Abstract
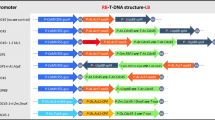
We transformed rice (Oryza sativa L. Japonica cv. Ilmi) calli with the Arabidopsis transcription factor gene AtMYB44 using Agrobacterium-mediated transformation. The T-DNA construct to be transformed contained tflA cDNA (encoding a toxoflavin lyase) as a selectable marker. Since toxoflavin is a photosensitizing phytotoxin, transgenic plantlets were selected based on their capacity for root development on medium containing this toxin in the light. Homozygous lines were selected by determining the segregation patterns, expression levels, and copy numbers of AtMYB44. Intergenic genomic locations of the inserted T-DNA in the three transgenic lines were confirmed by adaptor-ligation polymerase chain reaction and analysis using FSTVAL (http://bioinfo.mju.ac.kr/fstval/), an open-access web tool used to localize the flanking sequences of the transgene. Drought tolerance of young seedlings of the transgenic lines was determined based on the recovery of wilted leaves by re-watering after 3 days of water deprivation in a 105-well (35W × 35L × 45D mm/per well) plate. The three transgenic lines showed average survival rates of 80.4, 93.5, and 72.6%, respectively, whereas wild-type plants failed to recover after re-watering. Thus, the transgenic rice plants exhibited significantly enhanced tolerance to drought stress, as was shown previously in AtMYB44-overexpressing transgenic Arabidopsis and soybean. These results suggest that AtMYB44 activates a drought tolerance mechanism that is conserved in both monocotyledonous and dicotyledonous plants.




Similar content being viewed by others
References
Cook J, Oreskes N, Doran PT, Anderegg WRL, Verheggen B, Maibach EW, Carlton JS, Lewandowsky S, Skuce AG, Green SA, Nuccitelli D, Jacobs P, Richardson M, Winkler B, Painting R, Rice K (2016) Consensus on consensus: a synthesis of consensus estimates on human-caused global warming. Environ Res Lett 11:048002
Todaka D, Shinozaki K, Yamaguchi-Shinozaki K (2015) Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci 6:84
Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, Li Y, Zhang H, Ali J, Li Z (2014) Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 9:e92913
Silveira RD, Abreu FR, Mamidi S, McClean PE, Vianello RP, Lanna AC, Carneiro NP, Brondani C (2015) Expression of drought tolerance genes in tropical upland rice cultivars (Oryza sativa). Genet Mol Res 14:8181–8200
Dou M, Fan S, Yang S, Huang R, Yu H, Feng X (2016) Overexpression of Am Rosea1 gene confers drought and salt Tolerance in rice. Int J Mol Sci 18:2
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong J-J (2008) Overexpression of AtMYB44 enhances stomata closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146:623–635
Hieno A, Nazmin HA, Hyakumachi M, Higuchi-Takeuchi M, Matsui M, Yamamoto YY (2016) Possible involvement of MYB44-mediated stomatal regulation in systemic resistance induced by Penicillium simplicissimum GP17-2 in Arabidopsis. Microbes Environ 31:154–159
Seo JS, Sohn HB, Noh K, Jung C, An JH, Donovan CM, Somers DA, Kim DI, Jeong S-C, Kim C-G, Kim HM, Lee S-H, Choi YD, Moon TW, Kim CH, Cheong J-J (2012) Expression of the Arabidopsis AtMYB44 gene confers drought/salt-stress tolerance in transgenic soybean. Mol Breed 29:601–608
Jaradat MR, Feurtado JA, Huang D, Lu Y, Cutler AJ (2013) Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol 13:192
ZhaoY, Xing L, Wang X, Hou Y-J, Gao J, Wang P, Duan C-G, Zhu X, Zhu J-K (2015) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7(328): ra53
Xing L, Zhao Y, Gao J, Xiang C, Zhu J-K (2016) The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep 6:27177
Koh S, Kim H, Kim J, Goo E, Kim Y-J, Choi O, Jwa N-S, Ma J, Nagamatsu T, Moon JS, Hwang I (2011) A novel light-dependent selection marker system in plants. Plant Biotechnol J 9:348–358
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Pawar B, Kale P, Bahurupe J, Jadhav A, Kale A, Pawar S (2015) Proline and glutamine improve in vitro callus induction and subsequent shooting in rice. Rice Sci 22:283–289
Balzergue S, Dubreucq B, Chauvin S, Le-Clainche I, Le Boulaire F, De Rose R, Samson F, Biaudet V, Lecharny A, Cruaud C, Weissenbach J, Caboche M, Lepiniec L (2001) Improved PCR-walking for large-scale isolation of plant T-DNA borders. Biotechniques 30:496–504
Kim JS, Kim J, Lee T-H, Jun KM, Kim TH, Kim Y-H, Park H-M, Jeon J-S, An G, Yoon U-H, Nahm B-H, Kim Y-K (2012) FSTVAL—a new web tool to validate bulk flanking sequence tags. Plant Methods 8:19
Joo J, Choi HJ, Lee YH, Kim YK, Song SI (2013) A transcriptional repressor of the ERF family confers drought tolerance to rice and regulates genes preferentially located on chromosome 11. Planta 238:155–170
Jung C, Kim Y-K, Oh N-I, Shim JS, Seo JS, Choi YD, Nahm BH, Cheong J-J (2012) Quadruple 9-mer-based protein binding microarray analysis confirms AACnG as the consensus nucleotide sequence sufficient for the specific binding of AtMYB44. Mol Cells 34:531–537
Nguyen XC, Hoang MHT, Kim HS, Lee K, Liu X-M, Kim SH, Bahk S, Park HC, Chung WS (2012) Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem Biophys Res Commun 423:703–708
Persak H, Pitzschke A (2013) Tight interconnection and multi-level control of Arabidopsis MYB44 in MAPK cascade signaling. PLoS ONE 8:e57547
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF, Cutler SR (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068–1071
Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462:660–664
Tian X, Wang Z, Li X, Lv T, Liu H, Wang L, Niu H, Bu Q (2015) Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 8:28
Kuiper HA, Kleter GA, Noteborn HPJM, Kok EJ (2001) Assessment of the food safety issues related to genetically modified foods. Plant J 27:503–528
Gilbert N (2013) A hard look at GM crops. Nature 497:24–26
Ramessar K, Peremarti A, Gómez-Galera S, Naqvi S, Moralejo M, Muñoz P, Capell T, Christou P (2007) Biosafety and risk assessment framework for selectable marker genes in transgenic crop plants: a case of the science not supporting the politics. Trans Res 16:261–280
Kim M-S, Kim H, Moon JS, Hwang I, Joung H, Jeon J-H (2012) Toxoflavin lyase enzyme as a marker for selecting potato plant transformants. Biosci Biotechnol Biochem 76:2354–2356
Kim J, Kim JG, Kang Y, Jang JY, Jog GJ, Lim JY, Kim S, Suga H, Nagamatsu T, Hwang I (2004) Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol Microbiol 54:921–934
Latuasan HE, Berends W (1961) On the origin of the toxicity of toxoflavin. Biochim Biophys Acta 52:502–508
Nagamatsu T, Hashiguchi Y, Sakuma Y, Yoneda F (1982) Autorecycling oxidation of amines to carbonyl compound catalized by 3, 4-disubstituted 4-deazatoxoflavin derivatives. Chem Lett 11:1309–1312
Kim S-R, Lee J, Jun S-H, Park S, Kang Kang H-G, Kwon S, An G (2003) Transgene structures in T-DNA-inserted rice plants. Plant Mol Biol 52:761–773
Acknowledgments
The authors thank Dr. Ingyu Hwang of Seoul National University for kind donation of transformation vector and the research staffs of GreenGene Biotech Co. (Yongin, Korea) for technical assistances. This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (Grant Numbers 111076-5 and 115080-2) and in part by the National Research Foundation of Korea (Grant Number 2016R1A2B4012248).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Joo, J., Oh, NI., Nguyen, N.H. et al. Intergenic transformation of AtMYB44 confers drought stress tolerance in rice seedlings. Appl Biol Chem 60, 447–455 (2017). https://doi.org/10.1007/s13765-017-0297-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13765-017-0297-5