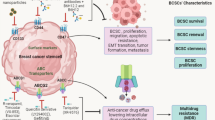
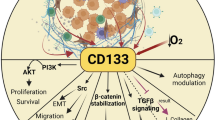
Abstract
Reactivation of the stem cell programme in breast cancer is significantly associated with persistent cancer progression and therapeutic failure. Breast cancer stem cells (BCSCs) are involved in the process of breast cancer initiation, metastasis and cancer relapse. Among the various important cues found in the formation and progression of BCSCs, microRNAs (miRNAs or miRs) play a pivotal role by regulating the expression of various tumour suppressor genes or oncogenes. Accordingly, there is evidence that miRNAs are associated with BCSC self-renewal, differentiation, invasion, metastasis and therapy resistance, and therefore cancer recurrence. miRNAs execute their roles by regulating the expression of stemness markers, activation of signalling pathways or their components and regulation of transcription networks in BCSCs. Therefore, a better understanding of the association between BCSCs and miRNAs has the potential to help design more effective and safer therapeutic solutions against breast cancer. Thus, an miRNA-based therapeutic strategy may open up new horizons for the treatment of breast cancer in the future. In view of this, we present the progress to date of miRNA research associated with stemness marker expression, signalling pathways and activation of transcription networks to regulate the self-renewal, differentiation and therapy resistance properties of BCSCs.



Similar content being viewed by others
References
Jemal A, Bray F, Centern MM, Ferlay J, Ward E, Forman D. Global cancer statistics 2011. CA Cancer J Clin. 2011;61:69–90.
Siegel RL, Miller KD, Jemal A. Cancer statistics 2015. CA Cancer J Clin. 2015;65:5–29.
Jeong H, Kim J, Lee Y, Seo JH, Hong SR, Kim A. Neuregulin-1 induces cancer stem cell characteristics in breast cancer cell lines. Oncol Rep. 2014;32:1218–24.
Das PK, Zahan T, AbdurRakib M, Khanam JA, Pillai S, Islam F. Natural compounds targeting cancer stem cells: a promising resource for chemotherapy. Anticancer Agents Med Chem. 2019;19:1.
Das PK, AbdurRakib M, Khanam JA, Pillai S, Islam F. Novel therapeutics against breast cancer stem cells by targeting surface markers and signaling pathways. Curr Stem Cell Res Ther. 2019;14:1.
Nakanishi T, Ross DD. Breast cancer resistance protein (BCRP/ABCG2): its role in multidrug resistance and regulation of its gene expression. Chin J Cancer. 2012;31(2):73–99.
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–26.
Charafe-Jauffret E, Ginestier C, Bertucci F, et al. ALDH1-positive cancer stem cells predict engraftment of primary breast tumors and are governed by a common stem cell program. Cancer Res. 2013;73(24):7290–300.
Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672–9.
Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57.
Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314.
Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–43.
Abdolvahabi Z, Nourbakhsh M, Hosseinkhani S, Hesari Z, Alipour M, Jafarzadeh M, Ghorbanhosseini SS, Seiri P, Yousefi Z, Yarahmadi S, Golpour P. MicroRNA-590-3P suppresses cell survival and triggers breast cancer cell apoptosis via targeting sirtuin-1 and deacetylation of p53. J Cell Biochem. 2018;120(6):9356–68.
Rahimi M, Sharifi-Zarchi A, Zarghami N, Geranpayeh L, Ebrahimi M, Alizadeh E. Down-regulation of miR-200c and up-regulation of miR-30c target both stemness and metastasis genes in breast cancer. Cell J. 2020;21(4):467–78.
Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12.
Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3–4):369–78.
Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One. 2010;5(9):e13067.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55.
Vahidian F, Mohammadi H, Ali-Hasanzadeh M, Derakhshani A, Mostaan M, Hemmatzadeh M, Baradaran B. MicroRNAs and breast cancer stem cells: potential role in breast cancer therapy. J Cell Physiol. 2019;234(4):3294–306.
Takahashi RU, Miyazaki H, Ochiya T. The roles of microRNAs in breast cancer. Cancers (Basel). 2015;7:598–616.
Shimono Y, Mukohyama J, Nakamura S-I, et al. MicroRNA regulation of human breast cancer stem cells. J Clin Med. 2015;5:2.
Troschel FM, Böhly N, Borrmann K, Braun T, Schwickert A, Kiesel L, Eich HT, Götte M, Greve B. miR-142-3p attenuates breast cancer stem cell characteristics and decreases radioresistance in vitro. Tumour Biol. 2018;40(8):1010428318791887.
Polytarchou C, Iliopoulos D, Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc Natl Acad Sci. 2012;109:14470–5.
Sin WC, Lim CL. Breast cancer stem cells-from origins to targeted therapy. Stem Cell Investig. 2017;4:96.
Collina F, Di Bonito M, Li Bergolis V, et al. Prognostic value of cancer stem cells markers in triple-negative breast cancer. Biomed Res Int. 2015;2015:158682.
Lee WJ, Kim SC, Yoon JH, et al. Meta-analysis of tumor stem-like breast cancer cells using gene set and network analysis. PLoS One. 2016;11(2):e0148818.
Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/β3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008;68(19):7711–7.
Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67.
Pham PV, Phan NL, Nguyen NT, et al. Differentiation of breast cancer stem cells by knockdown of CD44: promising differentiation therapy. J Transl Med. 2011;9:209.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–70.
Li Q, Yao Y, Eades G, Liu Z, Zhang Y, Zhou Q. Downregulation of miR-140 promotes cancer stem cell formation in basal-like early stage breast cancer. Oncogene. 2014;33:2589–600.
Wang X, Lu H, Li T, Yu L, Liu G, Peng X, Zhao J. Krüppel-like factor 8 promotes tumorigenic mammary stem cell induction by targeting miR-146a. Am J Cancer Res. 2013;3(4):356–73.
Takahashi RU, Miyazaki H, Takeshita F, Yamamoto Y, Minoura K, Ono M, Kodaira M, Tamura K, Mori M, Ochiya T. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat Commun. 2015;6:7318.
Lu Y, Qin T, Li J, Wang L, Zhang Q, Jiang Z, Mao J. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24(9):386–92.
Chen D, Dang BL, Huang JZ, Chen M, Wu D, Xu ML, Li R, Yan GR. MiR-373 drives the epithelial-to-mesenchymal transition and metastasis via the miR-373-TXNIP-HIF1α-TWIST signaling axis in breast cancer. Oncotarget. 2015;6(32):32701–12.
Huang Q, Gumireddy K, Schrier M. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10.
Stinson S, Lackner MR, Adai AT. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4(186):pt5.
Gwak JM, Kim HJ, Kim EJ, Chung YR, Yun S, Seo AN, Lee HJ, Park SY. MicroRNA-9 is associated with epithelial–mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat. 2014;147(1):39–49.
Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–51. https://doi.org/10.1038/onc.2010.215.
Hwang MS, Yu N, Stinson SY. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013;8:e66502.
Nassirpour R, Mehta PP, Baxi SM, Yin M-J. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One. 2013;8:e62170.
Feigin ME, Muthuswamy SK. Polarity proteins regulate mammalian cell–cell junctions and cancer pathogenesis. Curr Opin Cell Biol. 2009;21:694–700.
Cao M, Nie W, Li J, Zhang Y, Yan X, Guan X, Chen X, Zen K, Zhang CY, Jiang X, Hou D. MicroRNA-495 induces breast cancer cell migration by targeting JAM-A. Protein Cell. 2014;5(11):862–72.
Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X, Song Y, Meng Q, Yuan S, Luan L, Andl T, Feng X, Jiao B, Xu M, Plikus MV, Dai X, Lengner C, Cui W, Ren F, Shuai J, Millar SE, Yu Z. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat Commun. 2017;8(1):1036.
Harrison H, Farnie G, Brennan KR, Clarke RB. Breast cancer stem cells: something out of notching? Cancer Res. 2010;70:8973–6.
Hoey T, Yen WC, Axelrod F, et al. DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell. 2009;5:168–77.
Zheng A, Song X, Zhang L, Zhao L, Mao X, Wei M, Jin F. Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J Exp Clin Cancer Res. 2019;38(1):305.
El HR, Pinna G, Cabaud O, et al. miR-600 acts as a bimodal switch that regulates breast cancer stem cell fate through WNT signaling. Cell Rep. 2017;18:2256–68.
Drago-Ferrante R, Pentimalli F, Carlisi D, et al. Suppressive role exerted by microRNA-29b-1-5p in triple negative breast cancer through SPIN1 regulation. Oncotarget. 2017;8:28939–58.
Liu B, Du R, Zhou L, Xu J, Chen S, Chen J, Yang X, Liu DX, Shao ZM, Zhang L, Yu Z, Xie N, Guan JL, Liu S. miR-200c/141 regulates breast cancer stem cell heterogeneity via targeting HIPK1/β-catenin axis. Theranostics. 2018;8(21):5801–13.
Liu F, Liu Y, Shen J, Zhang G, Han J. MicroRNA-224 inhibits proliferation and migration of breast cancer cells by down-regulating Fizzled 5 expression. Oncotarget. 2016;7(31):49130–42.
Isobe T, Hisamori S, Hogan DJ, Zabala M, Hendrickson DG, Dalerba P, Cai S, Scheeren F, Kuo AH, Sikandar SS, Lam JS, Qian D, Dirbas FM, Somlo G, Lao K, Brown PO, Clarke MF, Shimono Y. miR-142 regulates the tumorigenicity of human breast cancer stem cells through the canonical WNT signaling pathway. Elife. 2014;3:e01977.
Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106.
McGowan PM, Simedrea C, Ribot EJ, et al. Notch1 inhibition alters the CD44hi/CD24lo population and reduces the formation of brain metastases from breast cancer. Mol Cancer Res. 2011;9:834–44.
Kang L, Mao J, Tao Y, et al. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci. 2015;106:700–8.
Zhang H, Li N, Zhang J, Jin F, Shan M, Qin J, Wang Y. The influence of miR-34a expression on stemness and cytotoxic susceptibility of breast cancer stem cells. Cancer Biol Ther. 2016;17(6):614–24.
Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, Maru DM, Hawke DH, Rak J, Mani SA, Zweidler-McKay P, Ellis LM. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23(2):171–85.
Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, Yang JY, Chang CJ. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Investig. 2014;124(7):3093–106.
Li CY, Miao KL, Chen Y, Liu LY, Zhao GB, Lin MH, Jiang C. Jagged2 promotes cancer stem cell properties of triple negative breast cancer cells and paclitaxel resistance via regulating microRNA-200. Eur Rev Med Pharmacol Sci. 2018;22(18):6008–14.
Mohammadi-Yeganeh S, Mansouri A, Paryan M. Targeting of miR9/NOTCH1 interaction reduces metastatic behavior in triple-negative breast cancer. Chem Biol Drug Des. 2015;86(5):1185–91.
Jacobsson H, Harrison H, Hughes É, Persson E, Rhost S, Fitzpatrick P, Gustafsson A, Andersson D, Gregersson P, Magnusson Y, Ståhlberg A, Landberg G. Hypoxia-induced secretion stimulates breast cancer stem cell regulatory signalling pathways. Mol Oncol. 2019;13(8):1693–705.
Leeman RJ, Lui VWY, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6(3):231–41.
Qin JJ, Yan L, Zhang J, Zhang WD. STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review. J Exp Clin Cancer Res. 2019;38(1):195.
Liu S, Patel SH, Ginestier C, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8:e1002751.
Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32(11):2858–68.
Muhammad N, Bhattacharya S, Steele R, Ray RB. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7(36):58595–605.
Zhang L, Li J, Wang Q, Meng G, Lv X, Zhou H, Li W, Zhang J. The relationship between microRNAs and the STAT3-related signaling pathway in cancer. Tumour Biol. 2017;39(7):1010428317719869.
Piasecka D, Braun M, Kordek R, Sadej R, Romanska H. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol. 2018;144(8):1401–11.
Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700.
Han M, Wang Y, Liu M, Bi X, Bao J, Zeng N, Zhu Z, Mo Z, Wu C, Chen X. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1α expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 2012;103(6):1058–64.
Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31(50):5162–71.
Li B, Lu Y, Wang H, et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother. 2016;79:93–101.
Bahena-Ocampo I, Espinosa M, Ceballos-Cancino G, Lizarraga F, Campos-Arroyo D, Schwarz A, Garcia-Lopez P, Maldonado V, Melendez-Zajgla J. miR-10b expression in breast cancer stem cells supports self-renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016;17(7):1081.
Yang Z, Han Y, Cheng K, et al. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587–95.
Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–57.
Li N, Xie C, Lu N. Crosstalk between Hippo signalling and miRNAs in tumour progression. FEBS J. 2017;284(7):1045–55.
Nandy SB, Arumugam A, Subramani R, Pedroza D, Hernandez K, Saltzstein E, Lakshmanaswamy R. MicroRNA-125a influences breast cancer stem cells by targeting leukemia inhibitory factor receptor which regulates the Hippo signaling pathway. Oncotarget. 2015;6(19):17366–78.
Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63.
Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J, Srovnal J, Hajduch M, Murray P, Kolar Z. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55.
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27.
Huang X, Tang F, Weng Z, Zhou M, Zhang Q. MiR-591 functions as tumor suppressor in breast cancer by targeting TCF4 and inhibits Hippo-YAP/TAZ signaling pathway. Cancer Cell Int. 2019;19:108.
Flanagan L, Kehoe J, Fay J, Bacon O, Lindner AU, Kay EW, Deasy J, McNamara DA, Prehn JH. High levels of X-linked inhibitor-of-apoptosis protein (XIAP) are indicative of radio chemotherapy resistance in rectal cancer. Radiat Oncol. 2015;10:131.
Wang X, Wang X, Gu J, Zhou M, He Z, Wang X, Ferrone S. Overexpression of miR-489 enhances efficacy of 5-fluorouracil-based treatment in breast cancer stem cells by targeting XIAP. Oncotarget. 2017;8(69):113837–46.
Roscigno G, Puoti I, Giordano I, Donnarumma E, Russo V, Affinito A, Adamo A, Quintavalle C, Todaro M, Vivanco MD, Condorelli G. MiR-24 induces chemotherapy resistance and hypoxic advantage in breast cancer. Oncotarget. 2017;8(12):19507–21.
Hsieh TH, Hsu CY, Tsai CF, Long CY, Wu CH, Wu DC, Lee JN, Chang WC, Tsai EM. HDAC inhibitors target HDAC5, upregulate microRNA-125a-5p, and induce apoptosis in breast cancer cells. Mol Ther. 2015;23(4):656–66.
Sun X, Li Y, Zheng M, Zuo W, Zheng W. MicroRNA-223 increases the sensitivity of triple-negative breast cancer stem cells to TRAIL-induced apoptosis by targeting HAX-1. PLoS One. 2016;11(9):e0162754.
Mandujano-Tinoco EA, Garcia-Venzor A, Muñoz-Galindo L, Lizarraga-Sanchez F, Favela-Orozco A, Chavez-Gutierrez E, Krötzsch E, Salgado RM, Melendez-Zajgla J, Maldonado V. miRNA expression profile in multicellular breast cancer spheroids. Biochim Biophys Acta Mol Cell Res. 2017;1864(10):1642–55.
Rahimi M, Sharifi-Zarchi A, Firouzi J, Azimi M, Zarghami N, Alizadeh E, Ebrahimi M. An integrated analysis to predict micro-RNAs targeting both stemness and metastasis in breast cancer stem cells. J Cell Mol Med. 2019;23(4):2442–56.
Tsai-Tsen L, Muh-Hwa Y. Revisiting epithelial–mesenchymal transition in cancer metastasis: the connection between epithelial plasticity and stemness. Mol Oncol. 2017;11(7):792–804.
Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial–mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–4.
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601.
Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907.
Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res. 2008;68:7846–54.
Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–9.
Kurashige J, Kamohara H, Watanabe M, Hiyoshi Y, Iwatsuki M, Tanaka Y, Kinoshita K, Saito S, Baba Y, Baba H. MicroRNA-200b regulates cell proliferation, invasion, and migration by directly targeting ZEB2 in gastric carcinoma. Ann Surg Oncol. 2012;19:S656–64.
Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell. 2010;39(5):761–72.
Duffy MJ, Synnott NC, McGowan PM, Crown J, O’Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014;40(10):1153–60.
Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–10.
Avtanski DB, Nagalingam A, Tomaszewski JE, Risbood P, Difillippantonio MJ, Saxena NK, Malhotra SV, Sharma D. Indolo-pyrido-isoquinolin based alkaloid inhibits growth, invasion and migration of breast cancer cells via activation of p53-miR34a axis. Mol Oncol. 2016;10(7):1118–32.
Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–5.
Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48.
Lee JT, Gu W. SIRT1: regulator of p53 deacetylation. Genes Cancer. 2013;4:112–7.
Wang ZQ, Chen WY. Emerging roles of SIRT1 in cancer drug resistance. Genes Cancer. 2013;4:82–90.
Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–5.
Ma W, Xiao GG, Mao J, Lu Y, Song B, Wang L, Fan S, Fan P, Hou Z, Li J, Yu X, Wang B, Wang H, Wang H, Xu F, Li Y, Liu Q, Li L. Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget. 2015;6(12):10432–44.
Dimri M, Kang M, Dimri GP. A miR-200c/141-BMI1 autoregulatory loop regulates oncogenic activity of BMI1 in cancer cells. Oncotarget. 2016;7(24):36220–34.
Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Investig. 2005;115:1503–21.
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603.
Yin J, Zheng G, Jia X, Zhang Z, Zhang W, Song Y, Xiong Y, He Z. A Bmi1-miRNAs cross-talk modulates chemotherapy response to 5-fluorouracil in breast cancer cells. PLoS One. 2013;8(9):e73268.
Dimri M, Cho JH, Kang M, Dimri GP. PLK1 inhibition down-regulates polycomb group protein BMI1 via modulation of the miR-200c/141 cluster. J Biol Chem. 2015;290(5):3033–44.
Ke J, Zhao Z, Hong SH, Bai S, He Z, Malik F, Xu J, Zhou L, Chen W, Martin-Trevino R, Wu X, Lan P, Yi Y, Ginestier C, Ibarra I, Shang L, McDermott S, Luther T, Clouthier SG, Wicha MS, Liu S. Role of microRNA221 in regulating normal mammary epithelial hierarchy and breast cancer stem-like cells. Oncotarget. 2015;6(6):3709–21.
Lee S, Hong S, Kim S, Kang S. Ataxin-1 occupies the promoter region of E-cadherin in vivo and activates CtBP2-repressed promoter. Biochim Biophys Acta. 2011;1813:713–22.
Klement IA, Skinner PJ, Kaytor MD, Yi H, Hersch SM, Clark HB, Zoghbi HY, Orr HT. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53.
Moody SE, Perez D, Pan TC, Sarkisian CJ, Portocarrero CP, Sterner CJ, Notorfrancesco KL, Cardiff RD, Chodosh LA. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197–209.
Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–28.
Reddy SD, Gajula RP, Pakala SB, Kumar R. MicroRNAs and cancer therapy: the next wave or here to stay? Cancer Biol Ther. 2010;9(7):479–82.
Ishida M, Selaru FM. miRNA-based therapeutic strategies. Curr Pathobiol Rep. 2013;1(1):63–70.
Loh HY, Norman BP, Lai KS, Rahman NM, Alitheen NBM, Osman MA. The regulatory role of MicroRNAs in breast cancer. Int J Mol Sci. 2019;20(19):E4940.
Mayoral-Varo V, Calcabrini A, Sánchez-Bailón MP, Martín-Pérez J. miR205 inhibits stem cell renewal in SUM159PT breast cancer cells. PLoS One. 2017;12(11):e0188637.
Sun X, Fan C, Hu LJ, Du N, Xu CW, Ren H. Role of let-7 in maintaining characteristics of breast cancer stem cells. Xi Bao Yu Fen ZiMian Yi XueZaZhi. 2012;28(8):789–92.
Deng ZQ, Yin JY, Tang Q, Liu FQ, Qian J, Lin J, Shao R, Zhang M, He L. Over-expression of miR-98 in FFPE tissues might serve as a valuable source for biomarker discovery in breast cancer patients. Int J Clin Exp Pathol. 2014;7(3):1166–71.
Li X, Xie W, Xie C, Huang C, Zhu J, Liang Z, Deng F, Zhu M, Zhu W, Wu R, Wu J, Geng S, Zhong C. Curcumin modulates miR-19/PTEN/AKT/p53 axis to suppress bisphenol A-induced MCF-7 breast cancer cell proliferation. Phytother Res. 2014;28(10):1553–60.
Guttilla IK, White BA. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem. 2009;284(35):23204–16.
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, Zur Hausen A, Brunton VG, Morton J, Sansom O, Schüler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–95.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
None.
Conflict of Interest
The authors (PKD, AS, SYA, SA, AR, JAK, SP and FI) declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Das, P.K., Siddika, M.A., Asha, S.Y. et al. MicroRNAs, a Promising Target for Breast Cancer Stem Cells. Mol Diagn Ther 24, 69–83 (2020). https://doi.org/10.1007/s40291-019-00439-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-019-00439-5