Abstract
Objectives
Antimicrobial resistance (AMR) represents a significant threat to patient and population health. The study aim was to develop and validate a model of AMR that defines and quantifies the value of new antibiotics.
Methods
A dynamic disease transmission and cost-effectiveness model of AMR consisting of three components (disease transmission, treatment pathway and optimisation) was developed to evaluate the health economic value of new antibiotics. The model is based on the relationship between AMR, antimicrobial availability and consumption. Model analysis explored the impact of different antibiotic treatment strategies on the development of AMR, patient and population estimates of health benefit, across three common treatment indications and pathogens in the UK.
Results
Population-level resistance to existing antimicrobials was estimated to increase from 10.3 to 16.1% over 10 years based on current antibiotic availability and consumption. In comparison, the diversified use of a new antibiotic was associated with significant reduction in AMR (12.8% vs. 16.1%) and quality-adjusted life year (QALY) gains at a patient (7.7–10.3, dependent on antimicrobial efficacy) and population level (3657–8197, dependent on antimicrobial efficacy and the prevalence of AMR). Validation across several real-world data sources showed that the model output does not tend to systematically under- or over-estimate observed data.
Conclusions
The development of new antibiotics and the appropriate use of existing antibiotics are key to addressing the threat of AMR. This study presents a validated model that quantifies the value of new antibiotics through clinical and economic outcomes of relevance, and accounts for disease transmission of infection and development of AMR. In this context, the model may be a useful tool that could contribute to the decision-making process alongside other potential models and expert advice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The prevalence of antimicrobial resistance (AMR) is increasing worldwide as a result of the misuse of antimicrobials and poor infection control practices, posing a serious threat to public health. The development of new antimicrobials and the appropriate use of existing antimicrobials are key to combating AMR. |
Progress in the development of new antimicrobials and appropriate use of existing antimicrobials can be supported by valuation frameworks and models that reflect the potential patient and population benefits of antimicrobials. This study presents a simplified validated model of AMR and outcomes that aims to quantify the potential value of new antimicrobials, accounting for transmission of infection and development of AMR. In this context, the model is designed to be a useful tool to inform clinical and policy decision-making, whereby it could contribute to the process alongside other potential models (which may use other methodology and/or provide somewhat different predictions) and expert advice. |
Model outputs emphasise the importance of stewardship strategies that allow infections to be treated effectively and, at the same time, manage resistance within the pool of available antimicrobials using diversity or cycling treatment strategies. The introduction of a new antimicrobial as part of a diversity approach offered the best balance between reducing infection and preventing the development of resistance, and was associated with significant quality-adjusted life year (QALY) gains at a patient and population level. |
1 Introduction
The prevalence of antimicrobial resistance (AMR) is increasing worldwide as a result of the misuse of antimicrobials and poor infection control practices [1, 2], posing a serious threat to public health. It is estimated that if no action is taken to optimise the use of current antimicrobials and accelerate research and development of new ones, by 2050, AMR could result globally in 10 million deaths per year and cumulatively incur at least US$100 trillion in hospital costs and productivity losses [3].
The 2015 Global Action Plan on AMR from the World Health Organization proposed that national and local stewardship programmes should be developed to monitor and promote appropriate antimicrobial use [4]. In the UK, there has been a government-commissioned review on AMR [5], and the National Institute for Health and Care Excellence (NICE) have issued guidelines on antimicrobial stewardship [6, 7]. However, despite the strong recommendations made for securing new drugs for future generations, progress in the development of new antimicrobials and appropriate use of existing antimicrobials may be thwarted unless supported by valuation frameworks and models that reflect the potential patient and population benefits of antimicrobials.
A challenge in demonstrating the clinical and cost-effectiveness of new antimicrobials concerns the available clinical data that would inform health technology assessment (HTA). In clinical trials, antimicrobials are typically assessed for non-inferiority, rather than superiority, in comparison with existing treatments [8]. Therefore, they may not prove cost-effective in traditional analyses, where the new antimicrobial is associated with an additional cost and a comparable health outcome. However, the value of antimicrobials goes beyond the demonstration of non-inferiority at patient-level efficacy and includes the population-level impact on AMR. The appropriate assessment of antimicrobial value has been discussed in detail in recent publicly available reports from the Office of Health Economics (OHE) [8] and the Policy Research Unit in Economic Evaluation of Health and Care Interventions Report (EEPRU) [9]. Briefly, the value of new antimicrobials to patients and health services is broad and may include the following: transmission value (the benefits of avoiding infection spread), insurance value (the benefits of having a treatment available as an insurance against future outbreaks), diversity value (the benefits of having multiple antibiotics available that may be used within treatment strategies aiming to reduce selection pressure and minimise resistance development), spectrum value (the benefits of replacing broad-spectrum antibiotics with narrow-spectrum ones that reduce collateral damage to the microbiome and minimise opportunities for resistant organisms to thrive), novel action value (the benefits associated with a new mechanism of action or structure, which may allow avoiding cross-resistance and boost the development of follow-on drugs, increasing diversity), and enablement value (the benefits associated with antibiotic use in the setting of surgical or medical procedures, which could not be safely undertaken were effective antibiotics not available to prevent or treat surgical site or post-procedure infections) [8, 9].
Reimbursement frameworks and health economic valuation methods should reflect the full value of new antimicrobials to patients and populations in order to promote antimicrobial development and stewardship. While frameworks for reimbursement of antimicrobials, including a de-linked payment model, have been discussed in the EEPRU report [9], they are beyond the scope of this publication. The broader aim of this study was to develop, build and validate a novel, dynamic, disease transmission and cost-effectiveness model of antibiotic treatment and resistance. Our aim was to capture the salient features defining AMR and outcomes in a simplified model to assess the potential value of the new antibiotics, accounting for transmission of infection and development of resistance, as well as antibiotic availability, efficacy and patterns of use. The model has the potential to be used both in the valuation of new antimicrobials and to support informed clinical and policy decision-making concerning the utilisation of new and existing antibiotics. While the model is capable of evaluating individual antibiotics based on their effectiveness, this paper presents an analysis that aimed to estimate the value—independent of a particular treatment—associated with the availability and appropriate consumption of new and existing antimicrobials.
2 Methods
2.1 Model Structure
2.1.1 Overview
A novel, dynamic, disease transmission and cost-effectiveness model was developed to support the valuation of antimicrobial treatment and resistance. This deterministic open-cohort model is informed by the findings of a recent systematic review by Drake et al. [10] and best practice recommendations from the International Society for Pharmacoeconomics and Outcomes Research (ISPOR) [11]. Best practice recommendations support the use of a dynamic model, as the introduction of new antimicrobials or changing the pattern of use of existing antimicrobials was anticipated to impact transmission within the modelled population and alter the distribution of patients infected with pathogens resistant or sensitive to treatment. Furthermore, modelled populations and subgroups were large, and as such, approximating a systems average behaviour was deemed more appropriate than a patient-level stochastic model.
In accordance with ISPOR best practice recommendations for model conceptualisation [12], clinical and methodological experts were involved in the design of the model to ensure that it captured the relevant dynamics of infection transmission and antimicrobial use, and that model inputs and data sources were appropriate. The experts were selected by the study sponsor based on their subject knowledge and experience across different aspects of the project and specialised in the fields of microbiology, infection control and modelling. Expert consultations took the form of unstructured group discussions, so as not to restrict the experts in expressing their opinion and judgment. No formal voting process was in place, and any disagreements were resolved by consensus. Guidance was provided through an iterative qualitative approach to model development and interpretation to ensure the model was methodologically appropriate and included all important aspects of transmission of infection, resistance development and patient management. Specifically, the experts were requested to comment on the appropriateness of model conceptualisation and completeness of the captured infection transmission/AMR components, assess model inputs and their application and guide research questions. Recommendations from clinical and methodological experts were incorporated within the model, and the changes were subsequently fed back to the experts. This process of refining the model based on expert guidance and re-assessment was repeated until the experts concluded that the final model captured the desired components appropriately and robustly, and that model output had clinical validity.
The model concept and structure are generic and can be parameterised to reflect different indications and pathogens of interest, investigating up to three lines of antimicrobial therapy and different treatment strategies. Model outputs reflect deterministic applications of the model (e.g. use of a new antimicrobial at first-line, last-line, or a mixing/diversity strategy), and an optimisation procedure allows ‘policy goals’ to be specified, such that a model solution (treatment strategy) is identified that minimises or maximises the policy goal (e.g. minimise overall resistance, maximise life years) subject to constraints on the system (e.g. overall costs do not increase, deaths do not increase). Analysis may be conducted from the perspective of a single healthcare provider, or from a national perspective accounting for heterogeneity in infection and antimicrobial incidence across the country.
The core model structure is based on interrelated disease transmission model and treatment pathway components. Additionally, model analysis is supported by a novel application of constrained optimisation that can identify and evaluate treatment strategies that satisfy antimicrobial policy objectives subject to relevant constraints on model input values. All model components were developed within Microsoft Excel.
2.1.2 Disease Transmission Component
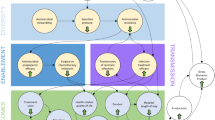
A compartmental multi-state disease transmission model estimates the prevalence of bacterial infections within a healthcare environment based on patient interactions and exposure to antimicrobials. During each model cycle, patients may move between discrete health states representing colonised, infected or susceptible health states, or death. Transitions between health states are controlled by the incidence and prevalence of infection and colonisation within the modelled environment, described using a series of difference equations. These transition parameters were derived from data reported by the English Surveillance Programme for Antimicrobial Utilisation and Resistance (ESPAUR) [13] and the Public Health Profiles Fingertips tool [14] published by Public Health England, which provides local indicators of AMR and infection incidence. Due to the lack of available published evidence, model inputs describing infection transmission dynamics and resistance development were derived empirically through calibration, such that the model reproduced observed data from Public Health England describing infection incidence and resistance development over time [13, 14]. These model settings are further discussed and summarised in the Electronic Supplementary Material (Table S1 and S2), and the model structure is presented in Fig. 1.
Overview of the model structure. a Disease transmission flow diagram and b treatment decision pathway. Upon becoming infected (a), patients enter the treatment pathway (shown in b). The dotted line and arrow around panel a highlight the interaction between the disease transmission diagram and the treatment pathway, while the dotted arrow in panel b allows patients to re-enter the treatment pathway of a subsequent treatment line (up to three) as required
2.1.3 Treatment Pathway Component
The multi-state disease transmission module is linked to a treatment pathway determining the health economic impact of a specified treatment strategy in the context of the modelled infectious environment. Patients in the infected health state receive antimicrobial treatment, described by an antimicrobial treatment pathway that represents up to three lines of treatment, where patients are either cured (successfully treated or infection resolves spontaneously) or die from infection. Subsequent lines of treatment are received only if a treatment is unsuccessful. As a result, patients’ progression through the treatment pathway and exposure to antimicrobials is a function of treatment efficacy (the likelihood that treatment will be successful conditional on susceptibility at initiation) and the prevalence of AMR in the infected population, with high efficacy or low resistance leading to more effective treatment and improved patient outcomes.
The treatment pathway component of the model allows for flexible treatment algorithms where different treatment pathways may be specified, including the number of treatment lines available or the order in which treatments are received. As such, the model can assess not only the impact of differing antimicrobial efficacy, but also that of various stewardship strategies. Furthermore, the effect of treatment mixing on outcomes may be examined, where combinations of different antimicrobial treatment pathways are used as part of (1) a diversity-based strategy, where multiple antimicrobials are used in the same line of the treatment pathway, (2) an antimicrobial cycling strategy where the treatment pathway is altered periodically, or (3) a combination of both.
The principle supporting treatment mixing is that the more frequently an antimicrobial agent is prescribed, the more likely it is that resistance to it will develop. Withdrawal, or reductions in use, of an antimicrobial agent will limit the selective pressures exerted by those agents (and hence the emergence of resistance), maintaining a higher relative efficacy than could have been expected if the antimicrobial had been in use continuously.
2.1.4 Optimisation Component
Combinatorial and mathematical optimisation can be used when the complexity of a problem makes a complete enumeration of the parameter space unfeasible. The optimisation module achieves this by changing input variables (within constraints) to produce a candidate solution, then using heuristic [15] or linear [16] programming methods to decide if it should replace the current solution with a new one, with the goal of achieving the best possible solution to the problem.
The optimisation module is used to describe the minimisation/maximisation of an objective function (e.g. treatment/policy objective) subject to relevant constraints (e.g. resistance, available treatments). Optimal solutions are sought using Microsoft Excel’s built in Solver application, with users able to select the optimisation algorithm utilised (generalised reduced gradient non-linear, simplex linear-programming, evolutionary). Given the non-linear nature of the model and the incorporation of integer variables (e.g. treatment sequence), the evolutionary method is employed as the default settings.
2.2 Resistance Gain and Loss
The development of resistance is a core feature of the model and is primarily a function of treatment exposure and the number of resistant patients at each model cycle. Each day a patient receives a given antimicrobial treatment, the exposed pathogen has a probability of developing resistance to that treatment. As such, the development of resistance is time-updated as a function of the number of patients requiring treatment, the antimicrobials used, and treatment efficacy. In the model, a resistant infection can arise via two mechanisms: resistance can be acquired during the course of an infection or a patient may become infected with an already resistant pathogen. The inclusion of de novo resistance development in addition to transmission of resistant pathogens is in line with previous work in the field [17]. Resistant infections acquired via either mechanism can be transmitted to others in the hospital cohort. There are two mechanisms by which patients may lose resistance. The first is following successful treatment, where a proportion of patients can enter the susceptible health state. The second mechanism is a consequence of the fitness cost. Mutations conferring antibiotic resistance can affect important cellular processes, rendering them less efficient and therefore impacting the “fitness” of the microorganism, i.e. its ability to survive and replicate in a given environment. In the presence of an antibiotic, resistant strains have a competitive advantage over susceptible strains, but when selection pressure from the antibiotic is removed (e.g. antibiotics are switched as part of a cycling treatment strategy), resistant pathogens may be outcompeted by susceptible organisms with improved fitness. As the model has been developed at a population level, it does not use a mechanistic approach to resistance loss over time as a result of selection pressure; instead, resistant infections and colonisation become sensitive to treatment at a constant rate.
2.3 Outputs
The model estimates infection incidence, life years and quality-adjusted life years (QALYs) lost due to infection, infection incidence, patient mortality due to infection and AMR levels. In addition, it provides standard health economic information, including estimates of total and incremental treatment and disease-related costs, cost per life year lost and cost per QALY lost.
In the context of this disease transmission model, probabilistic sensitivity analysis may produce spurious model outputs as the majority of model inputs are derived based on empirical calibration to observed data. As a result, no associated estimates of uncertainty or covariance are produced that would enable robust sensitivity analyses. When input parameters and associated uncertainty are unbounded, the model may result in uninformative or misleading conclusions. Given the number of input parameters and inter-dependencies, a systematic, transparent and justified approach was not considered achievable; therefore, probabilistic analysis was not undertaken; however, extensive scenario analysis is supported by the model to characterise the consequences associated with different permutations of input parameters and models settings. This approach to sensitivity analysis was considered to align to the relevant clinical practice scenarios and policy goals that would drive evaluations performed with this model.
2.4 Model Settings
The disease transmission model utilises up to a 10-year time horizon with a monthly cycle to evaluate model outputs. The model cycle length and time horizon were informed by a previously published antimicrobial cost-effectiveness evaluation [18] and expert opinion. Since the value of a new antimicrobial becomes more apparent as the time horizon is extended, a 10-year horizon was selected to balance the inherent uncertainty associated with long-term extrapolation and the underestimation of value that would arise from a short time horizon. Due to the monthly cycle length, all infections are assumed to be resolved within the incident cycle. Patients who die due to infection incur a loss of life expectancy and quality-adjusted life expectancy corresponding to that of the general population. Capturing life years and QALYs lost due to unsuccessful treatment allows the model to quantify the impact of infections avoided in addition to the benefits of successful treatment.
The model was populated with data reflective of the UK setting and adopted a UK payer (National Health Service [NHS]) perspective, considering direct healthcare resource utilisation and health effects. Model inputs reflect a hospitalised adult population with one of the following healthcare-associated infections (HCAI): complicated urinary tract infections (cUTI), complicated intra-abdominal infections (cIAI), or hospital-acquired pneumonia (HAP) (including ventilator-associated pneumonia [VAP]). For each indication, patients were infected with one of three bacterial pathogens: Escherichia coli, Klebsiella pneumoniae or Pseudomonas aeruginosa; within the disease transmission model, pathogens are further stratified by resistance status (susceptible or resistant). Baseline pathogen resistance levels were sourced from ESPAUR [13], with a full description of the baseline distribution of patients across the susceptible, colonised and infected health states stratified by pathogen and indication, and baseline resistance levels. These data are reported in Table S3 (see the Electronic Supplementary Material).
2.5 Model Validation
The model was subject to external validation exercises to demonstrate the rigour, relevance and value of the model in the context of real-world evidence. External validation exercises compared outputs from the model to publicly available data not specifically used to construct the disease transmission component, to verify the accuracy of projections for resistance. For each validation exercise, model transition parameters were calibrated to a training set of time-series data describing the prevalence of AMR; model performance was evaluated on a subsequent 3-year period comparing observed and modelled predictions. A 3-year period was chosen for evaluation of model validation results based on a qualitative assessment of the availability of validation data; a compromise was made with respect to the amount of time series data that could be used to calibrate the model while also leaving enough data to assess model goodness of fit. The specific validation studies included in this analysis were based on data reported by the British Society for Antimicrobial Chemotherapy (BSAC) [19], European Centre for Disease Prevention and Control (ECDC) [20], Public Health Profiles Fingertips tool [14], English Surveillance Programme for Antimicrobial Utilisation and Resistance (ESPAUR) [13], Public Health Wales (PHW) [21], Scottish Antimicrobial Prescribing Group (SAPG) [22] and Antimicrobial Testing Leadership and Surveillance (ATLAS) [23] and included the pathogens E. coli, K. pneumoniae and P. aeruginosa and the antimicrobials piperacillin/tazobactam, colistin, tigecycline and carbapenems. Mean absolute error (MAE) was used to assess goodness of fit between predicted and observed values, calculated by comparing predicted resistance levels from the model with observed resistance levels reported in the identified data sources. Scatter plots of observed versus predicted endpoints are presented along with the coefficient of determination (R2) and the results of linear regression analysis. We performed formal statistical null hypothesis testing to assess whether the model predictions are a reasonable approximation of observed data. Statistical significance was determined to be p < 0.05.
2.6 Scenario Analysis
The impact of different treatment strategies on the development of AMR to existing antimicrobials was explored, and patient and population outcomes were estimated in terms of mortality, QALYs, and healthcare resource utilisation. The antimicrobials considered in the analysis were piperacillin/tazobactam for the treatment of cUTI and cIAI, colistin/tigecycline for the treatment of HAP/VAP, meropenem and a theoretical new antimicrobial for the treatment of all indications. The treatment strategies considered were as follows:
-
1.
An extrapolation of the most probable current prescribing patterns, to be used as a reference point for the impact of other treatment strategies. This strategy utilised a treatment pathway of piperacillin/tazobactam (cUTI and cIAI) or colistin/tigecycline (HAP/VAP) at first line, with meropenem reserved for patients requiring a second line of treatment, due to the growing public health concerns around resistance to carbapenems [24].
-
2.
The addition of a new antimicrobial to be used at last line in the empirical treatment pathway, following treatment failures with piperacillin/tazobactam or colistin/tigecycline and meropenem.
-
3.
The addition of a new antimicrobial to be used at first line in the empirical treatment pathway. Infections not cured after treatment with the new antimicrobial receive piperacillin/tazobactam or colistin/tigecycline followed by meropenem.
-
4.
The addition of a new antimicrobial as part of a first-line diversity strategy, where half of newly infected patients receive the new antimicrobial at first line, and half receive piperacillin/tazobactam or colistin/tigecycline. Patients failing treatment at first line receive the other first-line treatment (i.e. infections not resolved by treatment with the antimicrobial may receive piperacillin/tazobactam at second line or vice versa), with meropenem available as a last-line therapy.
-
5.
The addition of the new antimicrobial as part of an all-line diversity strategy, where infected patients are treated in equal proportions with the new antimicrobial, piperacillin/tazobactam or colistin/tigecycline, and meropenem, with no patient receiving the same antimicrobial twice.
-
6.
The addition of the new antimicrobial as part of a cycling strategy, where empirical treatment pathways of either (1) the new antimicrobial followed by piperacillin/tazobactam or colistin/tigecycline and finally meropenem or (2) piperacillin/tazobactam or colistin/tigecycline followed by the new antimicrobial and finally meropenem are rotated every 12 months.
These treatment strategies were evaluated in modelled populations, with AMR prevalence levels of 0–30% at baseline and 60–100% efficacy of the new antibiotic in patients infected with susceptible microorganisms based on expert opinion of plausible ranges. Scenarios focused on antimicrobial stewardship (scenarios 4–6) were conducted in patient populations where all infectious microorganisms were resistant to one or more of the existing antibiotics. All scenarios utilised the same transition parameters; only starting resistance, treatment efficacy and treatment pathways were varied.
3 Results
3.1 Validation
The model was subject to external validation, the results of which are described in detail in the Electronic Supplementary Material. The overall R2 statistic (when measures of goodness of fit from different data sources, antimicrobials, and pathogens were combined) was 0.82, indicating a high degree of linear correlation and resulting in an overall MAE of 1.34 percentage points; however, the accuracy of model predictions varied somewhat between data sources, antibiotics and pathogens (Table S4 and Fig. S1, see the Electronic Supplementary Material). Linear regression analysis (Table S5) revealed that model output does not tend to systematically under- or over-estimate observed data.
3.2 Scenario Analysis
Figure 2 shows the development of AMR levels over time for scenarios incorporating the new antimicrobial as a third- and first-line treatment, and as part of both diversity- and cycling-based stewardship strategies, in comparison to forecast resistance development when maintaining the current antimicrobial prescribing strategy. Assuming no change in the current antimicrobial prescribing strategy, the proportion of infections caused by a pathogen resistant to one or more of the existing antimicrobials was forecasted to reach 16.1% over the next 10 years, or an increase of 5.8 percentage points from resistance levels today. Utilising the new antimicrobial at third line is predicted to increase resistance further by 0.4 percentage points to 16.5% total, while reducing exposure to existing antimicrobials in the first line, first-line diversity, all-line diversity, and first-line cycling scenarios may see resistance levels reduce over the next 10 years by 6.4 percentage points, 3.3 percentage points, 2.5 percentage points and 2.5 percentage points, respectively. In contrast, strategies that minimised resistance to the existing antimicrobials tended to see the most rapid increase in resistance to the new antimicrobial. Figure 3 shows resistance to the new antimicrobial increasing to 10.1% over the 10-year transmission horizon for the first-line treatment strategy, 1.8% for the third-line treatment strategy and to 7.4%, 6.1% and 6.6% for the first-line diversity, all-lines diversity and cycling stewardship strategies, respectively.
Proportion of infections resistant to at least one of the existing antimicrobials over time for strategies assessed through dynamic health economic evaluation. Baseline resistance values are representative of current population resistance to piperacillin/tazobactam (or colistin/tigecycline in patients with hospital-acquired pneumonia) and meropenem in England
A reduction in population resistance levels corresponds with a reduction in patient resistance to infections. As a result, substantial QALY gains are predicted at a patient level in comparison to current antimicrobial use. In those patients who are infected with pathogens resistant to one or more of the existing antimicrobial treatments, the impact of introducing a new antimicrobial as part of a diversity or cycling strategy resulted in QALY gains of between 7.7 and 10.3 QALYs per patient (Fig. 4) corresponding to new antimicrobial efficacy of 70% and 90%, respectively. Furthermore, the impact of reducing resistance to existing antimicrobials by diversifying treatment strategies and having additional treatment options for those patients who are infected with a multidrug resistant pathogen leads to significant QALY gains at a population level, with gains between 3657 and 8197 QALYs, depending on the levels of resistance in the population, the efficacy of the new antimicrobial and the stewardship strategy utilised (Fig. 5).
4 Discussion
We present a novel, dynamic, disease transmission and cost-effectiveness model aiming to address the challenge of appropriate estimation of value associated with new antimicrobials. The model represents a simplified account of the complex relationships between antibiotic availability, utilisation, resistance and consequent clinical and economic outcomes. The model has been subject to external validation exercises based on real-world evidence and extensive expert consultation throughout its conceptualisation, implementation and interpretation, to ensure and demonstrate the rigour, relevance and value of the model. Across several real-world data sources [13, 19,20,21,22], there was a high degree of correlation between model predictions and observed data, supporting the model for the assessment of antimicrobial treatment and resistance.
Population benefits over the 10-year transmission horizon were likely to be maximised by using the new antimicrobial at first line; however, our model predicts that resistance to the new antimicrobial would develop more rapidly than the reduction in resistance to existing antimicrobials, which should preclude the use of a new antimicrobial in first line for extended periods. This emphasises the importance of stewardship strategies that allow infections to be treated effectively and, at the same time, manage resistance within the pool of available antimicrobials. Indeed, model outputs support using diversity or cycling treatment strategies, since the introduction of a new antimicrobial as part of a diversity approach offered the best balance between reducing infection and preventing the development of resistance, and was associated with significant QALY gains at a patient and population level.
There are several limitations in relation to the presented model that should be considered in applications of the model and the interpretation of its results, including the scarcity of data to inform the model development. Similar paucity of data suitable to parametrise and validate AMR models was reported in a recent systematic review [25]. In the case of this model, limited available data did not permit accurate predictions around P. aeruginosa resistance. As a result, the majority of model parameters were required to be derived through calibration to the best available observed evidence; however, model validation confirmed its ability to predict trends and estimates of resistance suggesting that calibration was appropriate in this analysis. However, since the outputs of the model and the estimates of value are conditional upon the input parameters and analysis settings, a remaining challenge is to ensure that the model is initialised and parameterised with data that reflect its future applications and the decision problems that it seeks to address. As new data becomes available, the predictive validity of the model should be assessed and, if needed, the approach to model parametrisation refined to reflect the setting, contemporaneous data and the decision problem being addressed. The model’s sensitivity to input parameters can be explored deterministically for a particular model application and analysis, facilitating transparency in model output for decision-making purposes. However, including probabilistic sensitivity analysis was considered neither practical nor appropriate in the context of this dynamic transmission model, where individual and combinations of inputs may form spurious outputs, with inferences that are not clinically plausible or valid. Although the model is an important step towards including non-traditional value aspects in the assessment of antibiotics, not all non-traditional aspects of antimicrobial value defined by the OHE [8] and EEPRU [9] reports were considered: spectrum value was not captured, as no data from an appropriate setting were available to accurately characterise the relationship between antimicrobial spectrum and resistance development. Similarly, enablement value was not considered, because of the challenges associated with appropriately quantifying the impact of missed surgeries or medical treatments due to infection on costs and health outcomes. The inability to include certain value aspects in the model suggests that it is likely to provide a conservative (and therefore more credible) estimate of the value of new antibiotics. Additionally, the scenarios presented do not necessarily correspond to optimal treatment pathways, rather strategies thought most likely to be implemented in clinical practice; greater benefit could be achieved through the introduction of a new antimicrobial within a strategy that would minimise total resistance gain and overall burden of infection. Furthermore, the model only considered the impact of resistance in the hospital environment and not in the community. We did not determine interactions between different species of pathogens within an indication or between the same pathogen across indications. Therefore, the model may underestimate the rate of resistance spread as well as resistance levels. Also, if resistance to modelled treatments became too high, no effective treatment options remained in the model, which does not adequately reflect real-world scenarios where a broader set of treatment options is available, compared with a maximum of three antimicrobials as assessed in the model. Finally, as with all economic models, the model presented herein is subject to the uncertainty associated with two key factors: (1) extrapolation of outcomes beyond the available data and (2) necessary simplification of the underlying disease pathology and the between-patient variability in natural disease course, response to treatment, mechanisms of treatment failure, and other relevant phenomena.
Whilst it is important to be aware of the limitations and caveats surrounding its use, the dynamic disease transmission cost-effectiveness model presented here addresses a fundamental challenge of appropriately estimating the value of new antimicrobials. We recognise that models are not universally valid, and whilst the current model has undergone extensive validation, it is based upon a simplified representation of the complexities of AMR dynamics; we have aimed for a parsimonious model that can be supported by data and is useful for clinical and policy decision making, alongside other potential models (which may use other methodology and/or provide somewhat different predictions) and expert advice. The face validity of the model is supported by extensive expert clinical opinion during model development and the results of model validation exercises, which support the model’s usefulness as a tool for generating predicted outcomes. We recognise that current projections should be assessed against future data, and future applications of the model should be considered against contemporary data, clinical and policy settings. The appropriateness of the methods, data sources and outputs of this model was confirmed based on proposals within the EEPRU report [9], which advocate that population-level as well as patient-level benefits are captured within a dynamic disease transmission model, development of resistance is incorporated, new antibiotics should be evaluated alongside (not in replacement of) current antimicrobials, and a range of potential treatment strategies are considered. Consequently, the proposed model provides an assessment of potential treatment strategies and outcomes associated with the availability and use of antimicrobials for HCAI in the presence of AMR. Furthermore, the model has flexibility for future applications investigating other scenario analyses not explored in this study. It can be adapted and populated with data for various antimicrobials and can be used to consider a multitude of scenarios, including epidemics, high resistance levels or improved hospital infection control policies, directed compared with empiric treatment. Additionally, the optimisation component can be used to assess how inputs (e.g. treatment pathways/strategies) could be modified to achieve a certain policy goal (e.g. minimise resistance), while incorporating real-world constraints (e.g. no increase in healthcare resource utilisation), thus providing a valuable tool for decision-makers on how public health may be improved in the context of finite resources.
A recent systematic review of population-level mathematical models of AMR identified 117 modelling studies set across a range of pathogen (bacterial, viral, fungal, parasitic) and host (human, animal, plant) types [17]. Among these studies, modifications of treatment strategy were the second most commonly investigated type of intervention aimed at combating AMR (included in 46 studies [39%]), secondary only to improved hygiene and infection control [17]. The same review, however, reported that only 9% of the identified studies included a cost–benefit analysis [17]. The novelty of our study in light of previous research lies therefore in the combination of disease transmission and AMR modelling with a full economic model, capable of quantifying both the clinical and economic value of antibiotics in a transparent and rigorous manner that can support decision-making. Another important advantage of our model is its methodological quality and rigour, including detailed validation. In contrast, a recent systematic review of mathematical models in AMR highlighted insufficient adherence to good modelling practice guidelines among AMR models, including limited or no validation being reported [25].
Importantly, the model described in this study represents a ‘middle ground’ between two other economic modelling exercises in the area of antimicrobials and AMR recently published by EEPRU [9] and the OECD [26]. The EEPRU model [9] includes two components: a mechanistic model of infection transmission dynamics and a Markov model for patient outcomes. The model is based on a small healthcare setting of an ICU with 15 beds and includes a single pathogen (Acinetobacter baumannii) and four treatment scenarios. Outcomes (incremental costs and QALYs) are estimated over a 10-year time horizon from the UK NHS perspective. In contrast, the OECD three-component microsimulation Markov model [26] predicts a range of outcomes (number of deaths, disability-adjusted life years, extra hospital days, healthcare costs at point of delivery and costs of implementing policy programmes) arising from healthcare- and community-acquired infections with eight pathogens. The time horizon of the model spans 35 years, with projections up to 2050 for multiple countries, and rather than evaluating treatment strategies, the analysis is focused on programmes aiming to reduce AMR such as improved hygiene. Consequently, the EEPRU model appears most relevant for antimicrobial value assessments at the local commissioning level, and the OECD model for long-term international policy discussions. The model described in this publication fills the resulting gap for a national-level model which could be used both for estimating the value of antimicrobials for the purpose of reimbursement decisions, and for national-level discussions around optimising antimicrobial stewardship and related drug policies.
5 Conclusions
The development of new antimicrobials and the appropriate use of existing antimicrobials are key to combating AMR. This study presents a validated model that quantifies the value of new antimicrobials through clinical and economic outcomes of relevance, and accounts for transmission of infection and development of AMR. In this context, the model is designed to be a useful tool to inform clinical and policy decision-making, whereby it could contribute to the process alongside other potential models (which may use other methodology and/or provide somewhat different predictions) and expert advice.
References
Mladenovic-Antic S, Kocic B, Velickovic-Radovanovic R, Dinic M, Petrovic J, Randjelovic G, et al. Correlation between antimicrobial consumption and antimicrobial resistance of Pseudomonas aeruginosa in a hospital setting: a 10-year study. J Clin Pharm Ther. 2016;41(5):532–7.
Schwartz KL, Morris SK. Travel and the spread of drug-resistant bacteria. Curr Infect Dis Rep. 2018;20(9):29.
O’Neill J. Tackling drug-resistant infections globally: Final report and recommendations. 2016. https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf. Accessed 22 Feb 2019.
World Health Organization. Global action plan on antimicrobial resistance. 2015. https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/. Accessed 22 Feb 2019.
The Review on Antimicrobial Resistance. Tackling drug-resistant infections globally: final report and recommendations. May 2016. https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf. Accessed 22 Feb 2019.
National Institute for Health and Care Excellence. NICE guideline [NG15]: Antimicrobial stewardship: systems and processes for effective antimicrobial medicine use. 2015. https://www.nice.org.uk/guidance/ng15. Accessed 22 Feb 2019.
National Institute for Health and Care Excellence. NICE guideline [NG63]: Antimicrobial stewardship: changing risk-related behaviours in the general population. 2017. https://www.nice.org.uk/guidance/ng63. Accessed 22 Feb 2019.
Karlsberg Schaffer S, West P, Towse A, Henshall C, Mestre-Ferrandiz J, Masterton R et al. Assessing the Value of New Antibiotics: Additional Elements of Value for Health Technology Assessment Decisions. Office for Health Economics, London. 2017. https://www.ohe.org/news/assessing-value-new-antibiotics-additional-elements-value-health-technology-assessment. Accessed 22 Feb 2019.
Rothery C, Woods B, Schmitt L, Claxton K, Palmer S, Sculpher M. Framework for value assessment of new antimicrobials. implications of alternative funding arrangements for NICE appraisal. Policy Research Unit in Economic Evaluation of Health and Care Interventions. 2018. https://www.eepru.org.uk/wp-content/uploads/2017/11/eepru-report-amr-oct-2018-059.pdf. Accessed 22 Feb 2019.
Drake TL, Devine A, Yeung S, Day NP, White LJ, Lubell Y. Dynamic transmission economic evaluation of infectious disease interventions in low- and middle-income countries: a systematic literature review. Health Econ. 2016;25(Supp 1):124–39.
Pitman R, Fisman D, Zaric GS, Postma M, Kretzschmar M, Edmunds J, et al. Dynamic transmission modeling: a report of the ISPOR-SMDM Modeling Good Research Practices Task Force–5. Value Health. 2012;15(6):828–34.
Roberts M, Russell LB, Paltiel AD, Chambers M, McEwan P, Krahn M. Conceptualizing a model: a report of the ISPOR-SMDM Modeling Good Research Practices Task Force-2. Med Decis Mak Int J Soc Med Decis Mak. 2012;32(5):678–89.
Public Health England. English surveillance programme for antimicrobial utilisation and resistance (ESPAUR) report. 2017. https://www.gov.uk/government/publications/english-surveillance-programme-antimicrobial-utilisation-and-resistance-espaur-report. Accessed 22 Feb 2019.
Public Health England. AMR local indicators - Fingertips profile. 2018. https://fingertips.phe.org.uk/profile/amr-local-indicators. Accessed 22 Feb 2019.
Gendreau M, Potvin J-Y. Metaheuristics in combinatorial optimization. Ann Oper Res. 2005;140(1):189–21313.
Sierksma G, Zwols Y. Linear and integer optimization: theory and practice. Third Edition ed. New York: Chapman and Hall/CRC Press; 2015.
Niewiadomska AM, Jayabalasingham B, Seidman JC, Willem L, Grenfell B, Spiro D, et al. Population-level mathematical modeling of antimicrobial resistance: a systematic review. BMC Med. 2019;17(1):81.
Jansen JP, Kumar R, Carmeli Y. Cost-effectiveness evaluation of ertapenem versus piperacillin/tazobactam in the treatment of complicated intraabdominal infections accounting for antibiotic resistance. Value Health. 2009;12(2):234–44.
The British Society for Antimicrobial Chemotherapy (BSAC). Resistance Surveillance Project. The British Society for Antimicrobial Chemotherapy (BSAC). 2018. https://www.bsacsurv.org/. Accessed 22 Feb 2019.
European Centre for Disease Prevention and Control. Surveillance ATLAS of infectious diseases 2018. https://ecdc.europa.eu/en/surveillance-and-disease-data. Accessed 22 Feb 2019.
Public Health Wales. Antibiotic resistance: AMR Programme Reports. 2018. https://www.wales.nhs.uk/sitesplus/888/page/94136. Accessed 22 Feb 2019.
Scottish Antimicrobial Prescribing Group (SAPG). Scottish Antimicrobial Prescribing Group (SAPG) Report on Antimicrobial Use and Resistance in Humans. 2018. https://www.isdscotland.org/Health-Topics/Prescribing-and-Medicines/SAPG/AMR-Annual-Report/. Accessed 22 Feb 2019.
Pfizer Anti-Infectives. ATLAS - Antimicrobial Testing Leadership and Surveillance. 2018. https://atlas-surveillance.com. Accessed 22 Feb 2019.
Codjoe FS, Donkor ES. Carbapenem Resistance: A Review. Med Sci (Basel). 2017;6(1):1.
Birkegård AC, Halasa T, Toft N, Folkesson A, Græsbøll K. Send more data: a systematic review of mathematical models of antimicrobial resistance. Antimicrob Resist Infect Control. 2018;7:117.
OECD. Stemming the Superbug Tide. 2018. https://doi.org/10.1787/9789264307599-en.
Acknowledgements
The authors would like to thank Professor Mary Slack for providing expert guidance during the model development process and Rachel Russell of Pfizer UK Health & Value for her insights and review of the manuscript. Medical writing and editorial assistance with the development of this publication was provided by Karolina Badora, PhD of Health Economics and Outcomes Research Ltd. and was funded by Pfizer.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Author contributions
All authors contributed to the model conception and design, and to drafting this manuscript. OD developed the model under supervision from JG and PME. Expert guidance was provided by SG, JO and MW throughout the model development process to ensure modelled relationships were appropriate and observed infection dynamics captured. NH provided expert guidance on modelling methods and approaches. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole, and have given final approval to the version to be published. As the corresponding author, JG takes full responsibility for the work as a whole and the decision to submit and publish the manuscript.
Funding
This study was funded by Pfizer.
Conflict of interest
JG, OD and PM are employees of Health Economics and Outcomes Research Ltd., which received funding from Pfizer to undertake the research outlined in this study. AB, AT and CC are employees and stockholders of Pfizer, which provided funding for this study. MH was an employee and stockholder of Pfizer at the time this study was conducted and is now an employee of Johnson & Johnson. ML was an employee and stockholder of Pfizer at the time this study was conducted and is now an employee and stockholder of Moderna Therapeutics. NH received consulting fees related to this study. SG received consulting fees from Astellas, MSD, Shionogi and Pfizer. JO received consulting fees from Pfizer and Gama Healthcare. MW received consulting fees from AiCuris, Astra-Zeneca, Bayer, Cerexa, Durata, The Medicines Company, Menarini, Motif Biosciences, Nabriva, Paratek and Pfizer; lecture fees from Allergan, Astra-Zeneca and Pfizer; and grant support from Motif Biosciences, Nabriva, Paratek, Pfizer, Qpex Biopharma and VenatoRx.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Gordon, J., Darlington, O., McEwan, P. et al. Estimating the Value of New Antimicrobials in the Context of Antimicrobial Resistance: Development and Application of a Dynamic Disease Transmission Model. PharmacoEconomics 38, 857–869 (2020). https://doi.org/10.1007/s40273-020-00906-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40273-020-00906-6