. These results as compared to other state-of-the-art methods are better. The model outperforms all other models not being tailored to a certain kind of segmentation.
Observations A special type of neural network namely pixel classifier is used in the model. The specialty of the project lies in the approach to neuronal membrane segmentation. A strong pixel classifier was designed using deep neural network trained by online back propagation.
5.13 Malignant lésion segmentation [22]
Dataset used Neoadjuvant chemotherapy data of several patients were used as dataset.
Results obtained Segmentation performed by the proposed method was compared with radiologist generated results, and it was found that the overlap ratio was 63.3% and 58.5%, respectively. Overall VOR was found to 62.6 ± 9.1% (mean ± SD), 61.0 ± 11.3%, and 64.3 ± 10.4%, respectively, for computer and first radiologist, computer and second radiologist, and both the radiologists.
Observations The proposed method proves to be reproducible, effective, and fast marker-controlled watershed algorithm. It carries out semi-automatic segmentation of the tumor in contrast-enhanced T1-weighted MR images. In case of irregular-shaped tumors, better results were generated.
5.14 Unsupervised clustering with “adaptive resolution” [23]
Dataset used MRI dataset of three volumetric dataset representing T1-, T2-, and proton density-weighted MRI was used.
Results obtained The result of the proposed method is highly stable. The system proves to be interactive and allows user to display and modify data during processing and to set the parameters.
Observation The proposed process used the following steps in analysis sequence: reduction of dimensionality, unsupervised data clustering, voxel classification, and interactive post-processing refinement.
5.15 Deep residual learning for image recognition [24]
Dataset used CIFAR-10 and CIFAR-100 datasets are used for training purpose.
Results obtained On testing with COCO detection dataset, 28% relative improvement was marked.
Observations VGG-16 is replaced with proposed ResNet-101. ResNet-101 has greater object detection mAP% on both VOC 7 and VOC 12 test data used with 07 + 12 and 07 ++12 training data, respectively. The observation holds good on COCO validation set also.
5.16 Lung pattern classification for interstitial lung diseases using a deep neural network [25]
Dataset used Training set consisted of 14,696 image patches collected from 120 CT scans.
Results obtained The classification performance was marked approximately to be 85.5%, which is higher as compared to previous methods that were deployed.
Observations A convolutional neural network comprising of 5 convolutional layers with leaky ReLu activation and 2*2 kernels accompanied by average pooling and 3 dense layers is proposed.
5.17 Deep features, SVM classifiers with fully connected convolutional layers (FC-CNN), and fisher vector encoded convolutional layer (FV-CNN) [26]
Dataset used Historical document dataset like CSG18, CSG863, and CB55, G. Washington, is used.
Results obtained Document segmentation was carried out using the proposed model, and 95% pixel accuracy and 93% mean accuracy were marked on G. Washington dataset on using proposed model with FV-CNN. 97% pixel accuracy and 76% mean accuracy were marked on G. Parzival dataset on using proposed model with FV-CNN. 94% pixel accuracy and 71% mean accuracy were marked on CSG863 dataset on using proposed model with FV-CNN. 99% pixel accuracy and 91% mean accuracy were marked on St. Gall dataset on using proposed model with FV-CNN.
Observations The proposed model works more efficiently with FV-CNN as compared to FC-CNN. Better feature representation is showcased as compared to other methods. Pre-trained CNNs are used for implementation.
6 Conclusion
In this work, the overview of various segmentation methodologies based on hard and soft computing approaches explained briefly with the description of observation of the implemented work, dataset used, and the results obtained. The aim of this paper was to provide a simple guide to the scientists and researchers of the world who are working in the field of medical image segmentation. As we have seen, there are several approaches for medical image segmentation such as thresholding, edge-based, graph cut-based, active contour-based, and fuzzy-based. We have observed that nowadays, researchers are moving toward the soft computing approaches such as genetic, fuzzy-based and based on artificial neural network as of the efficiency of such techniques. In spite of numerous years of investigation up to now to the knowledge of authors, there is no unanimously acknowledged process for image segmentation, as the outcome of image segmentation is pretentious by lots of reasons, such as similarity of several objects present in the images, longitudinal physiognomies of the image texture, continuity, and content of the image. Hence, there is no single method, which can be measured good for neither entirely category of images nor all approaches correspondingly respectable for a precise type of image. By considering all the above factors, segmentation of images remnants a thought-provoking issue in image processing and computer vision and is motionless and undecided problem in the world.
References
Chabrier S, Emile B, Laurent H, Rosenberger C, Marche P (2004) Unsupervised evaluation of image segmentation application to multi-spectral images. In: Proceedings of the 17th international conference on pattern recognition, 2004. ICPR 2004
Coquin D (2008) Quantitative evaluation of color image segmentation results using fuzzy neural network. In: Optomechatronic technologies
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2016) Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging 35(5):1299–1312
Suzuki K, Shiraishi J, Abe H, Macmahon H, Doi K (2005) False-positive reduction in computer-aided diagnostic scheme for detecting nodules in chest radiographs by means of massive training artificial neural network1. Acad Radiol 12(2):191–201
Dong C, Loy CC, He K, Tang X (2014) Learning a deep convolutional network for image super-resolution. In: Computer vision—ECCV 2014 Lecture notes in computer science, pp 184–199
Ganin Y, Lempitsky V (2015) N4-fields: neural network nearest neighbor fields for image transforms. In: Computer vision—ACCV 2014 Lecture notes in computer science, pp 536–551
Cui Z, Chang H, Shan S, Zhong B, Chen X (2014) Deep network cascade for image super-resolution. In: Computer vision—ECCV 2014 Lecture notes in computer science, pp 49–64
Saxena S, Sharma N, Sharma S, Singh S, Verma A (2016) An automated system for atlas based multiple organ segmentation of abdominal CT images. Br J Math Comput Sci 12(1):1–14
ChenY-J, Hua K-L, Hsu C-H, Cheng W-H, Hidayati SC (2015) Computer-aided classification of lung nodules on computed tomography images via deep learning technique. In: OncoTargets and therapy, p 2015
Wang G et al (2018) Interactive medical image segmentation using deep learning with image-specific fine tuning. IEEE Trans Med Imaging 37(7):1562–1573
Wang G, Zuluaga MA, Li W, Pratt R, Patel PA, Aertsen M, Doel T, David AL, Deprest J, Ourselin S, Vercauteren T (2008) DeepIGeoS: a deep interactive geodesic framework for medical image segmentation. In: IEEE transactions on pattern analysis and machine intelligence, pp 1–1
Wang C, Mauch L, Guo Z, Yang B (2016) On semantic image segmentation using deep convolutional neural network with shortcuts and easy class extension. In: 2016 Sixth international conference on image processing theory, tools and applications (IPTA)
Wang Q, Yuan C, Liu Y (2016) Deep conditional neural network for image segmentation. In: 2016 IEEE international conference on multimedia and expo (ICME)
Mekhmoukh A, Mokrani K (2015) Improved fuzzy C-means based particle swarm optimization (PSO) initialization and outlier rejection with level set methods for MR brain image segmentation. Comput Methods Programs Biomed 122(2):266–281
Chen M, Yan Q, Qin M (2017) A segmentation of brain MRI images utilizing intensity and contextual information by Markov random field. Comput Assist Surg 22(sup1):200–211
Sucharitha M, Geetha KP (2015) Brain tissue segmentation using fuzzy clustering techniques. Technol Health Care 23(5):571–580
Li X, Li L, Lu H, Liang Z (2005) Partial volume segmentation of brain magnetic resonance images based on maximum a posteriori probability. Med Phys 32(7Part1):2337–2345
Le T-N, Bao PT, Huynh HT (2016) Liver tumor segmentation from MR images using 3D fast marching algorithm and single hidden layer feedforward neural network. Biomed Res Int 2016:1–8
Giannakidis A, Kamnitsas K, Spadotto V, Keegan J, Smith G, Glocker B, Rueckert D, Ernst S, Gatzoulis MA, Pennell DJ, Babu-Narayan S, Firmin DN (2016) Fast fully automatic segmentation of the severely abnormal human right ventricle from cardiovascular magnetic resonance images using a multi-scale 3D convolutional neural network. In: 2016 12th international conference on signal-image technology and internet-based systems (SITIS)
Milletari F, Ahmadi S-A, Kroll C, Plate A, Rozanski V, Maiostre J, Levin J, Dietrich O, Ertl-Wagner B, Bötzel K, Navab N (2017) Hough-CNN: deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput Vis Image Underst 164:92–102
Dong C, Loy CC, Tang X (2016) Accelerating the super-resolution convolutional neural network. Computer vision—ECCV 2016 Lecture notes in computer science, pp 391–407
Cui Y, Tan Y, Zhao B, Liberman L, Parbhu R, Kaplan J, Theodoulou M, Hudis C, Schwartz LH (2009) Malignant lesion segmentation in contrast-enhanced breast MR images based on the marker-controlled watershed. Med Phys 36(10):4359–4369
Schenone A, Firenze F, Acquarone F, Gambaro M, Masulli F, Andreucci L (1996) Segmentation of multivariate medical images via unsupervised clustering with ‘adaptive resolution’. Comput Med Imaging Graph 20(3):119–129
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: 2016 IEEE conference on computer vision and pattern recognition (CVPR)
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35(5):1207–1216
Razzak MI, Naz S, Zaib A (2017) Deep learning for medical image processing: overview, challenges and the future. In: Lecture notes in computational vision and biomechanics classification in BioApps, pp 323–350
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All author states that there is no conflict of interest.
Human and animal rights
Humans/animals are not involved in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sinha, P., Tuteja, M. & Saxena, S. Medical image segmentation: hard and soft computing approaches. SN Appl. Sci. 2, 159 (2020). https://doi.org/10.1007/s42452-020-1956-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-1956-4