Abstract
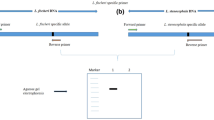
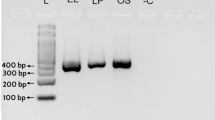
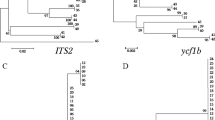
Swertia is ethno-medicinally an important genus belonging to family Gentianaceae. Swertia chirayita is used as imperative medicinal plant in Indian system of medicine. However, this species has been frequently adulterated due to its high demand and scarcity. Authentication of this species was needed to protect consumers and conservation measures and to find out the alternative source. Deoxyribose nucleic acid (DNA) extracted from fifteen samples of six species belonging to different localities, were used as templates. Four candidate barcodes were amplified by polymerase chain reaction and sequence analysis was executed by Z-Big Dye Terminator Cycle Sequencing v.3. Sequenced products were analyzed on automated applied biosystems 3730XI analyzer. Identification was performed by using molecular evolutionary genetics analysis 5 software (version 5.1). The amplification efficiency of all DNA barcodes [megakaryocyte-associated tyrosine kinase (matK), ribulose-bisphosphate carboxylase (rbcL), photosystem II protein D1- stuctural RNA- His tRNA (psbA-trnH) and internal transcribed spacer region (ITS)] was 100 %. Here, the highest interspecific divergence provided by ITS is 11.87 % and intraspecific by psbA-trnH being 10.22 % as compared to matK (5.04 %) and rbcL (0.99 %). ITS region proves robust molecular marker for differing the S. chirayita from its related adulterant species. All barcoding regions indicate that S. chirayita and S. minor both are more closely related than other Swertia species. Findings showed that DNA barcoding is an efficient tool for identification and authentication of S. chirayita. Use of S. minor as substitute to S. chirayita can be advocated.
Similar content being viewed by others
References
Ernst E (2005) The efficacy of herbal medicine—an overview. Fundam Clin Pharmacol 19:405–409
Tindle HA, Davis RB, Phillips RS, Eisenberg DM (2005) Trends in use of complementary and alternative medicine by US adults: 1997–2002. Altern Ther Health Med 11:42–49
Karan M, Vashisht K, Handa SS (1999) Antihepatotoxic activity of Swertia chirata on carbon tetrachloride induced hepatotoxicity in rats. Phytother Res 13:24–30
Banerjee S, Sur TP, Das PC, Sikdar S (2000) Assessment of the anti-inflammatory effects of S. chirata in acute and chronic experimental models in male albino rats. Indian J Pharmacol 32:21–24
Airi S, Bhatt A, Dhar U (2002) Characterization of Swertia chirayita (Roxb. ex Fleming) Karsten and S. angustifolia Ham. ex D. Don. using SDS-PAGE. PGR Newslett (FAO Biodiversity) 147:72–77
Rai MB (2003) Medicinal plants of Tehrathum district, Eastern Nepal. Our Nature 1:42–48
Gao-Feng SR, Run-Hua Y, Yunh-Shang L, Chun-Lei Y, Ai-Mei Li-Xiang (2004) Isolation and crystal structure of xanthones from Swertia chirayita. Chin J Struct Chem 23:1164–1168
Saha P, Mandal S, Das A, Das PC, Das S (2004) Evaluation of anti-carcinogenic activity of Swertia chirata Buch. Ham, an Indian medicinal plant on DMBA-induced mouse skin carcinogenesis model. Phytother Res 18:373–378
Joshi P, Dhawan V (2005) Swertia chirayita—an overview. Curr Sci 89(4):635–638
Phoboo S, Jha PK (2010) Trade and sustainable conservation of Swertia chirayita (Roxb. ex Fleming) h. Karst in Nepal. Nepal J Sci Technol 11:125–132
Ghosal S, Sharma PV, Chaudhur RK (1975) Tetra and pentaoxygenated xanthones of Swertia lawii. Phytochem 14:1393–1396
Heinrich M (2007) The identification of medicinal plants. A handbook of the morphology of botanicals in commerce, W. Applequist. American Botanical Council, Austin, TX, USA and Univ. Missouri Botanical Garden Press, St. Louis, USA. J Ethnopharmacol 111:440
Cheung KS, Kwan HS, But PP, Shaw PC (1994) Pharmacognostical identification of American and oriental ginseng roots by genomic fingerprinting using arbitrarily primed polymerase chain reaction (AP-PCR). J Ethnopharmacol 42:67–69
Sucher NJ, Carles MC (2008) Genome based approaches to the authentication of medicinal plants. Planta Med 74:603–623
Mizukami H, Ohbayashi K, Kitamura Y, Ikenaga T (1993) Restriction fragment length polymorphisms (RFLPs) of medicinal plants and crude drugs. I. RFLP probes allow clear identification of Duboisia interspecific hybrid genotypes in both fresh and dried tissues. Biol Pharm Bull 16:388–390
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321
Gregory TR (2005) DNA barcoding does not compete with taxonomy. Nature 434:1067
Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, Maurin O, Duthoit S, Barraclough TG, Savolainen V (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105:2923–2928
CBOL Plant Working Group (2009) A DNA Barcode for land Plants. Proc Natl Acad Sci USA 106:12794–12797
Kress WJ, Wurdack KJ, Zimmer EA, Weight LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102:8369–8374
Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, Zhu YJ, Ma XY, Gao T, Pang XH, Luo K, Li Y, Li XW, Jia XC, Lin YL, Leon C (2010) Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5:e8613
Sass C, Little DP, Stevenson DW, Specht CD (2007) DNA barcoding in the cycadales: testing the potential of proposed barcoding markers for species identification of cycads. PLoS One 11:e1154
Newmaster SG, Fazekas AJ, Steeves RA, Janovec J (2008) Testing candidate plant barcode regions in the Myristicaceae. Mol Ecol Resour 8(3):480–490
Song J, Yao H, Li Y, Li X, Lin Y, Liu C, Han J, Xie C, Chen S (2009) Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J Ethnopharmacol 124:434–439
Al-Qurainy F, Khan S, Tarroum M, Al-Hemaid FM, Ali MA (2011) Molecular authentication of the medicinal herb Ruta graveolens (Rutaceae) and an adulterant using nuclear and chloroplast DNA markers. Genet Mol Res 10(4):2806–2816
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of leaf tissue. Phytochem Bull 19:11–15
Thompson JD, Gibson TJ, Plewnik F, Jeanmougin F, Higgins DG (1997) The clustal windows interface: flexible strategies for multiple sequence alignment aided by quality analysistools. Nucl Acids Res 24:4876–4882
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Srirama R, Senthilkumar U, Sreejayan N, Ravikanth G, Gurumurthy BR, Shivanna MB, Sanjappa M, Ganeshaiah KN, Uma Shaanker R (2010) Assessing species admixtures in raw drug trade of Phyllanthus, a hepato-protective plant using molecular tools. J Ethanopharmacol 130:208–215
Gao T, Hui Y, Song J, Liu C, Zhu Y, Ma X, Pang X, Xu H, Chen S (2010) Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. J Ethanopharmacol 130:116–121
Hao D, Chen S, Xiao P, Peng Y (2010) Authentication of medicinal plants by DNA-based markers and genomics. Chin Herb Med 2(4):250–261
Wangchuk P, Pyne SG, Keller PA (2011) Ethnobotanical authentication and identification of Khrog-sman (lower elevation medicinal plants) of Bhutan. J Ethanopharmacol 134:813–823
Heubl G (2010) New aspects of DNA based authentication of Chinese medicinal plants by molecular biological techniques. Planta Med 76:1963–1974
Tamhankar S, Ghate V, Raut A, Rajpur B (2009) Moleculer profiling of Chirayat complex using Inter simple sequence repeat (ISSR) markers. Planta Med 75:1266–1270
Xue CY, Li DZ, Lu JM, Yang JB, Liu JQ (2006) Molecular authentication of the traditional Tibetan medicinal plant Swertia mussotii. Planta madica 72:1223–1226
Yu MT, Wong KL, Zong YY, Shaw PC, Che CT (2008) Identification of Swertia mussotii and its adulterant Swertia species by 5S rRNA gene spacer. Zhongguo Zhong Yao Za Zhi 33(5):502–504
Li M, Cao H, But PP, Shaw PC (2011) Identification of herbal medicinal materials by using DNA barcodes. J System Evol 49(3):271–283
Chassot P, Nemomissa S, Yuan YM, Kupfer P (2001) High paraphyly of Swertia L. (Gentianaceae) in the Gentianella-lineage as revealed by nuclear and chloroplast DNA sequence variation. Plant Syst Evol 229:1–21
Acknowledgments
Authors are indebted to Prof. S. R. Yadav Head, Department of Botany, Shivaji University, Kolhapur for providing necessary lab facilities and also thankful to Dr. Mansingraj Nimbalkar for their extended help and guidance.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kshirsagar, P., Umdale, S., Chavan, J. et al. Molecular Authentication of Medicinal Plant, Swertia chirayita and its Adulterant Species. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 87, 101–107 (2017). https://doi.org/10.1007/s40011-015-0556-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-015-0556-3