Abstract
Background
SB12 is being developed as a proposed biosimilar to eculizumab reference product (RP), a humanized monoclonal antibody (IgG2/4 kappa immunoglobulin) that binds to the human C5 complement protein. Binding to this protein inhibits complement-mediated intravascular hemolysis by blocking its cleavage into C5a and C5b. Eculizumab RP is indicated for the treatment of patients with paroxysmal nocturnal hemoglobinuria (PNH) to reduce hemolysis, atypical hemolytic uremic syndrome (aHUS) to inhibit complement-mediated thrombotic microangiopathy, generalized myasthenia gravis who are anti-acetylcholine receptor antibody-positive, and neuromyelitis optica spectrum disorder in adult patients who are anti-aquaporin-4 antibody-positive.
Objective
The objective of this study was to demonstrate structural, physicochemical, and biological similarity between eculizumab RP and SB12 using various state-of-the-art analytical methods.
Methods
Comprehensive analytical characterization was conducted with side-by-side comparison of SB12 with European Union (EU) and United States (US) eculizumab RPs using various analytical methods (more than 40 state-of-the-art assays). Comparisons included purity, product-related impurity, charge heterogeneity, primary structure, post-translational modification, higher-order structure, quantity, Fab-related biological activities (potency and binding activity), and Fc-related biological activities.
Results
Based on the analytical similarity assessment, the structural, physicochemical, and biological characterization results demonstrated that SB12 is highly similar to the EU and US eculizumab RP. In the structural aspects, it was confirmed that there is no difference between post-translational modification profiles and higher-order structures of SB12 compared with the eculizumab RP. Product-related impurities in the form of aggregates and charge variants were also confirmed to be similar. Mechanism of action (MoA)-related biological activities showed that SB12 is highly similar to the EU and US eculizumab RP with respect to overall critical and non-critical quality attributes analyzed. Moreover, similarity of comparative binding tendency of SB12 and eculizumab RP to Fc gamma receptors and C1q was confirmed through additional characterization methods. Based on these results, SB12 is expected to have highly similar safety and efficacy compared with eculizumab RP.
Conclusion
In summary, the overall analytical characterization and similarity assessment results show that SB12 is highly similar to the EU and US eculizumab RP in terms of structural, physicochemical, biophysical, and biological attributes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
SB12 has been developed as a proposed biosimilar to the eculizumab reference product. |
SB12 demonstrated high similarity with the European Union and United States eculizumab reference products in terms of structural, physicochemical, and biological properties. |
1 Introduction
SB12 is being developed as a biosimilar of the originator product Soliris, by Alexion Pharmaceuticals, Inc., which contains eculizumab as the active ingredient. Eculizumab is a humanized monoclonal antibody (mAb; IgG2/4 kappa immunoglobulin) that binds to the human C5 complement protein. Binding to this protein inhibits complement-mediated intravascular hemolysis by blocking its cleavage into C5a and C5b. Eculizumab is composed of two 448 amino acid heavy chains (HCs) and two 214 amino acid light chains (LCs) linked by disulfide bonds with molecular weight (MW) of approximately 148 kDa. Each HC contains 13 cysteine (Cys) residues, and each LC contains 5 Cys residues linked to intra- and inter-chain disulfide bonds [1, 7].
While the eculizumab reference product (RP) is manufactured using murine myeloma cells (NS0), SB12 is produced by recombinant DNA technology in a Chinese hamster ovary (CHO) mammalian cell expression system purified by various affinity/ion exchange chromatography steps that include specific viral inactivation and removal procedures. The product is presented as a vial containing 300 mg of eculizumab administered via intravenous infusions [4, 11, 12].
Biosimilar development requires a stepwise approach to demonstrate analytical similarity between a proposed biosimilar and an authorized original biological medicine (RP) and has no clinically meaningful differences in terms of structure, function, pharmacokinetics (PK), pharmacodynamics (PD), immunogenicity, clinical efficacy, and safety [3, 5]. As outlined in the International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH) guideline for clinical studies, a risk-based approach was taken to the clinical evaluation of SB12. Based on the previous clinical trial results (phase I), it was confirmed that the clinical profile of SB12 was similar to that of eculizumab RP (data not shown) [7]. In order to assess the structural and functional similarity of SB12 to eculizumab RP, a comprehensive analysis was conducted using state-of-the-art analytical methods, and similarity was established between eculizumab RP and SB12.
The purpose of this work was to confirm the structural, physicochemical and biological similarity between the eculizumab biosimilar product, SB12, and the eculizumab RP. Through the predefined quality range approach and side-by-side comparison, the analytical similarity between SB12 and the European Union (EU) and United States (US) eculizumab RPs was verified [2,3,4,5].
2 Materials and Methods
2.1 Reference Products
The EU- and US-sourced eculizumab RP, which expired from January 2016 to October 2022, was purchased from local distributors and then stored according to the manufacturer’s instructions.
Up to 49 EU-sourced RPs and 25 US-sourced RPs, which expired from January 2016 to October 2022, were purchased from local distributors and analyzed (refer to similarity assessment plan). Multiple lots of EU- and US-sourced RPs were procured over time and stored, handled according to the manufacturer’s instructions.
2.2 Intact Mass
Intact mass analysis was performed to determine MW by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS).
Glycosylated and deglycosylated whole protein mass, glycosylated and deglycosylated HC mass, and LC mass of eculizumab (SB12 and RP) were measured using a mass spectrometer after treatment or non-treatment of 1 M 1,4-Dithiothreitol (DTT) and/or PNGase F (NEB). The digested samples were loaded on a BEH300 C4 reverse-phase column (Waters) connected to Waters ultra performance liquid chromatography (UPLC)-MS. Data acquisition and processing was performed using MassLynx v4.1 (Waters) and/or BiopharmaLynx v1.2 software (Waters).
2.3 Peptide Mapping
Peptide mapping, which involves enzymatic digestion and tandem MS, was performed to analyze the overall amino acid sequence, post-translational modifications (oxidation and deamidation, etc.), and disulfide bond by LC-ESI-MS.
Eculizumab (SB12 and RP) samples were solubilized with 8 M urea. The denatured samples were reduced by 1 M DTT, and 1 M iodoacetamide (IAA) was then added for alkylation. The samples were 50 mM Tris-HCl buffer (pH 7.8) exchanged for enzyme digestion and digested with the endoprotease trypsin (Roche). For the disulfide linkage analysis, the reducing step was not performed and the non-reduced samples were also digested with trypsin. The digested samples were loaded on a BEH300 C18 reverse-phase column (Waters) connected to Waters UPLC-MS. Data acquisition and processing was performed using MassLynx v4.1 (Waters) and/or BiopharmaLynx v1.2 software (Waters).
2.4 Size Exclusion Chromatography Coupled with Multi-Angle Light Scattering
Size exclusion chromatography coupled with multi-angle light scattering (SEC-MALS) is used to determine the molar mass and extinction coefficient of the soluble proteins in eculizumab (SB12 and RP). Molar mass calculation was performed on a portion of the data channels near the peak apex. Extinction coefficient was determined by refractive index signal in MALS.
2.5 Circular Dichroism Spectroscopy
For far-ultraviolet (UV) and near-UV circular dichroism (CD) studies, eculizumab (SB12 and RP) samples were diluted with formulation buffer. A Chirascan Q100 (Applied Photophysics) with a 0.1 mm path length cell for far-UV and 10 mm path length cell for near-UV was used. The observed CD signal within a 200–260 range for a far-UV CD scan and a 250–350 range for a near-UV scan was blank-substracted and the average of the triplicate scans was used to make the CD plot. The CDNN algorithm was used to fit the far-UV CD data for prediction of the secondary structure.
2.6 Fourier Transform Infrared Spectroscopy
Fourier transform infrared (FT-IR) spectra were recorded using a Nicolet iS50 FT-IR spectrophotometer (Thermo Scientific) equipped with a Smart Orbit diamond attenuated total reflection (ATR) accessory and the MONIC software package for spectrophotometric control and data analysis.
2.7 Differential Scanning Calorimetry
Nano-differential scanning calorimetry (DSC; TA Instruments) was used to analyze the melting temperature and thermodynamics of eculizumab (SB12 and RP). For DSC analysis, eculizumab was diluted with formulation buffer. The diluted sample and corresponding formulation buffer were loaded into the analyzer. Thermodynamic scanning was performed from 15 to 100 °C and the scan rate was 1.5 °C/min. Data processing was performed by NanoAnalyze software (TA Instruments).
2.8 Sedimentation Velocity-Analytical Ultra Centrifugation
The sedimentation velocity-analytical ultra centrifugation (SV-AUC; Beckman) method is used to identify and characterize monomer content, the presence of aggregates and fragments, and the MW of the main molecular species in a protein solution. Eculizumab (SB12 and RP) samples were diluted to 0.4 mg/mL with formulation buffer. Centrifugation was carried out at 20 °C at a speed of 45,000 rpm. Radial scans of the concentration profile were collected sequentially by absorbance at 280 nm until no further sedimentation was observed. The resulting data sets were analyzed using the SEDFIT program, with a continuous c(s) distribution model, yielding best-fit distributions for the number of sedimenting species and the effective MWs.
2.9 Hydrogen/Deuterium Exchange-Mass Spectrometry
Hydrogen/deuterium exchange-MS (H/DX-MS) was initiated by dilution of eculizumab (SB12 and RP) in D2O buffer for 10 s, 1 min, 10 min, 1 h and 4 h. After labeling, the solution was mixed with quenching buffer and injected into the Waters nano LC system. Loaded protein was digested on an immobilized pepsin column (Waters) and the digested peptide fragments was separated on an analytical C18 column (Waters). The analyte was then introduced to MS with MSE mode. Mass spectra were analyzed using PLGS software (Waters) for peptide identification, and DynamX software (Waters) for deuterium uptake calculation and generation of the butterfly plot and uptake plots.
2.10 Size Exclusion-High Performance Liquid Chromatography
For SEC analysis, eculizumab (SB12 and RP) was injected onto a TSK-GEL G3000 SWXL analytical column (Tosoh) connected to a Waters HPLC system. The mobile phase (100 mM sodium phosphate, 300 mM sodium chloride, pH 6.8) was used as the isocratic gradient. Monomer and high MW (HMW) were detected by UV signal at 280 nm. Data were acquired and processed by Empower®3 software (Waters).
2.11 Capillary Electrophoresis-Sodium Dodecyl Sulfate
Capillary electrophoresis-sodium dodecyl sulfate (CE-SDS) was performed using the capillary electrophoresis system PA800 plus Pharmaceutical Analysis (SCIEX). Eculizumab (SB12 and RP) samples were mixed with 250 mM IAA, 10 kDa internal standard and the SDS-MW sample buffer (Beckman) under non-reducing or reducing conditions. For the reduced condition, IAA was replaced with 14.3 M 2-mercaptoethanol. The mixture was incubated in a heated condition for denaturation and loaded for separation in the capillary cartridge. Electrophoresis was performed and the signals were monitored at 220 nm using 32 Karat software (SCIEX).
2.12 Imaged Capillary Isoelectric Focusing
The imaged capillary isoelectric focusing (icIEF) system performs free solution isoelectric focusing in a capillary column and detects focused protein zones using a whole-column UV absorption detector. Eculizumab (SB12 and RP) samples were mixed with pharmalyte 3-10, methyl cellulose, urea, distilled water, and pI marker. The mixtures were loaded and analyzed onto an iCE3 instrument (Protein Simple).
2.13 Anion Exchange Chromatography
Eculizumab (SB12 and RP) was injected onto a weak anion exchange (WAX) analytical column (Thermo Scientific) connected to a Waters HPLC system. 2-(N-morpholino)ethanesulfonic acid (MES) buffer and MES containing sodium chloride buffer were used as the mobile phases A and B, respectively, for anion exchange chromatography. Charge variants were detected by UV signal at 280 nm. Data were acquired and processed using Empower®3 software (Waters).
2.14 Glycan Profile
The identification and relative percentage of N-linked glycan in eculizumab was determined by hydrophilic interaction UPLC (HILIC-UPLC). Eculizumab (SB12 and RP) samples were treated with PNGase-F enzyme to release N-glycan after denaturation. The released N-glycan was separated from proteins using cold ethanol, and dried completely. 2-aminobenzamide (2-AB)-labeled N-glycan was gradually separated using an Acquity UPLC BEH glycan column (Waters), and quantified on a fluorescence detector connected to a Waters UPLC system. For identification, N-glycan was labeled with procainamide and identified on LC-ESI-MS connected to a Waters UPLC system.
2.15 Reverse Phase-Ultra Performance Liquid Chromatography
The hydrophobicity profile of eculizumab (SB12 and RP) samples was determined using 2.15 reverse phase-UPLC (RP-UPLC). Eculizumab (SB12 and RP) samples were loaded on a BEH300 C4 reverse-phase column (Waters) and separated by reverse-phase mode on UPLC (Waters) with UV detection at 280 nm. Data were acquired and processed using Empower®3 software (Waters).
2.16 Protein Concentration by UV A280
Protein concentrations of eculizumab (SB12 and RP) were determined by UV measurement at A280. Eculizumab (SB12 and RP) samples were diluted with 0.9% sodium chloride to 1.0 mg/mL based on the expected concentration of each sample. Absorbance was measured at a wavelength of 280 nm after normalizing with 0.9% sodium chloride (blank solution).
2.17 C5 Inhibition Assay
Eculizumab (SB12 and RP) inhibits the formation of the membrane attack complex (MAC) by blocking C5 cleavage, and prevents B-cell lysis promoted by the anti-CD20 antibody. The blockage effect of eculizumab on the terminal complement complex is evaluated through the detection of cell lysis induced by complement dependent cytotoxicity (CDC) activation. To determine the effect of eculizumab (SB12 and RP), CDC activation is induced through human serum as a complement and anti-CD20 antibody, and the effect of blocking cell lysis is measured through incubation with Raji cells (ATCC), which are CD20-positive B cells.
2.18 Anti-Hemolytic Assay
Eculizumab (SB12 and RP) inhibits cleavage of C5 to C5a and C5b and prevents generation of the terminal complement complex C5b-9 (MAC) responsible for the lysis of paroxysmal nocturnal hemoglobinuria/atypical hemolytic uremic syndrome (PNH/aHUS) red blood cells (RBCs) lacking cell surface terminal complement inhibitor CD59. The anti-hemolytic assay using the luminescence system measured the inhibition of hemolysis of RBCs (Innovative Research) by eculizumab (SB12 and RP)
2.19 C5 Binding Assay
Human complement protein C5 is the first material adsorbed onto the solid phase. Blocking of non-specific binding to the plate surface is followed using BSA-containing buffer. After blocking, eculizumab (SB12 and RP) binds to C5, and a peroxidase-linked secondary antibody then binds to eculizumab (SB12 and RP). Finally, appropriate chromogenic substrate is added, producing colored product. The test plate is read on a spectrophotometer at 450 nm, yielding the optical density that is directly proportional to the concentration of eculizumab (SB12 and RP). Finally, relative C5 binding activity of eculizumab (SB12 and RP) is calculated by analyzing the four-parameter curve plotted from the data in the parallel-line analysis (PLA) software (Stegmann Systems GmbH).
2.20 FcRn Binding Assay by Surface Plasmon Resonance (SPR)
Purified FcRn is immobilized on a CM5 sensor chip (Cytiva) using N-Hydroxysuccinimide/1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide. Various concentrations (31.7–507 nM) of SB12 and eculizumab (SB12 and RP) samples were prepared by serial dilution with Hepes buffered saline–EDTA, Surfactant P20 buffer. Samples were injected into the flow cell for 120 s association at a flow rate of 30 μL/min, and 240 s dissociation at a flow rate of 30 µL/min. Binding affinity was calculated for the sensorgrams using BIAevaluationTM software’s 1:1 binding model.
2.21 Additional Biological Characterization
In order to measure Fc gamma receptor affinity for eculizumab (SB12 and RP; by surface plasmon resonance [SPR]), the His-tagged Fc gamma receptor is captured by the immobilized anti-His antibody on the sensor chip surface, and a series of diluted eculizumab in free solution is then injected onto the captured Fc gamma receptor. The association is followed by running buffer and dissociation. Afterwards, regeneration solution is injected, thus bound eculizumab and Fc gamma receptor are washed out for the sequential cycle. The association and dissociation of serially diluted eculizumab are evaluated to the appropriate binding model.
3 Results
To demonstrate the similarity between the eculizumab biosimilar product, SB12 and the EU/US-sourced eculizumab RP, analytical characterization of the primary structure, higher-order structure, physicochemical attributes, and biological activities were performed using state-of-the-art methods. Analysis of all produced SB12 batches was carried out and all drug products produced from individual drug substances were analyzed; however, in the case of a drug substance batch that did not produce drug products, only the corresponding drug substance was analyzed.
For the similarity assessment, all batches were analyzed together in a side-by-side manner using all testing methods. The summary of quality attributes and the respective analytical methods for eculizumab (SB12 and RP) is presented in Table 1.
3.1 Primary Structure
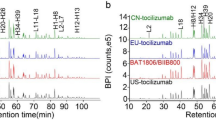
The peptide chromatograms of trypsin-digested SB12 and eculizumab RP showed a highly similar peak profile without missing or additional new peaks with comparative retention time (Fig. 1). In addition, the data confirmed that the amino acid sequences of SB12 and eculizumab RP were identical and that the sequence coverage by MS/MS was 100% (data not shown). Based on the results of intact mass and peptide mapping analysis, the MWs of SB12 and eculizumab RP were identical. Sequence variants such as N-terminal pyroglutamate form and C-terminal lysine variants form were both identified in SB12 and eculizumab RP. Post-translational modifications such as oxidation and deamidation were also identified with comparative level (data not shown).
Peptide mapping profiles of a SB12, b EU eculizumab, and c US eculizumab in a trypsin-digested condition. EU eculizumab eculizumab reference product sourced from the European Union, US eculizumab eculizumab reference product sourced from the United States, TOF MS ES+ time-of-flight mass spectrometry electrospray (positive), BPI base peak intensity
3.2 Higher-Order Structure
The higher-order structures of SB12 and eculizumab RP were compared using CD, FT-IR spectroscopy, DSC, and H/DX-MS. The far-UV CD spectrum of SB12 and eculizumab RP showed comparable profiles throughout much of the far-UV region, especially at the local minimum at around 208 nm, which strongly corresponds with the secondary structure of protein folding (Fig. 2a). In addition, the samples showed comparable near-UV CD profile maxima at around 295, 266, and 260 nm, respectively. No significant differences between the samples were observed, suggesting the samples share common tertiary structure characteristics (Fig. 2b). The secondary structure was further identified by FT-IR spectroscopy. The second derivative spectra of SB12 and eculizumab RP in the amide I band (1600–1700 cm-1) and their distribution into amide regions support high similarity in secondary structures (Fig. 2c). The DSC thermograms of samples showed highly similar thermal profiles and thermal transition midpoint temperatures, indicating that the thermal stability and conformation of SB12 are highly similar to those of the eculizumab RP (Fig. 2d). The tertiary structure was also determined using H/DX-MS. The dynamics of deuterium uptake over time (10 s to 4 h) revealed high symmetry between SB12 and eculizumab RP (Fig. 2e, f).
Comparison of higher-order structure profiles of SB12 and eculizumab RP: a far-UV CD spectra; b near-UV CD spectra; c second derivative FT-IR spectra; d DSC thermograms; e, f H/DX-MS comparison deterium uptake profiles of SB12 and eculizumab RP; (e) butterfly plot of SB12 and EU eculizumab for heavy chain (top) and light chain (bottom); f butterfly plot of SB12 and US eculizumab for heavy chain (top) and light chain (bottom). EU eculizumab eculizumab reference product sourced from the European Union, US eculizumab eculizumab reference product sourced from the United States, UV-CD ultraviolet circular dichroism, FT-IR fourier-transform infrared spectroscopy, DSC differencial scanning calorimetry, H/DX-MS hydrogen/deuterium exchange mass spectrometry
3.3 Purity and Impurity
The purity of the products was analyzed via product monomer content and amount of related impurities using SE-HPLC and CE-SDS, respectively, and the results are described in Fig. 3. In SE-HPLC analysis, all samples showed prominent monomer peaks. The level of HMW (%HMW) species of SB12 and eculizumab RP was comparably low (Fig. 3a). The results were confirmed by orthogonal analyses using SEC-MALS and sedimentation velocity analytical ultracentrifugation (SV-AUC) [data not shown]. In CE-SDS analysis, the results showed that the electrophoretic profiles of SB12 and eculizumab RP were similar (Fig. 3b, c). The level of IgG species (%main IgG of non-reduced CE-SDS), HC (%HC of reduced CE-SDS), LC (%LC of reduced CE-SDS), two heavy chains, one light chain (2H1L; %2H1L of non-reduced CE-SDS), and non-glycosylated heavy chain (NGHC; %NGHC of reduced CE-SDS) in SB12 were similar to those of eculizumab RP.
Comparison of purity and impurity profiles of SB12 and eculizumab RP a overlay of SE-HPLC profiles for monomer and HMW; b overlay of CE-SDS profiles under non-reduced conditions for main IgG and 2H1L; c overlay of CE-SDS profiles under reduced conditions for heavy chain, light chain and NGHC. EU eculizumab eculizumab reference product sourced from the European Union, US eculizumab eculizumab reference product sourced from the United States, HMW high molecular weight, CE-SDS capillary electrophoresis-sodium dodecyl sulfate, 2H1L two heavy chains, one light chain, NGHC non-glycosylated heavy chain, STD standard, Ig immunoglobulin, SE-HPLC size exclusion-high performance liquid chromatography
3.4 Charge Heterogeneity
To assess charge variants of SB12 and eculizumab RP, icIEF and AEX analyses were performed. The results of icIEF showed that the isoelectric point (pI) values of all samples were identical and their electropherograms were similar (data not shown). The results of %Main, %Acidic, and %Basic of all samples were comparable. These results have been confirmed using the orthogonal analytical method, i.e. AEX analysis (data not shown).
3.5 Glycan Profile
A total number of 32 peaks of N-glycan were identified in SB12 and eculizumab RP using LC-ESI-MS (data not shown). The identification of N-glycans, except HM/GlcNAc/α-gal/sialic acid-attached forms, was identical between SB12 and eculizumab RP, and the profiles were similar with physicochemical analysis using HILIC-FLD on 2-AB labeling (Fig. 4). The relative amount of N-glycan species is described in Table 2.
Identification of the GlcNAc-attached glycan form varied in SB12 and eculizumab RP, and α-gal/NGNA-attached glycan forms were confirmed in eculizumab RP but not in SB12. These differences were due to the difference in the expression cell-line system. SB12 is produced by recombinant DNA technology in a CHO mammalian cell line and eculizumab RP is produced in an NS0 murine cell line (Table 2). mAbs produced in murine myeloma cells such as NS0 contain glycans such as α-gal and N-glycolylneuraminic acid that are not naturally present in humans and can be immunogenic when used as therapeutics [13].
Additionally, there were some level differences in high mannose species and galactosylated species, but major N-glycan species were commonly identified and these differences were negligible because there is no Fc effector function in eculizumab RP.
3.6 Biological Activity
The similarity of biological activity between SB12 and eculizumab RP was measured by mechanism of action (MoA)-based C5 inhibition potency assay, hemolysis assay using chicken red blood cells, C5-binding enzyme-linked immunosorbent assay (ELISA), and FcRn-binding affinity using SPR (Table 3). Additionally, binding affinity of Fc gamma receptors between SB12 and eculizumab RP were compared and analyzed through the additional assays. Each lot of the EU and US eculizumab RP was analyzed before its respective expiry date. The similarity between SB12 and eculizumab RP was confirmed using three representative batches and the results are shown in Figs. 5 and 6.
Similarity assessment of the biological activity between SB12 and eculizumab RP. Analysis was conducted on three representative lots each for SB12 and EU/US eculizumab. a C5-binding activity; b C5 inhibition potency; c anti-hemolytic potency; d FcRn binding affinity. EU eculizumab eculizumab reference product sourced from the European Union, US eculizumab eculizumab reference product sourced from the United States, FcRn neonatal Fc receptor
Fc gamma receptor binding affinity by SPR. Compared with the binding affinities of the IgG1-positive control, the Fc gamma receptor binding affinities of SB12 are similar to those of eculizumab RP using a side-by-side comparison. a FcγRIa; b FcγRIIa; c FcγRIIb; d FcγRIIIa. EU eculizumab eculizumab reference product sourced from the European Union, US eculizumab eculizumab reference product sourced from the United States, SPR surface plasmon resonance, FcγIa CD64; Fc gamma receptor Ia, FcγIIa CD32a; Fc gamma receptor IIa, FcγIIb CD32b; Fc gamma receptor IIb, FcγIIIa CD16a; Fc gamma receptor IIIa, Ig immunoglobulin
3.6.1 Fab-Related Biological Activities
The relative potency profiles of SB12 and eculizumab RP were highly similar. In the case of relative C5-binding activity using the ELISA method, no meaningful differences among SB12 and EU/US eculizumab were observed, and the C5-binding activities of SB12 were within the similarity range (Fig. 5a). The individual results value of the C5 inhibition and anti-hemolytic assays met the similarity range for EU and US eculizumab RP, as shown in Fig. 5b, c.
3.6.2 Fc-Related Biological Activities (FcRn-Binding Assay by SPR)
SPR is used to determine SB12 FcRn-binding affinity. In order to assess the similarity between SB12 and eculizumab RP, the relative FcRn binding affinities of SB12 materials were compared with the predefined similarity ranges for the EU and US eculizumab RP, individually. The relative FcRn binding affinities of SB12 met both similarity ranges established by eculizumab RP, as shown in Fig. 5d.
3.6.3 Additional Biological Activities
Eculizumab is an IgG2/4 hybrid mAb having weak binding affinity to the Fc receptor. The binding specificity of SB12 and eculizumab RP to Fc receptors was determined using SPR. The Fc gamma receptor binding affinities measured by SPR are summarized in Fig. 6. Compared with the binding affinities of the IgG1-positive control, it was confirmed that SB12 and eculizumab RP weakly bound to FcγRIa, FcγRIIa, FcγRIIb, and FcγRIIIa with comparable binding affinities. Binding affinities of SB12 and eculizumab RP to FcγRIIIb were not detected, while the IgG1-positive control was bound to FcγRIIIb (data not shown). As no meaningful difference between SB12 and eculizumab RP was observed with a side-by-side comparison, the Fc gamma receptor binding affinities of SB12 are similar to those of eculizumab RP.
4 Discussion
A biosimilar needs to demonstrate a high level of similarity with the RPs in terms of the structural, physicochemical and biological properties as well as clinical outcomes. Compared with ABP 959 and Elizaria®, which were previously disclosed as eculizumab proposed biosimilars, the novelty of the study is that it clearly suggested similarity with a more objective and accurate similarity evaluation by analyzing a relatively large number of EU and US eculizumab RPs [10, 14]. A similarity range that can represent the characteristics of the RP was secured by reflecting the various lots and production times of the reference drug, and through this, the similarity between SB12 and the RP could be more accurately demonstrated. The similarity in PK, PD, safety, and immunogenicity between SB12 and eculizumab RP has been shown in our previously published phase I study [7]. Since our study focuses on the biosimilarity demonstration with respect to structural and functional characterization, other clinical aspects (efficacy, safety, PK, and immunogenicity) of SB12 in a phase III study have not been reported and discussed. To assess the analytical similarity, a study on quality risk assessment, structural and physicochemical properties, and biological activity was performed in accordance with the regulatory guidelines [8, 9, 12]. For each quality attribute, predefined quality ranges were established on the basis of the analysis of a sufficient number of RP lots over a long period. Furthermore, the similarity range was statistically established through quality monitoring.
In-depth analytical characterization was conducted by side-by-side comparison of SB12 with the EU and US eculizumab RPs. The similarity between SB12 and eculizumab RP was demonstrated using various state-of-the-art analytical methods evaluating purity, product-related impurity, charge heterogeneity, primary structure, post-translational modification, higher-order structure, and biological properties (potency by cell-cased assay and binding activity). To demonstrate structural integrity, SB12 and eculizumab RP showed very similar secondary and tertiary structure profiles. Under impurity analysis, for aggregates and charge variants, there were no significant differences in the chromatographic and electrophoretic profiles. In N-glycan profiles, there was an inevitable difference caused by the differences in expression cell line, but this was not expected to affect the other biological activities. Since Fc-effector function (e.g. antibody-dependent cellular cytotoxicity [ADCC], complement-dependent cytotoxicity [CDC]) is not related to the MoA of eculizumab, the glycan species are not expected to affect efficacy. Moreover, similar FcRn binding affinity and similar PK profiles in clinical trials were observed between SB12 and RPs. In addition, similar immunogenicity and safety profiles in clinical trials were observed between SB12 and RPs. Consequently, the difference in N-glycan contents between SB12 and RPs was not considered significant in terms of efficacy, PK, immunogeneity, and safety. Therefore, the overall results can support the similarity in the structural and physicochemical properties between SB12 and eculizumab RP. Lastly, in terms of biological activities, no significant differences were observed [6, 10].
Most of the structural and biological characterization results demonstrated that eculizumab biosimilar SB12 materials are highly similar to the EU and US eculizumab RPs by showing qualities that met the criteria for analytical similarity assessment set with the eculizumab RP. Most importantly, the high-risk quality attributes were within the EU and US similarity range. For some of the low-risk quality attributes, especially in N-glycan profiles, minor differences were observed. However, as shown in the discussions above, these differences are negligible and have been shown to not cause clinically meaningful differences in terms of safety, purity and potency.
In summary, the overall characterization results show that SB12 is highly similar to the EU and US eculizumab RPs in terms of structural, physicochemical, biophysical and biological attributes.
5 Conclusion
The present study demonstrates that a structural and functional similarity was established between SB12 and both the EU- and US-sourced eculizumab RPs. Extensive analytical characterization was conducted on the structural, physicochemical and biological properties to support the biosimilarity of SB12 with respect to the EU/US eculizumab RPs. These results represent an important contribution to the totality of evidence for supporting the biosimilarity of SB12 to its RP, following the demonstration of analytical and functional similarity by conducting extensive quality risk assessments.
References
Zuber J, Fakhouri F, Roumenina LT, et al. Use of eculizumab for atypical haemolytic uraemic syndrome and C3 glomerulopathies. Nat Rev Nephrol. 2012;8:643–57.
US FDA. Scientific considerations in demonstrating biosimilarity to a reference product. https://www.fda.gov/media/82647/download. Accessed 26 Sep 2022.
US FDA. Development of therapeutic protein biosimilars: comparative analytical assessment and other quality-related considerations. https://www.fda.gov/media/159261/download. Accessed 26 Sep 2022.
European Medicines Agency. Soliris. Summary of product characteristics. 2020. europa.eu. https://www.ema.europa.eu/en/medicines/human/EPAR/soliris. Accessed 8 Aug 2022.
European Medicines Agency. Guideline on similar biological medicinal products (revision 1). 2014. europa.eu. https://www.ema.europa.eu/en/similar-biological-medicinal-products-scientific-guideline. Accessed 26 Sept 2022.
Kim E, Han J, Chae Y, et al. Evaluation of the structural, physicochemical, and biological characteristics of SB11, as Lucentis® (Ranibizumab) biosimilar. Ophthalmol Ther. 2022;11(2):639–52.
Lee HA, Jang H, Jeong D, et al. A randomized, double-blind, three-arm, parallel group, single-dose phase I study to evaluate the pharmacokinetic similarity between SB12 (a proposed eculizumab biosimilar) and eculizumab (Soliris) in healthy subjects. Int J Clin Pharmacol Ther. 2022;60(6):269–79.
European Medicines Agency. Guidance for similar biological medicinal products containing monoclonal antibodies: non-clinical and clinical issues. 2015. europa.eu. https://www.ema.europa.eu/en/similar-biological-medicinal-products-containing-monoclonal-antibodies-non-clinical-clinical-issues. Accessed 26 Sep 2022.
European Medicines Agency. Guidance for similar medicinal products containing biotechnology-derived proteins as active substance: quality issues. 2014. europa.eu. https://www.ema.europa.eu/en/similar-biological-medicinal-products-containing-biotechnology-derived-proteins-active-substance. Accessed 26 Sept 2022.
Hutterer KM, Ip A, Kuhns S, et al. Analytical similarity assessment of ABP 959 in comparison with eculizumab reference product. BioDrugs. 2021;35:563–77.
Dhara VG, Naik HM, Majewska NI, et al. Recombinant antibody production in CHO and NS0 cells: differences and similarities. BioDrugs. 2018;32:571–84.
Pharmaceutical and Medical Devices Agency (PMDA). Soliris review report. Review Reports: Drugs. www.pmda.go.jp. Accessed 8 Aug 2022
Liu L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J Pharm Sci. 2015;104:1866–84.
Gusarova V, Degterev M, Lyagoskin I, et al. Analytical and functional similarity of biosimilar Elizaria® with eculizumab reference product. J Pharm Biomed Anal. 2022;220: 115004.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
This work was funded by Samsung Bioepis Co., Ltd, Incheon, Republic of Korea.
Conflict of interest
Hyunsoo Kim, Eunkyoung Hong, Jungmin Lee, Seokku Hong, Jihye Kim, Miju Cho, Yikwon Kim, and Taekyung Yoo are employees of either Samsung Bioepis Co., Ltd, or an organization contracted by Samsung Bioepis Co., Ltd. For the present study, there are no other relationships or activities that could appear to influence the submitted work.
Ethics
This article does not contain any studies involving human participants or animals. All laboratory health and safety procedures have been complied within the course of conducting this experimental work.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Data availability statement
The data sets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
Author contributions
HK and EH contributed to data collection, analysis, and interpretation, and drafting and revision of the manuscript. JL contributed to data discussion and reviewed the manuscript. SH, JK, and MC contributed to method development and testing. YK and TY contributed to study design and assessment plan, and development and authoring of the manuscript.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Kim, H., Hong, E., Lee, J. et al. Characterization for the Similarity Assessment between Proposed Biosimilar SB12 and Eculizumab Reference Product Using a State-of-the-Art Analytical Method. BioDrugs 37, 569–581 (2023). https://doi.org/10.1007/s40259-023-00591-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40259-023-00591-9