Abstract
Cucumber mosaic virus (CMV), a member of the Cucumovirus genus in the family Bromoviridae, is a frequently occurring virus in pepper worldwide. A broad host range, which includes many weed species and a large number of insects, means it is difficult to control CMV in the field. CMV resistance has been reported in Capsicum spp. In Korea, the C. annuum ‘Bukang’ cultivar is resistant to the CMVFNY and CMVKorean strains; however, a new strain (CMVP1) is able to infect the resistant cultivars. Therefore, it is necessary to pursue CMVP1 resistance in pepper breeding. In this study, we surveyed the quantitative trait loci (QTL) for resistance to CMVP1 using a single-seed-descent (SSD) F3 segregating population derived from a cross between a resistant line, ‘A1’, and a susceptible line, ‘2602.’ A total of 174 plants were evaluated for resistance to CMVP1 at 30 days after mechanical inoculation, with phenotypes confirmed by reverse transcription PCR. The number of resistant and susceptible plants was 88 and 86, respectively. Of these, 96 individuals including 48 resistant lines and 48 susceptible lines were used for a genotyping-by-sequencing (GBS) analysis. Approximately 19,000 single-nucleotide polymorphism (SNP) genotypes were obtained. Using the SNP data, a pepper genetic linkage map consisting of 906 SNP markers was constructed. The map showed 12 linkage groups with a total linkage distance of 1,272.9 cM. QTL analysis using a composite interval mapping (CIM) method revealed that two QTLs, cmvP1-5.1 and cmvP1-10.1, located on chromosomes 5 and 10, respectively, had R 2 (coefficient of determination) values of 17.81% and 22.78%. This information will be helpful for developing SNP markers linked to CMVP1-resistant QTLs and for developing new CMVP1-resistant pepper cultivars.
Similar content being viewed by others
Literature Cited
Ben Chaim A, Grube RC, Lapidot M, Jahn M, Paran I (2001) Identification quantitative trait loci associated with resistance to cucumber mosaic virus in Capsicum annuum. Theor Appl Genet 102:1213–1220
Bielenberg DG, Rauh B, Fan S, Gasic K, Abbott AG, Reighard GL, Okie WR, Wells CE (2015) Genotyping by sequencing for SNP-based linkage map construction and QTL analysis of chilling requirement and bloom date in peach [Prunus persica (L.) Batsch]. PLoS One 10:e0139406
Boutet G, Carvalho SA, Falque M, Peterlongo P, Lhuillier E, Bouchez O, Lavaud C, Pilet-Nayel ML, Rivière N, et al (2016) SNP discovery and genetic mapping using genotyping by sequencing of whole genome genomic DNA from a pea RIL population. BMC Genomics 17:121
Caranta C, Palloix A, Lefebvre V, Daubèze AM (1997) QTLs for a component of partial resistance to cucumber mosaic virus in pepper: restriction of virus installation in host-cells. Theor Appl Genet 94:431–438
Caranta C, Pflieger S, Lefebvre V, Daubèze AM, Thabuis A, Palloix A (2002) QTLs involved in the restriction of cucumber mosaic virus (CMV) long-distance movement in pepper. Theor Appl Genet 104:586–591
Choi GS, Kim JH, Lee DH, Kim JS, Ryu KH (2005) Occurrence and distribution of viruses infecting pepper in Korea. Plant Pathol J 21:258–261
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 142:169–196
Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11:485
Deschamps S, Llaca V, May GD (2012) Genotyping-by-sequencing in plants. Biology 1:460–483
Doolittle SP (1916) A new infectious mosaic disease of cucumber. Phytopathol 6:145–147
Grube RC, Radwanski ER, Jahn M (2000) Comparative genetics of disease resistance within the Solanaceae. Genetics 155:873–887
Grube RC, Zhang Y, Murphy JF, Loaiza-Figueroa F, Lackney VK, Provvidenti R, Jahn MK (2000) New source of resistance to Cucumber mosaic virus in Capsicum frutescens. Plant Dis 84:885–891
Jaganathan D, Thudi M, Kale S, Azam S, Roorkiwal M, Gaur PM, Kishor PBK, Nguyen H, Sutton T, et al (2015) Genotyping-bysequencing based intra-specific genetic map refines a “QTL-hotspot” region for drought tolerance in chickpea. Mol Genet Genomics 290:559–571
Kang WH, Hoang NH, Yang HB, Kwon JK, Jo SH, Seo JK, Kim KH, Choi D, Kang BC (2010) Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor Appl Genet 120:1587–1596
Kim JE, Oh SK, Lee JH, Lee BM, Jo SH (2014a) Genome-wide SNP calling using next generation sequencing data in tomato. Mol Cells 37:36–42
Kim JS, Choi GS (2002) Viruses infecting pepper and their characteristics. J. Kor. Capsicum Res Coop 7:23–47
Kim S, Park M, Yeom SI, Kim YM, Lee JM, Lee HA, Seo E, Choi J, Cheong K, et al (2014b) Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46:270
Kosambi DD (1943) The estimation of map distance from recombination value. Ann Eug 12:172–175
Lapidot M, Paran I, Ben-Joseph R, Ben-Harush S, Pilowsky M, Cohen S, Shifris C (1997) Tolerance to cucumber mosaic virus in pepper: development of advanced breeding lines and evaluation of virus level. Plant Dis 81:185–188
Lefebvre V, Daubèze AM, van der Voort JR, Peleman J, Bardin M, Palloix A (2003) QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor Appl Genet 107:661–666
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaher B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Min WK, Rye JH, Ahn SH (2014) Developmental changes of recessive genes-mediated Cucumber mosaic virus (CMV) resistance in peppers (Capsicum annuum L.). Korean J Hortic Sci Technol 32:235–240
Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RIB (1992) Cucumber mosaic virus. In: Advances in Virus Research (Maramorosch, K., F.A. Murphy, and A.J. Shatkin, eds). San Diego, CA: Academic Press, Vol.41, pp.281–348
Pflieger S, Lefebvre V, Caranta C, Blattes A, Goffinet B, Palloix A (1999) Disease resistance gene analogs as candidates for QTLs involved in pepper-pathogen interactions. Genome 42:1100–1110
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253
Poland JA, Rife TW (2012) Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 5:92–102
Ro NY, Hur OS, Ko HC, Kim SG, Rhee JH, Gwag JG, Kwon JK, Kang BC (2012) Evaluation of resistance in pepper germplasm to Cucumber mosaic virus by high resolution melting analysis. Res Plant Dis 18:290–297
Roossinck MJ (2001) Cucumber mosaic virus, a model for RNA virus evolution. Mol Plant Pathol 2:59–63
Shin JE, Xu SJ, Kim JY, Woo JH, Kim HG, Park YJ, Hong SJ, Kim BS (2013) CMV-P1 resistance evaluation using enzyme-linked immunosorbent assay of pepper genetic sources (Capsicum spp.). Korean J Hortic Sci Technol 31:764–771
Suzuki K, Kuroda T, Miura Y, Murai J (2003) Screening and field traits of virus resistant sources in Capsicum spp. Plant Dis 87:779–783
Voorrips RE (2002) MAPCHART: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang S, Basten CJ, Gaffney P, Zeng ZB (2007) Windows QTL Cartographer. Bioinformatics Research Center, North Carolina State University, Raleigh, NC
Yao M, Li N, Wang F, Ye Z (2013) Genetic analysis and identification of QTLs for resistance to Cucumber mosaic virus in chili pepper (Capsicum annuum L.). Euphytica 193:135–145
Zhuang YL, Chen L, Sun L, Cao JX (2012) Bioactive characteristics and antioxidant activities of nine peppers. J Funct Foods 4:331–338
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
13580_2016_128_MOESM1_ESM.xlsx
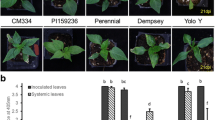
Appearances of plants resistant (A) and susceptible (B and C) to the CMVP1 strain. A, normal and non-infected leaves; B, mosaic symptoms; C, severe mosaic symptoms.
Rights and permissions
About this article
Cite this article
Eun, M.H., Han, JH., Yoon, J.B. et al. QTL mapping of resistance to the Cucumber mosaic virus P1 strain in pepper using a genotyping-by-sequencing analysis. Hortic. Environ. Biotechnol. 57, 589–597 (2016). https://doi.org/10.1007/s13580-016-0128-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13580-016-0128-3