Abstract
In metabolism processes, enzymes are involved in a cooperative manner of step by step mechanisms trying to guarantee cell survival. Besides, every change in a biological system and its environment within a cell could lead to metabolism alterations due to natural cell response. So, it is reasonable to find metabolism variations within a cell in different conditions. Such enzymes behaviors are very similar to a competitive game where different players try to play for reaching a specific outcome, either by a cooperative or selfish strategy. In this study, we have explored an assumption in which there exists a shift in playing strategy of enzymes from normal to a different biological condition (here cancer) by investigation of pattern alterations in the metabolic subsystem of four cancer cell types (in four cancer stages) using flux calculations and Gene Ontology analysis. Based on the results, we found some metabolic subsystems which contained less flux variation during the cancer progression. The results indicate there might be a game-theoretic strategy in which changing from a beneficial (cooperative) to a selfish strategy happened during cancer stages. It also could be interesting since picking a similar game strategy for all cells would eventually lead the system toward a tragedy of commons (i.e., lower overall fitness for a group or population), in the case of cancer cells, cause the system to be finally disrupted, means death.
Similar content being viewed by others
1 Introduction
Noise is an inescapable fact of life, which usually considers as an undesired disturbance in useful information that harms the quality. Although, in most cases, noise assumes to be destructive, it could sometimes be beneficial and constructive, especially in biology. For cells, noise meaning variability ranging from stochastic processes within such as gene expression to randomness in an outside environment like food sources, deadly toxins, and pathogens would be a matter of life and death [1]. Noise has been difficult to study in biology since most techniques average the output of many experimental results and erase individual variability. Recently, the development of high-resolution technologies for single cells let scientists be able to capture noise at this level and to explore how it influences the emergence of a system [2]. Single-cell studies revealed that a cell is a nondeterministic machine and could choose different strategies leading to a specific cellular decision based on its conditions [3,4,5]. Hence, noise measurement seems to create an opportunity to find discoveries in biological systems. Strictly speaking, the exact determination of noise effects could shed light on treatments for infections, diseases, and even cancer. Cancer modeling has been counted as a complementary tool next to experimental studies and developed in recent years by the advent of genome-scale models [6]. In addition to the genetic aspect of cancer for modeling approaches, a metabolic point of view is also informative. It is now accepted that cancer cells reprogram their metabolism to attain abnormal cell growth requirements. So, genome-scale metabolic (GEM) modeling of cancer is still a novice with great potential for various analysis methods to deal with [6].
Furthermore, it is also possible to explore genotype–phenotype relationships with metabolic systems biology approaches. However, modeling at the genome-scale confronts some limitations due to the large size of a system. Therefore, dynamic methods such as kinetic modeling are hard to apply because of an insufficient number of parameters. Instead, a metabolic flux overview of a system could be driven using the flux analysis method [7]. In a flux analysis, a system of stoichiometric matrix kernel is calculated under a steady-state using a linear programming approach to obtain an optimum, which satisfies all system constraints. For example, a set of flux values of all reactions involved in a metabolic model could be obtained where maximization of growth rate is considered as an objective function. Since the calculated flux set may not be unique, it is more realistic to obtain a flux range for every reaction, which still keeps the value of an objective function optimized. This method is called flux variability analysis (FVA) that determines a flux ranges (min and max values) for every reaction involved in a metabolic model instead of a fixed flux value [8]. Hence, the flux range (or flux capacity) for each part of the system might change through different GEM cancer models and could consider as a criterion emphasizing variation of every part of the system. The ability to measure variation (noise) in every reaction of a GEM model using an FVA approach would make an opportunity to explore some biological assumptions related to metabolism and cancer development. In a metabolic system, all enzymes are involved in the division of labor. It means each enzyme performs some vital function needed for the next step in a pathway, which finally leads to the survival of the cell as a whole. It is very similar to a game-theoretic approach in which every element (enzyme) work together in a cooperative behavior to reach a specific goal (cell survival). Therefore, there would be a reasonable size of metabolism variation within cell metabolism due to the cooperativity of enzymes. But the situation would not be the same in a cancer system in which each cell tries to grow as fast as it is possible. So, there is a change in the strategy of the players and the division of labor strategy does not work. Instead, every enzyme tries to maximize its benefit. Hence, the cooperative scenario could not be guaranteed and a decrease in variation size of metabolism is now expected.
To explore this assumption, we have performed flux variability and Gene Ontology (GO) analysis on various stages of four different cancer cell types and studied flux variations together with their distributions in the GO metabolic process tree for metabolic subsystems.
2 Materials and methods
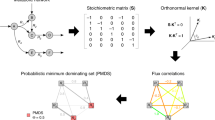
A summary of the workflow of this study has been demonstrated in Fig. 1. Here, we describe the methods in detail.
Schematic overview of this study; Integration of gene expression data into a generic metabolic model, performing FVA on reconstructed cancer models, detecting subsystems with decrement or increment in flux variations, performing a GO analysis on selected subsystems and exploring the distribution of metabolic pathways through the GO metabolic process tree
2.1 Gene expression data
Whole genome-based microarray data were downloaded from the Gene Expression Omnibus (GEO) database and summarized in Table 1. All datasets selected from the same platform (GPL570) and related to the four different types of cancer, including colon, lymphoma, neuroblastoma, and ovarian.
Microarray data were preprocessed and analyzed using the LIMMA package in R [9], which was originally developed for differential expression analysis of microarray data. The detail definition of each cancer stage is provided in the Supplementary file S1. More detailed descriptions of the methods can be found in the original studies on colon cancer [10, 11], lymphoma [12, 13], neuroblastoma [14], and ovarian cancer [15].
2.2 Model reconstruction
We have used the corrected human metabolic model [16], which contains 1905 genes, 2766 metabolites, and 3748 reactions. Gene expression values related to metabolic genes have been mapped into the human metabolic model using the E-Flux algorithm for constraining the possible flux through the reactions [17]. The E-Flux method does not reduce gene expression levels to binary states. Instead, it constrains the upper and lowers bound of each reaction according to its corresponding gene expression level. In summary, for integration of the expression data, gene expression values have been mapped into the Recon1_corrected model using the E-Flux method. The E-Flux approach tries to map gene expression data into a GEM by constraining the maximum possible flux through the reactions. However, this method does not reduce gene expression levels to binary states, as some other algorithms (such as GIMME, iMAT, and MADE) do. E-Flux constrains the upper and lower bound of a reaction according to its corresponding gene expression level. Using this approach, we have constructed four cancer tissue-specific metabolic models based on their corresponding gene expression data. We have written a Mathematica script to apply the E-Flux method to the human metabolic model (supplementary file S1). This script reads gene expression data, performs the E-Flux algorithm, and changes the upper and lower bounds of each reaction according to its Gene-Protein-reactions Relationships (GPRs) Matrix available through the human metabolic model. Then, the RPMI-1640 medium and biomass equation have been added to the models. The biomass and the medium compositions are given in the supplementary file S1 (Tables 1, 2), respectively. All cancer metabolic models are available in the supplementary file S2 as MATLAB structure files.
2.3 FVA for metabolic models
We have used the COBRA toolbox (Constraints Based Reconstruction and Analysis) [18] for calculating fluxes of the metabolic models through the maximization of the biomass equation. The glpk solver has been used for linear programming problems on a computer with the Intel Core-i7-3770 k CPU and 8 GB RAM. For FVA, the fluxVariability function has been performed by just considering solutions that give at least 90 percent of the optimal solution. We wanted to compare the behavior of a cancer system (in term of its noise) while the system tries to keep its objective value in the maximum value. This means the system always keeps to have the maximum biomass (by maximizing the objective function) although it is in different cancer stages. So, the flux variability of each reaction in such condition (while a system always reaches to its maximum objective value) could resemble the narrowness of the flux in various situations. Therefore, using FVA, we could be sure about the system to keep its biomass value in the maximum value, and now we could deal with the noise of the system through the allowance of reactions fluxes (i.e., the range of the fluxes). In another words, we want to keep the objective value in its maximum value in all calculations to resemble the biological aspect of a cell and now looking at the noise effect in the system through its reactions flux ranges. The FVA output would be two vectors, including minimum and maximum fluxes for each reaction. So, the flux range for every reaction could be calculated using corresponding minimum and maximum values. Then, we have calculated flux ranges for 101 different subsystems by averaging over their corresponding reactions. The subsystems information is available through the vector called subSystems in the MATLAB model structure. The biological definition of 101 subsystems is based on metabolic pathway classification [19]. All FVA results related to different cancer stages are available in the supplementary file S3.
2.4 Gene Ontology (GO) analysis
To identify flux variations together with their distributions in the GO metabolic process, we detect all genes that participate in the subsystems (974 genes). Then, we analyzed the genes using the Biological Networks Gene Ontology tool (BiNGO) app [20] that is a tool to define which Gene Ontology (GO) terms are considerably overrepresented in a set of genes. Also, the GO (biological process) analysis was repeated using two gene lists; one list includes subsystems in which flux variations were decreased in four cancer types (282 genes), and the other contains subsystems with decrement in flux variations for two cancer types and increment in flux variations for two other cancer types (194 genes). The results of the two lists were highlighted through the tree that was reconstructed via 974 genes (Fig. 2).
Distribution of genes/enzymes in a metabolic process branch of a GO analysis represented as a hierarchical tree. Orange nodes are metabolic genes related to all subsystems. Violet nodes are genes corresponding to 17 metabolic subsystems with a decrement in flux ranges during the tumor development of four cancer types. These genes are gathered in six clusters through the biological process tree. Blue nodes are genes which include increment in flux variation of two cancer cell types and decrement in two others. The inline figure is the cluster number 3, which was zoomed in for better representation (see text)
3 Results
The flux ranges of each subsystem have been calculated for four different stages of cancer cell types (including ovarian, colon, lymphoma, and neuroblastoma). To better distinguish the results, the outputs of stages 1 and 2 have been merged as well as stages 3 and 4. Then, the flux range comparison between initial stages (stages 1 and 2) and more developing stages (stages 3 and 4) have been performed for all cancers. As it is shown in Table 2, there are 17 subsystems in which ranges of the fluxes are decreased. It means flux capacity or variation of the subsystems have been reduced, and the subsystems confronted with a lack of variation through the cancer development. Flux ranges of the remaining subsystems do not satisfy a reasonable pattern. We have chosen one of these cases (increment in two cancer cell types, whereas decrement in two others) to be assumed as random behavior of subsystems among cancer cells (Table 3). It should be noted that surprisingly, there is not any subsystem in which flux capacity is increased in four cell types during cancer progression. For example, for glycolysis sub-system, there were increment in flux ranges in four type of cancers (as expected) but in terms of variations (i.e., standard variation values) there were increment in three cancer types, but a decrement in ovarian cancer. It has been reported that drug resistance in ovarian cancer is directly associated with a decrease in glycolysis ability [21]. So, it seems cancer cells might use such mechanism to increase the chance of being drug-resistance. A complete list of flux variations for all subsystems is available in the supplementary file S3.
The GO tree was reconstructed to investigate flux variations with their distributions in the GO metabolic process, and the results were visualized using Cytoscape [22]. The result showed 820 biological processes for the whole imported gene list. Besides, the result of analysis represented 141 biological processes for the first gene subsets (contains subsystems with a decrement in flux variations for two cancer types and increment in flux variations for two other cancer types). Moreover, the result illustrated 196 biological processes for the second subsets (in which flux variations were decreased in four cancer types) that overrepresented in the original tree (Fig. 2). Finally, 144 GO are common for both gene subsets. All GO results have been provided in the supplementary file S4.
4 Discussion and conclusion
The noise could be both an obstacle for some kinds of cellular behaviors along with a beneficial feature for others. Noise study in biology started in 2002 when Elowitz and his collaborators mapped random gene activity in single cells and demonstrated that a cell is a nondeterministic machine [23]. After that, many studies have been performed to figure out whether noise might influence important cellular decisions. It seems noise could control cell fate decisions, and this makes some cells to switch fates randomly [5]. In 2009, Cagatay et al. also demonstrated that noise could influence the cells’ survival [3]. The consequences of noise aren’t limited to microbes. Noise might also play an important role in the development of more complex organisms, including humans [24]. Understanding how noise works in the cell would help scientists to clarify a confusing occurrence of random fluctuations in gene activity, which possibly plays a role in some diseases [25, 26]. Such fluctuations could lead to generating diversity through a system. However, In contrast to the production of diversion, nature has been developed influential ways to control it. In other words, each cell contains some best-fitted variations which try to attain benefits for the whole system, including growth and cell division. For example, in the metabolism process, all enzymes are supposed to be involved in the division of labor in a step by step mechanism, which goes to cell survival. The division of labor situation enforces variation increment since each set of enzymes concentrates on a specific subsystem different from the others. This is similar to a game-theoretic method in which different players would play in a special way to increase the system benefits, leading to a cooperative state. If a condition of a cell is changed, enzymes can resemble a non-cooperative behaviour in which all enzymes pick a selfish strategy to increase their benefits. In such case, the selection of a selfish strategy would lead to extortion, not cooperation. Studying game-theoretic mathematical models in cancer metabolism seems to be an interesting phenomenological modeling area and some studies tries to deal with it in previous works [27, 28]. To pursue a game-theoretic approach of noise in metabolism, we have explored flux variations through cancer cell stages. We have assumed there will be a decrement in flux variation for essential metabolic enzymes in a cancer cell because of a replacement in playing strategy from cooperative to selfish. Therefore, we are expected some metabolic subsystems to contain less flux variation during the progression of a cancer cell.
The flux balance analysis relies on two assumptions; steady-state approximation and optimality. The steady-state approximation reduces the system to a set of linear equations, that is then solved to catch a flux distribution which satisfies the steady-state condition subject to the specified constraints while maximizing the value of an objective function. A standard approach to solve such system is to apply linear programming. In this study, by integration of gene expression data of different cancer stages to the Recon1_corrected human metabolic model using the E-Flux algorithm, the only thing that changes through the models would be the ability to carry flux for each reaction. So, any changes in the FVA results (which uses the same objective function to be maximized) would be the flux ranges. Therefore, this removes any other sources of causes such as environmental or availability of new metabolites or reactions which might create uncertainty about the resource of the noise in the system. Thus, we could be almost sure that we fixed all other parameters and just focusing of the noise caused by the flux ranges. So, we have performed FVA to calculate flux ranges for metabolic subsystems for four different cancer cell types in 4 stages to examine the hypothesis. Comparing the flux ranges of stages 1–2 with 3–4 for all cancer cell types demonstrates there are some metabolic subsystems in which changes in flux ranges do not show a specific trend (decrement in 1, 2, or 3 cancers whereas increment in others). But, as it is demonstrated in Table 2, 17 metabolic subsystems are common through 4 cancers in which flux ranges are decreased during tumor development. Figure 2 shows the distribution of corresponding genes/enzymes (violet nodes) in a metabolic process branch of a GO analysis represented as a hierarchical tree. These genes/enzymes represent players with a selfish strategy in the game. We have also repeated the analysis for the metabolic genes/enzymes (blue nodes), which include increment in flux variation of two cancer cell types and decrement in two others. These genes/enzymes have been picked up to show players with cooperative strategies. This means we considered the most ideal one (in which flux variations were decreased in two of four cancer types and increased in the two others) as a cooperative behavior. It is clear that there are other options for such strategy (for instance decrement in one or three cancers whereas increment in the others). We considered the scenario of two decrement-two increment because it was the most ideal cases and could better rely on a balanced (and not preferable) strategy among different types of cancers. Now, It is possible to explore distribution of different players (cooperative vs. non-cooperative) through the entire system by comparing the violet nodes with the blue ones (i.e., all metabolic genes/enzymes of the cancer models available in the metabolic process of GO analysis tree). It could be realized that while cooperative players are distributed randomly, the non-cooperative ones do not satisfy a similar behavior. Instead, they are collected in sub-grouped locations of the metabolic process tree (Fig. 2). In other words, the variation reduction, which is common to all cancer cells, has been happened to clusters that seem to be essential for cancer development. As has been illustrated in Fig. 2, there are six clusters, including a well-known process related to the cancer progression (supplementary file S5). Many types of research showed that cancer cells are dependent on various enzymatic reactions to survive, so metabolic reprogramming must meet their requirement [29]. For instance, the cluster number 4 shows the overrepresentation of fatty acid process during cancer progression. However, previous study showed that fatty acid oxidation (FAO) pathway cannot be activated independent of the microenvironment of the cancer cell [29]. In ovarian cancer, FAO (β-oxidation) pathway in interaction with adipocytes promotes ovarian cancer metastasis and provides energy for rapid tumor growth [30]. The regulation of fatty acid pathway and its effect on cancer cells is summarized on [29]. Moreover, recent findings indicate that altered metabolism in cancer cells relies on the preferential use of alternative isoforms or genomic amplification of enzymes involved in glucose and amino acid metabolism [31]. As Fig. 2 shows, cluster numbers 3 and 4 indicate the overrepresentation of the amino acid alteration process, which confirms previous studies [31, 32]. Besides, clusters 2 and 3 indicate activation of the glucose metabolism process, which is crucial for cancer cells based on previous researches [31, 33]. Furthermore, malate-aspartate over-represents in cluster number 6, which the previous study indicated the occurrence of the malate-aspartate shuttle in various tumor types [34, 35].
Cancer cells try to grow as fast as it is possible. At this point, cells try to concentrate on critical metabolic pathways for cell division and growth. In the game theory language, this tendency goes to change in the game strategy of elements (here metabolic enzymes) from a division of labor (cooperative) to maximization of benefit [36]. Hence, it is expected to see a decrement of metabolic flux ranges associated with the significant subsystems, as it is shown in this study. It should also be noted that picking a similar game strategy for all cells would eventually lead to a tragedy of commons (TOC). The tragedy of commons is considered as a situation in which individual players reduce the resources over which they compete, leading to lower overall fitness for a group or population. This concept has been largely studied in evolutionary biology [37, 38]. There is also a study related to tragedy of the commons for cooperators in cancer [39]. However, the TOC, in our case (cancer cells), would cause the system to be finally disrupted, which means death.
However, in this study we just used some basic concept of the game theory that might be responsible to obtain such biological results. So, further theoretical and experimental works needed to find more about the relationship between game theory and noise effect in a metabolic system. In addition, there are still lots of questions regarding noise effect in cells and application of game theory in biology to be answered since these fields have been revived in this decade. For example, how would noise influence a community as a whole? Do cells have an awareness of each other’s noise? Would a cell become noisy if its neighbors are very noisy? Which strategy should we enforce to a cancer system to turn selfish into a cooperative strategy? These questions remain for further study in the future.
Code availability
All analysis has been performed in MATLAB (R2016b) and Mathematica (v. 10.3) software.
References
Tsimring LS (2014) Noise in biology. Rep Prog Phys 77(2):26601
Raj A, van Oudenaarden A (2008) Stochastic gene expression and its consequences. Cell 135(2):216–226
Cagatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM, Çağatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM (2009) Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell 139(3):512–522
Sanchez A, Golding I (2013) Genetic determinants and cellular constraints in noisy gene expression. Science 342(6163):1188–1193
Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB (2006) An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440(7083):545–550
Yizhak K, Chaneton B, Gottlieb E, Ruppin E (2015) Modeling cancer metabolism on a genome scale. Mol Syst Biol 11(6):817
Palsson BØ (2015) Systems biology: constraint-based reconstruction and analysis. Cambridge University Press, Cambridge
Mahadevan R, Schilling CH (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5(4):264–276
Smyth GK (2005) Limma: linear models for microarray data. In: Huber W (ed) in Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer-Verlag, New York, pp 397–420
Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL, Eschrich SA, Yeatman TJ, Deane NG, Beauchamp RD (2012) Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of beta-catenin. Gastroenterology 142(3):562–571
Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, Eschrich S, Kis C, Levy S, Washington MK, Heslin MJ, Coffey RJ, Yeatman TJ, Shyr Y, Beauchamp RD (2010) Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138(3):958–968
Cardesa-Salzmann TM, Colomo L, Gutierrez G, Chan WC, Weisenburger D, Climent F, Gonzalez-Barca E, Mercadal S, Arenillas L, Serrano S, Tubbs R, Delabie J, Gascoyne RD, Connors JM, Mate JL, Rimsza L, Braziel R, Rosenwald A, Lenz G, Wright G, Jaffe ES, Staudt L, Jares P, Lopez-Guillermo A, Campo E (2011) High Microvessel density determines a poor outcome in patients with diffuse large B-Cell lymphoma treated with rituximab plus chemotherapy. Haematologica 96(7):996–1001
Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, Vose J, Bast M, Fu K, Weisenburger DD, Greiner TC, Armitage JO, Kyle A, May L, Gascoyne RD, Connors JM, Troen G, Holte H, Kvaloy S, Dierickx D, Verhoef G, Delabie J, Smeland EB, Jares P, Martinez A, Lopez-Guillermo A, Montserrat E, Campo E, Braziel RM, Miller TP, Rimsza LM, Cook JR, Pohlman B, Sweetenham J, Tubbs RR, Fisher RI, Hartmann E, Rosenwald A, Ott G, Muller-Hermelink HK, Wrench D, Lister TA, Jaffe ES, Wilson WH, Chan WC, Staudt LM (2008) Project lymphoma/leukemia molecular profiling “stromal gene signatures in large-b-cell lymphomas.” N Engl J Med 359(22):2313–2323
Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, Valteau-Couanet D, Frebourg T, Michon J, Lyonnet S, Amiel J, Delattre O (2008) Somatic and germline activating mutations of the alk kinase receptor in neuroblastoma. Nature 455(7215):967–970
Wu YH, Chang TH, Huang YF, Huang HD, Chou CY (2014) COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene 33(26):3432–3440
Shlomi T, Benyamini T, Gottlieb E, Sharan R, Ruppin E (2011) Genome-Scale Metabolic Modeling Elucidates the Role of Proliferative Adaptation in Causing the Warburg Effect. PLoS Comput Biol 7(3):e1002018
Colijn C, Brandes A, Zucker J, Lun DS, Weiner B, Farhat MR, Cheng TY, Moody DB, Murray M, Galagan JE (2009) Interpreting expression data with metabolic flux models: predicting mycobacterium tuberculosis mycolic acid production. PLoS Comput Biol 5(8):e1000489
Schellenberger J, Que R, Fleming RM, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson BO (2011) Quantitative prediction of cellular metabolism with constraint-based models: the cobra toolbox v2.0. Nat Protoc 6(9):1290–1307
Nelson DL, Cox MM (2005) Lehninger principles of biochemistry. Freeman, W. H
Maere S, Heymans K, Kuiper M (2005) BiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21(16):3448–3449
Jin L, Huo Y, Zheng Z, Jiang X, Deng H, Chen Y, Lian Q, Ge R, Deng H (2014) Down-regulation of ras-related protein rab 5C-dependent endocytosis and glycolysis in cisplatin-resistant ovarian cancer cell lines. Mol Cell Proteomics 13(11):3138–3151
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Elowitz MB, Levine AJ, Siggia ED, Swain PS (2002) Stochastic gene expression in a single cell. Science 297(5584):1183–1186
Chazaud C, Yamanaka Y, Pawson T, Rossant J (2006) Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell 10(5):615–624
Raj A, Rifkin SA, Andersen E, van Oudenaarden A (2010) Variability in gene expression underlies incomplete penetrance. Nature 463(7283):913–918
Taheri M, Nouri K, Asgari Y, Zabihinpour Z, Sadeghi M (2019) The significance of noise in the evolution of negative and positive cooperativity in protein complexes. Match Commun Math Comput Chem 81(1):177–192
Archetti M (2014) Evolutionary dynamics of the warburg effect: glycolysis as a collective action problem among cancer cells. J Theor Biol 341:1–8
Archetti M (2016) Cooperation among cancer cells as public goods games on voronoi networks. J Theor Biol 396:191–203
Carracedo A, Cantley LC, Pandolfi PP (2013) Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer 13(4):227–232
Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, Lengyel E (2011) Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 17(11):1498–1503
Cheong H, Lu C, Lindsten T, Thompson CB (2012) Therapeutic targets in cancer cell metabolism and autophagy. Nat Biotechnol 30(7):671–678
Leichtle AB, Nuoffer JM, Ceglarek U, Kase J, Conrad T, Witzigmann H, Thiery J, Fiedler GM (2012) Serum amino acid profiles and their alterations in colorectal cancer. Metabolomics 8(4):643–653
Hamanaka RB, Chandel NS (2012) Targeting glucose metabolism for cancer therapy. J Exp Med 209(2):211–215
Greenhouse WV, Lehninger AL (1976) Occurrence of the malate-aspartate shuttle in various tumor types. Cancer Res 36(4):1392–1396
Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, Zhao S, Yang Y, Ling ZQ, Guan KL, Xiong Y, Ye D (2015) SIRT3-dependent GOT2 acetylation status affects the malate-aspartate nadh shuttle activity and pancreatic tumor growth. EMBO J 34(8):1110–1125
Stewart AJ, Plotkin JB (2014) Collapse of cooperation in evolving games. Proc Natl Acad Sci U S A 111(49):17558–17563
Press WH, Dyson FJ (2012) Iterated prisoner’s dilemma contains strategies that dominate any evolutionary opponent. Proc Natl Acad Sci 109(26):10409–10413
Rankin DJ, Bargum K, Kokko H (2007) The tragedy of the commons in evolutionary biology. Trends Ecol Evol 22(12):643–651
West J, Hasnain Z, Mason J, Newton PK (2016) The prisoner’s dilemma as a cancer model. Converg Sci Phys Oncol 2(3):035002
Acknowledgement
The authors thank Prof. M. Sadeghi for comments that greatly improved the manuscript.
Author information
Authors and Affiliations
Contributions
YA designed the study; YA and PK wrote and edited the manuscript. YA performed FVA studies. PK performed GO analysis. YA and PK generated all figures and tables. All authors have read and approved the final version of the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Availability of data and material
All data have been provided as supplementary materials.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Asgari, Y., Khosravi, P. Flux variability analysis reveals a tragedy of commons in cancer cells. SN Appl. Sci. 2, 1966 (2020). https://doi.org/10.1007/s42452-020-03762-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-03762-3