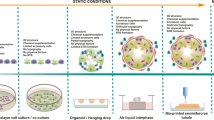
Abstract
Background:
Human testicular cells are greatly valuable to the research community as tools for studying testicular physiology and the effects of environmental pollutants. Because adult testicular cells have a limited self-organization capacity and life span, we investigated whether human pluripotent stem cells (hPSCs) can be used together with testicular cells to move a step closer toward making an optimal model of the human testis.
Methods:
We used in vitro culture of donor testicular cells under serum-containing and chemically defined conditions. CRISPR-Cas9 technology was applied to introduce fluorescent transgenes (mCherry2 and EGFP) into hPSCs and testicular cells. hPSC-derived spheroids were co-cultured with human testicular cells in mini-spin bioreactors.
Results:
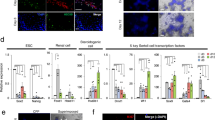
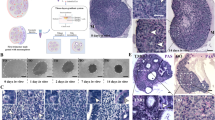
Traditional cell culture conditions used for maintenance of testicular somatic cells generally contain serum and pose limitations on evaluating the role of active molecules on cell functions. We established that chemically defined culture conditions can be used to maintain testicular cells without the loss of proliferative activity. These cultures demonstrate marker expression which is characteristic of common testicular cell types: Sertoli, Leydig, endothelial, myoid cells, and macrophages. In order to model testicular physiology, it is important to be able to perform live cell microscopy. Thus, we generated fluorescent protein-expressing human testicular cells and hPSCs and demonstrated that these cell types can be successfully co-cultured for prolonged periods of time in a three-dimensional microenvironment.
Conclusion:
Our research extends the possible applications of human testis-derived somatic cells and shows that they can be used together with hPSCs for further studies of human male reproductive biology.




Similar content being viewed by others
References
Messerlian C, Williams PL, Ford JB, Chavarro JE, Mínguez-Alarcón L, Dadd R, et al. The environment and reproductive health EARTH study: a prospective preconception cohort. Hum Reprod Open. 2018;2018:hoy001.
Levine H, Jørgensen N, Martino-Andrade A, Mendiola J, Weksler-Derri D, Mindlis I, et al. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update. 2017;23:646–59.
Skakkebaek NE, Rajpert-De Meyts E, Buck Louis GM, Toppari J, Andersson A-M, Eisenberg ML, et al. Male reproductive disorders and fertility trends: influences of environment and genetic susceptibility. Physiol Rev. 2016;96:55–97.
Ho SM, Cheong A, Adgent MA, Veevers J, Suen AA, Tam NNC, et al. Environmental factors, epigenetics, and developmental origin of reproductive disorders. Reprod Toxicol. 2017;68:85–104.
Pendergraft SS, Sadri-Ardekani H, Atala A, Bishop CE. Three-dimensional testicular organoid: a novel tool for the study of human spermatogenesis and gonadotoxicity in vitro. Biol Reprod. 2017;96:720–32.
Sakib S, Uchida A, Valenzuela-Leon P, Yu Y, Valli-Pulaski H, Orwig K, et al. Formation of organotypic testicular organoids in microwell culture†. Biol Reprod. 2019;100:1648–60.
Baert Y, De Kock J, Alves-Lopes JP, Söder O, Stukenborg JB, Goossens E. Primary human testicular cells self-organize into organoids with testicular properties. Stem Cell Reports. 2017;8:30–8.
Richer G, Baert Y, Goossens E. In-vitro spermatogenesis through testis modelling: towards the generation of testicular organoids. Andrology. 2019. https://doi.org/10.1111/andr.12741.
Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–97.
Lancaster MA, Huch M. Disease modelling in human organoids. Dis Model Mech. 2019;12:dmm039347.
Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125.
Dedhia PH, Bertaux-Skeirik N, Zavros Y, Spence JR. Organoid models of human gastrointestinal development and disease. Gastroenterology. 2016;150:1098–112.
Rodríguez Gutiérrez D, Eid W, Biason-Lauber A. A human gonadal cell model from induced pluripotent stem cells. Front Genet. 2018;9:498.
Hou YP, Zhang ZY, Xing XY, Zhou J, Sun J. Direct conversion of human fibroblasts into functional Leydig-like cells by SF-1, GATA4 and NGFI-B. Am J Transl Res. 2018;10:175–83.
Sepponen K, Lundin K, Knuus K, Väyrynen P, Raivio T, Tapanainen JS, et al. The role of sequential BMP signaling in directing human embryonic stem cells to bipotential gonadal cells. J Clin Endocrinol Metab. 2017;102:4303–14.
Svingen T, Koopman P. Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev. 2013;27:2409–26.
Rey R, Josso N, Racine C. Sexual Differentiation. In: Feingold KR, Anawalt B, Boyce A, Chrousos G, Dungan K, Grossman A, et al., editors. Endotext. South Dartmouth : MDText.com, Inc.; 2000.
Irie N, Surani MA. Efficient induction and isolation of human primordial germ cell-like cells from competent human pluripotent stem cells. Methods Mol Biol. 2017;1463:217–26.
Mitsunaga S, Odajima J, Yawata S, Shioda K, Owa C, Isselbacher KJ, et al. Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs. Proc Natl Acad Sci U S A. 2017;114:E9913–22.
Hu YC, Nicholls PK, Soh YQ, Daniele JR, Junker JP, van Oudenaarden A, et al. Licensing of primordial germ cells for gametogenesis depends on genital ridge signaling. PLoS Genet. 2015;11:e1005019.
Fang F, Li Z, Zhao Q, Li H, Xiong C. Human induced pluripotent stem cells and male infertility: an overview of current progress and perspectives. Hum Reprod. 2018;33:188–95.
Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling zikv exposure. Cell. 2016;165:1238–54.
Przepiorski A, Sander V, Tran T, Hollywood JA, Sorrenson B, Shih JH, et al. A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Reports. 2018;11:470–84.
Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev. 2012;92:577–95.
Yoshida S. Open niche regulation of mouse spermatogenic stem cells. Dev Growth Differ. 2018;60:542–52.
Wen L, Yuan Q, Sun M, Niu M, Wang H, Fu H, et al. Generation and characteristics of human Sertoli cell line immortalized by overexpression of human telomerase. Oncotarget. 2017;8:16553–70.
Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming GL. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat Protoc. 2018;13:565–80.
Pryzhkova MV, Aria I, Cheng Q, Harris GM, Zan X, Gharib M, et al. Carbon nanotube-based substrates for modulation of human pluripotent stem cell fate. Biomaterials. 2014;35:5098–109.
Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X, et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28:1141–57.
Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9.
Teerds KJ, Huhtaniemi IT. Morphological and functional maturation of Leydig cells: from rodent models to primates. Hum Reprod Update. 2015;21:310–28.
Guo Y, Hai Y, Yao C, Chen Z, Hou J, Li Z, et al. Long-term culture and significant expansion of human Sertoli cells whilst maintaining stable global phenotype and AKT and SMAD1/5 activation. Cell Commun Signal. 2015;13:20.
Mori H, Hiromoto N, Nakahara M, Shiraishi T. Stereological analysis of Leydig cell ultrastructure in aged humans. J Clin Endocrinol Metab. 1982;55:634–41.
Zhu F, Gamboa M, Farruggio AP, Hippenmeyer S, Tasic B, Schüle B, et al. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 2014;42:e34.
Brennan J, Capel B. One tissue, two fates: molecular genetic events that underlie testis versus ovary development. Nat Rev Genet. 2004;5:509–21.
Yang Y, Workman S, Wilson M. The molecular pathways underlying early gonadal development. J Mol Endocrinol. 2018.https://doi.org/10.1530/JME-17-0314.
Kumar SV, Er PX, Lawlor KT, Motazedian A, Scurr M, Ghobrial I, et al. Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Development. 2019;146:dev172361.
Taguchi A, Nishinakamura R. Higher-Order Kidney Organogenesis from Pluripotent Stem Cells. Cell Stem Cell. 2017;21:730-46.e6.
Hiramatsu R, Harikae K, Tsunekawa N, Kurohmaru M, Matsuo I, Kanai Y. FGF signaling directs a center-to-pole expansion of tubulogenesis in mouse testis differentiation. Development. 2010;137:303–12.
de Santa Barbara P, Moniot B, Poulat F, Berta P. Expression and subcellular localization of SF-1, SOX9, WT1, and AMH proteins during early human testicular development. Dev Dyn. 2000;217:293–8.
Croft B, Ohnesorg T, Hewitt J, Bowles J, Quinn A, Tan J, et al. Human sex reversal is caused by duplication or deletion of core enhancers upstream of SOX9. Nat Commun. 2018;9:5319.
DeFalco T, Takahashi S, Capel B. Two distinct origins for Leydig cell progenitors in the fetal testis. Dev Biol. 2011;352:14–26.
Liu C, Rodriguez K, Yao HH. Mapping lineage progression of somatic progenitor cells in the mouse fetal testis. Development. 2016;143:3700–10.
Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun. 2018;9:2164.
Yu C, Liu Y, Ma T, Liu K, Xu S, Zhang Y, et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–7.
Liu M, Rehman S, Tang X, Gu K, Fan Q, Chen D, et al. Methodologies for improving HDR efficiency. Front Genet. 2018;9:691.
Easley CA 4th, Phillips BT, McGuire MM, Barringer JM, Valli H, Hermann BP, et al. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440–6.
Lim JJ, Shim MS, Lee JE, Lee DR. Three-step method for proliferation and differentiation of human embryonic stem cell (hESC)-derived male germ cells. PLoS One. 2014;9:e90454.
Durruthy Durruthy J, Ramathal C, Sukhwani M, Fang F, Cui J, Orwig KE, et al. Fate of induced pluripotent stem cells following transplantation to murine seminiferous tubules. Hum Mol Genet. 2014;23:3071–84.
Oliver E, Stukenborg JB. Rebuilding the human testis in vitro. Andrology. 2019. https://doi.org/10.1111/andr.12710.
Young JC, Wakitani S, Loveland KL. TGF-β superfamily signaling in testis formation and early male germline development. Semin Cell Dev Biol. 2015;45:94–103.
Jiang X, Skibba M, Zhang C, Tan Y, Xin Y, Qu Y. The roles of fibroblast growth factors in the testicular development and tumor. J Diabetes Res. 2013;2013:489095.
Acknowledgements
We thank Immanuel Rasool from WRTC for coordinating the acquisition of deidentified human testis samples used for this study, Stephen Wellard for technical assistance, Ran Brosh for providing the EF1α promoter sequence, and Barry Zirkin and JinYong Chung for discussion. This work was supported by a KY Cha Award In Stem Cell Technology from the American Society for Reproductive Medicine and a research grant from National Institute of General Medical Sciences (R01GM11755), both awarded to P.W.J.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical statement
The use of human ESC line was approved by the JHU institutional stem cell research oversight committee (protocol ISCRO00000089). Deidentified donor testes tissues were designated as “not human subjects research” by Johns Hopkins University (JHU IRB No: 00006700).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pryzhkova, M.V., Jordan, P.W. Adaptation of Human Testicular Niche Cells for Pluripotent Stem Cell and Testis Development Research. Tissue Eng Regen Med 17, 223–235 (2020). https://doi.org/10.1007/s13770-020-00240-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13770-020-00240-0