Abstract
Background
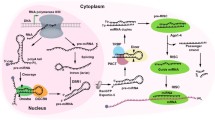
Cancer is one of the leading causes of mortality. The neoplastic transformation of normal cells to cancer cells is caused by a progressive accumulation of genetic and epigenetic alterations in oncogenes, tumor suppressor genes and epigenetic regulators, providing cells with new properties, collectively known as the hallmarks of cancer. During the process of neoplastic transformation cells progressively acquire novel characteristics such as unlimited growth potential, increased motility and the ability to migrate and invade adjacent tissues, the ability to spread from the tumor of origin to distant sites, and increased resistance to various types of stresses, mostly attributed to the activation of genetic stress-response programs. Accumulating evidence indicates a crucial role of microRNAs (miRNAs or miRs) in the initiation and progression of cancer, acting either as oncogenes (oncomirs) or as tumor suppressors via several molecular mechanisms. MiRNAs comprise a class of small ~22 bp long noncoding RNAs that play a key role in the regulation of gene expression at the post-transcriptional level, acting as negative regulators of mRNA translation and/or stability. MiRNAs are involved in the regulation of a variety of biological processes including cell cycle progression, DNA damage responses and apoptosis, epithelial-to-mesenchymal cell transitions, cell motility and stemness through complex and interactive transcription factor-miRNA regulatory networks.
Conclusions
The impact and the dynamic potential of miRNAs with oncogenic or tumor suppressor properties in each stage of the multistep process of tumorigenesis, and in the adaptation of cancer cells to stress, are discussed. We propose that the balance between oncogenic versus tumor suppressive miRNAs acting within transcription factor-miRNA regulatory networks, influences both the multistage process of neoplastic transformation, whereby normal cells become cancerous, and their stress responses. The role of specific tumor-derived exosomes containing miRNAs and their use as biomarkers in diagnosis and prognosis, and as therapeutic targets, are also discussed.


Similar content being viewed by others
References
D. Hanahan, R.A. Weinberg, Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011)
S.I. Ellenbroek, J. van Rheenen, Imaging hallmarks of cancer in living mice. Nat Rev Cancer 14, 406–418 (2014)
L.A. Torre, F. Bray, R.L. Siegel, J. Ferlay, J. Lortet-Tieulent, A. Jemal, Global cancer statistics, 2012. CA Cancer J Clin 65, 87–108 (2015)
A.L. Gartel, E.S. Kandel, miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol 18, 103–110 (2008)
K. Ruan, X. Fang, G. Ouyang, MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 285, 116–126 (2009)
A. Ventura, T. Jacks, MicroRNAs and cancer: short RNAs go a long way. Cell 136, 586–591 (2009)
P.M. Voorhoeve, MicroRNAs: Oncogenes, tumor suppressors or master regulators of cancer heterogeneity? Biochim Biophys Acta 1805, 72–86 (2010)
M.V. Iorio, C.M. Croce, microRNA involvement in human cancer. Carcinogenesis 33, 1126–1133 (2012)
P.M. Voorhoeve, R. Agami, Classifying microRNAs in cancer: the good, the bad and the ugly. Biochim Biophys Acta 1775, 274–282 (2007)
M. Hatziapostolou, C. Polytarchou, D. Iliopoulos, miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol Metab 24, 361–373 (2013)
J. Winter, S. Jung, S. Keller, R.I. Gregory, S. Diederichs, Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11, 228–234 (2009)
V.N. Kim, J. Han, M.C. Siomi, Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126–139 (2009)
S.L. Ameres, P.D. Zamore, Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14, 475–488 (2013)
M. Ha, V.N. Kim, Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524 (2014)
S. Lin, R.I. Gregory, MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15, 321–333 (2015)
V. Taucher, H. Mangge, J. Haybaeck, Non-coding RNAs in pancreatic cancer: challenges and opportunities for clinical application. Cell Oncol 39, 295–318 (2016)
M. Vitiello, A. Tuccoli, L. Poliseno, Long non-coding RNAs in cancer: implications for personalized therapy. Cell Oncol 38, 17–28 (2015)
A. Kozomara, S. Griffiths-Jones, miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39, D152–D157 (2011)
D.P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004)
R.I. Gregory, R. Shiekhattar, MicroRNA biogenesis and cancer. Cancer Res 65, 3509–3512 (2005)
M. Esteller, Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874 (2011)
L. He, G.J. Hannon, MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5, 522–531 (2004)
J.T. Mendell, MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle 4, 1179–1184 (2005)
G.A. Calin, C.M. Croce, MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866 (2006)
M. Inui, G. Martello, S. Piccolo, MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11, 252–263 (2010)
G. Tiscornia, J.C. Izpisua Belmonte, MicroRNAs in embryonic stem cell function and fate. Genes Dev 24, 2732–2741 (2010)
V. Ambros, The functions of animal microRNAs. Nature 431, 350–355 (2004)
A.K. Leung, P.A. Sharp, MicroRNA functions in stress responses. Mol Cell 40, 205–215 (2010)
F.d.A. di Fagagna, A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol 24, 171–178 (2014)
M.J. Bueno, M. Malumbres, MicroRNAs and the cell cycle. Biochim Biophys Acta 1812, 592–601 (2011)
A. Esquela-Kerscher, F.J. Slack, Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6, 259–269 (2006)
S.M. Hammond, MicroRNAs as tumor suppressors. Nat Genet 39, 582–583 (2007)
C.M. Croce, Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10, 704–714 (2009)
M. Negrini, M.S. Nicoloso, G.A. Calin, MicroRNAs and cancer--new paradigms in molecular oncology. Curr Opin Cell Biol 21, 470–479 (2009)
M.S. Nicoloso, R. Spizzo, M. Shimizu, S. Rossi, G.A. Calin, MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer 9, 293–302 (2009)
R. Spizzo, M.S. Nicoloso, C.M. Croce, G.A. Calin, SnapShot: MicroRNAs in Cancer. Cell 137, 586–586 e581 (2009)
A. Lujambio, S.W. Lowe, The microcosmos of cancer. Nature 482, 347–355 (2012)
C. Baer, R. Claus, C. Plass, Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res 73, 473–477 (2013)
S. Babashah, M. Soleimani, The oncogenic and tumour suppressive roles of microRNAs in cancer and apoptosis. Eur J Cancer 47, 1127–1137 (2011)
N. Weinhold, A. Jacobsen, N. Schultz, C. Sander, W. Lee, Genome-wide analysis of noncoding regulatory mutations in cancer. Nat Genet 46, 1160–1165 (2014)
O. Hobert, Gene regulation by transcription factors and microRNAs. Science 319, 1785–1786 (2008)
N.J. Martinez, A.J. Walhout, The interplay between transcription factors and microRNAs in genome-scale regulatory networks. BioEssays 31, 435–445 (2009)
A.M. Gurtan, P.A. Sharp, The role of miRNAs in regulating gene expression networks. J Mol Biol 425, 3582–3600 (2013)
H. Hermeking, p53 enters the microRNA world. Cancer Cell 12, 414–418 (2007)
H. Hermeking, MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 12, 613–626 (2012)
P.Y. Lui, D.Y. Jin, N.J. Stevenson, MicroRNA: master controllers of intracellular signaling pathways. Cell Mol Life Sci 72, 3531–3542 (2015)
K.D. Taganov, M.P. Boldin, K.J. Chang, D. Baltimore, NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103, 12481–12486 (2006)
D. Iliopoulos, H.A. Hirsch, K. Struhl, An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009)
X. Ma, L.E. Becker Buscaglia, J.R. Barker, Y. Li, MicroRNAs in NF-kappaB signaling. J Mol Cell Biol 3, 159–166 (2011)
M.P. Boldin, D. Baltimore, MicroRNAs, new effectors and regulators of NF-kappaB. Immunol Rev 246, 205–220 (2012)
L.A. Macfarlane, P.R. Murphy, MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics 11, 537–561 (2010)
C.P. Bracken, H.S. Scott, G.J. Goodall, A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet 17, 719–732 (2016)
S.A. Melo, S. Ropero, C. Moutinho, L.A. Aaltonen, H. Yamamoto, G.A. Calin, S. Rossi, A.F. Fernandez, F. Carneiro, C. Oliveira, B. Ferreira, C.G. Liu, A. Villanueva, G. Capella, S. Schwartz Jr., R. Shiekhattar, M. Esteller, A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 41, 365–370 (2009)
D.A. Hill, J. Ivanovich, J.R. Priest, C.A. Gurnett, L.P. Dehner, D. Desruisseau, J.A. Jarzembowski, K.A. Wikenheiser-Brokamp, B.K. Suarez, A.J. Whelan, G. Williams, D. Bracamontes, Y. Messinger, P.J. Goodfellow, DICER1 mutations in familial pleuropulmonary blastoma. Science 325, 965 (2009)
S.A. Melo, C. Moutinho, S. Ropero, G.A. Calin, S. Rossi, R. Spizzo, A.F. Fernandez, V. Davalos, A. Villanueva, G. Montoya, H. Yamamoto, S. Schwartz Jr., M. Esteller, A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18, 303–315 (2010)
S.A. Melo, H. Sugimoto, J.T. O'Connell, N. Kato, A. Villanueva, A. Vidal, L. Qiu, E. Vitkin, L.T. Perelman, C.A. Melo, A. Lucci, C. Ivan, G.A. Calin, R. Kalluri, Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 (2014)
P. Carninci, T. Kasukawa, S. Katayama, J. Gough, M.C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, R. Kodzius, K. Shimokawa, V.B. Bajic, S.E. Brenner, S. Batalov, A.R. Forrest, M. Zavolan, M.J. Davis, L.G. Wilming, V. Aidinis, J.E. Allen, A. Ambesi-Impiombato, R. Apweiler, R.N. Aturaliya, T.L. Bailey, M. Bansal, L. Baxter, K.W. Beisel, T. Bersano, H. Bono, A.M. Chalk, K.P. Chiu, V. Choudhary, A. Christoffels, D.R. Clutterbuck, M.L. Crowe, E. Dalla, B.P. Dalrymple, B. de Bono, G. Della Gatta, D. di Bernardo, T. Down, P. Engstrom, M. Fagiolini, G. Faulkner, C.F. Fletcher, T. Fukushima, M. Furuno, S. Futaki, M. Gariboldi, P. Georgii-Hemming, T.R. Gingeras, T. Gojobori, R.E. Green, S. Gustincich, M. Harbers, Y. Hayashi, T.K. Hensch, N. Hirokawa, D. Hill, L. Huminiecki, M. Iacono, K. Ikeo, A. Iwama, T. Ishikawa, M. Jakt, A. Kanapin, M. Katoh, Y. Kawasawa, J. Kelso, H. Kitamura, H. Kitano, G. Kollias, S.P. Krishnan, A. Kruger, S.K. Kummerfeld, I.V. Kurochkin, L.F. Lareau, D. Lazarevic, L. Lipovich, J. Liu, S. Liuni, S. McWilliam, M. Madan Babu, M. Madera, L. Marchionni, H. Matsuda, S. Matsuzawa, H. Miki, F. Mignone, S. Miyake, K. Morris, S. Mottagui-Tabar, N. Mulder, N. Nakano, H. Nakauchi, P. Ng, R. Nilsson, S. Nishiguchi, S. Nishikawa, F. Nori, O. Ohara, Y. Okazaki, V. Orlando, K.C. Pang, W.J. Pavan, G. Pavesi, G. Pesole, N. Petrovsky, S. Piazza, J. Reed, J.F. Reid, B.Z. Ring, M. Ringwald, B. Rost, Y. Ruan, S.L. Salzberg, A. Sandelin, C. Schneider, C. Schonbach, K. Sekiguchi, C.A. Semple, S. Seno, L. Sessa, Y. Sheng, Y. Shibata, H. Shimada, K. Shimada, D. Silva, B. Sinclair, S. Sperling, E. Stupka, K. Sugiura, R. Sultana, Y. Takenaka, K. Taki, K. Tammoja, S.L. Tan, S. Tang, M.S. Taylor, J. Tegner, S.A. Teichmann, H.R. Ueda, E. van Nimwegen, R. Verardo, C.L. Wei, K. Yagi, H. Yamanishi, E. Zabarovsky, S. Zhu, A. Zimmer, W. Hide, C. Bult, S.M. Grimmond, R.D. Teasdale, E.T. Liu, V. Brusic, J. Quackenbush, C. Wahlestedt, J.S. Mattick, D.A. Hume, C. Kai, D. Sasaki, Y. Tomaru, S. Fukuda, M. Kanamori-Katayama, M. Suzuki, J. Aoki, T. Arakawa, J. Iida, K. Imamura, M. Itoh, T. Kato, H. Kawaji, N. Kawagashira, T. Kawashima, M. Kojima, S. Kondo, H. Konno, K. Nakano, N. Ninomiya, T. Nishio, M. Okada, C. Plessy, K. Shibata, T. Shiraki, S. Suzuki, M. Tagami, K. Waki, A. Watahiki, Y. Okamura-Oho, H. Suzuki, J. Kawai, Y. Hayashizaki, The transcriptional landscape of the mammalian genome. Sci 309, 1559–1563 (2005)
M.V. Iorio, C.M. Croce, MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4, 143–159 (2012)
K. Jazdzewski, S. Liyanarachchi, M. Swierniak, J. Pachucki, M.D. Ringel, B. Jarzab, A. De la Chapelle, Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA 106, 1502–1505 (2009)
L.J. Chin, E. Ratner, S. Leng, R. Zhai, S. Nallur, I. Babar, R.-U. Muller, E. Straka, L. Su, E.A. Burki, A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non–small cell lung cancer risk. Cancer Res 68, 8535–8540 (2008)
G.A. Calin, C.D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, C.M. Croce, Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99, 15524–15529 (2002)
N. Samuel, G. Wilson, M. Lemire, B.I. Said, Y. Lou, W. Li, D. Merino, A. Novokmet, J. Tran and K.E. Nichols, Genome-wide DNA methylation analysis reveals epigenetic dysregulation of MicroRNA-34A in TP53-associated cancer susceptibility. J Clin Oncol JCO676940 (2016)
S.K. Botla, S. Savant, P. Jandaghi, A.S. Bauer, O. Mücke, E.A. Moskalev, J.P. Neoptolemos, E. Costello, W. Greenhalf and A. Scarpa, Early epigenetic down-regulation of microRNA-192 expression promotes pancreatic cancer progression. Cancer Res 0390.2015 (2016)
V. Davalos, C. Moutinho, A. Villanueva, R. Boque, P. Silva, F. Carneiro, M. Esteller, Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062–2074 (2012)
Y. Saito, G. Liang, G. Egger, J.M. Friedman, J.C. Chuang, G.A. Coetzee, P.A. Jones, Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443 (2006)
A. Lujambio, S. Ropero, E. Ballestar, M.F. Fraga, C. Cerrato, F. Setien, S. Casado, A. Suarez-Gauthier, M. Sanchez-Cespedes, A. Git, I. Spiteri, P.P. Das, C. Caldas, E. Miska, M. Esteller, Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67, 1424–1429 (2007)
M. Hatziapostolou, C. Polytarchou, E. Aggelidou, A. Drakaki, G.A. Poultsides, S.A. Jaeger, H. Ogata, M. Karin, K. Struhl, M. Hadzopoulou-Cladaras, D. Iliopoulos, An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell 147, 1233–1247 (2011)
S. Venkataraman, I. Alimova, R. Fan, P. Harris, N. Foreman, R. Vibhakar, MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One 5, e10748 (2010)
G. Romano, M. Acunzo, M. Garofalo, G. Di Leva, L. Cascione, C. Zanca, B. Bolon, G. Condorelli, C.M. Croce, MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci USA 109, 16570–16575 (2012)
H. He, K. Jazdzewski, W. Li, S. Liyanarachchi, R. Nagy, S. Volinia, G.A. Calin, C.-g. Liu, K. Franssila, S. Suster, The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 102, 19075–19080 (2005)
C. le Sage, R. Nagel, D.A. Egan, M. Schrier, E. Mesman, A. Mangiola, C. Anile, G. Maira, N. Mercatelli, S.A. Ciafre, M.G. Farace, R. Agami, Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 26, 3699–3708 (2007)
C. le Sage, R. Nagel, R. Agami, Diverse ways to control p27Kip1 function: miRNAs come into play. Cell Cycle 6, 2742–2749 (2007)
M. Kedde, M. van Kouwenhove, W. Zwart, J.A. Oude Vrielink, R. Elkon, R. Agami, A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol 12, 1014–1020 (2010)
P. Pineau, S. Volinia, K. McJunkin, A. Marchio, C. Battiston, B. Terris, V. Mazzaferro, S.W. Lowe, C.M. Croce, A. Dejean, miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA 107, 264–269 (2010)
M. Garofalo, G. Di Leva, G. Romano, G. Nuovo, S.S. Suh, A. Ngankeu, C. Taccioli, F. Pichiorri, H. Alder, P. Secchiero, P. Gasparini, A. Gonelli, S. Costinean, M. Acunzo, G. Condorelli, C.M. Croce, miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509 (2009)
G. Roscigno, C. Quintavalle, E. Donnarumma, I. Puoti, A. Diaz-Lagares, M. Iaboni, D. Fiore, V. Russo, M. Todaro, G. Romano, R. Thomas, G. Cortino, M. Gaggianesi, M. Esteller, C.M. Croce, G. Condorelli, MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget 7, 580–592 (2016)
G.A. Calin, C.M. Croce, MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 66, 7390–7394 (2006)
F. Meng, R. Henson, H. Wehbe-Janek, K. Ghoshal, S.T. Jacob, T. Patel, MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 (2007)
M.E. Hatley, D.M. Patrick, M.R. Garcia, J.A. Richardson, R. Bassel-Duby, E. van Rooij, E.N. Olson, Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 18, 282–293 (2010)
P. Olson, J. Lu, H. Zhang, A. Shai, M.G. Chun, Y. Wang, S.K. Libutti, E.K. Nakakura, T.R. Golub, D. Hanahan, MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev 23, 2152–2165 (2009)
C.T. Dickman, R. Towle, R. Saini, C. Garnis, Molecular characterization of immortalized normal and dysplastic oral cell lines. J Oral Pathol Med 44, 329–336 (2015)
N.K. Cervigne, P.P. Reis, J. Machado, B. Sadikovic, G. Bradley, N.N. Galloni, M. Pintilie, I. Jurisica, B. Perez-Ordonez, R. Gilbert, P. Gullane, J. Irish, S. Kamel-Reid, Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum Mol Genet 18, 4818–4829 (2009)
A.J. Granados Lopez, J.A. Lopez, Multistep model of cervical cancer: participation of miRNAs and coding genes. Int J Mol Sci 15, 15700–15733 (2014)
Q. Ren, J. Liang, J. Wei, O. Basturk, J. Wang, G. Daniels, L.L. Gellert, Y. Li, Y. Shen, I. Osman, J. Zhao, J. Melamed, P. Lee, Epithelial and stromal expression of miRNAs during prostate cancer progression. Am J Transl Res 6, 329–339 (2014)
G.S. Markopoulos, E. Roupakia, M. Tokamani, G. Vartholomatos, T. Tzavaras, M. Hatziapostolou, F.O. Fackelmayer, R. Sandaltzopoulos, C. Polytarchou, E. Kolettas, Senescence-associated microRNAs target cell cycle regulatory genes in normal human lung fibroblasts. Exp Gerontol 96, 110–122 (2017)
P.S. Linsley, J. Schelter, J. Burchard, M. Kibukawa, M.M. Martin, S.R. Bartz, J.M. Johnson, J.M. Cummins, C.K. Raymond, H. Dai, N. Chau, M. Cleary, A.L. Jackson, M. Carleton, L. Lim, Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol 27, 2240–2252 (2007)
S. Arora, R. Rana, A. Chhabra, A. Jaiswal, V. Rani, miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics 288, 77–87 (2013)
M.R. Stratton, P.J. Campbell, P.A. Futreal, The cancer genome. Nature 458, 719–724 (2009)
B. Vogelstein, N. Papadopoulos, V.E. Velculescu, S. Zhou, L.A. Diaz Jr., K.W. Kinzler, Cancer genome landscapes. Science 339, 1546–1558 (2013)
L.A. Garraway, E.S. Lander, Lessons from the cancer genome. Cell 153, 17–37 (2013)
H. Shen, P.W. Laird, Interplay between the cancer genome and epigenome. Cell 153, 38–55 (2013)
F. Castro-Giner, P. Ratcliffe, I. Tomlinson, The mini-driver model of polygenic cancer evolution. Nat Rev Cancer 15, 680–685 (2015)
T.A. Ince, A.L. Richardson, G.W. Bell, M. Saitoh, S. Godar, A.E. Karnoub, J.D. Iglehart, R.A. Weinberg, Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell 12, 160–170 (2007)
R.A. Weinberg, Mechanisms of malignant progression. Carcinogenesis 29, 1092–1095 (2008)
K. Gross, A. Wronski, A. Skibinski, S. Phillips, C. Kuperwasser, Cell fate decisions during breast cancer development. J Dev Biol 4, 4 (2016)
P.A. Futreal, L. Coin, M. Marshall, T. Down, T. Hubbard, R. Wooster, N. Rahman, M.R. Stratton, A census of human cancer genes. Nat Rev Cancer 4, 177–183 (2004)
T.J. Hudson, W. Anderson, A. Artez, A.D. Barker, C. Bell, R.R. Bernabe, M.K. Bhan, F. Calvo, I. Eerola, D.S. Gerhard, A. Guttmacher, M. Guyer, F.M. Hemsley, J.L. Jennings, D. Kerr, P. Klatt, P. Kolar, J. Kusada, D.P. Lane, F. Laplace, L. Youyong, G. Nettekoven, B. Ozenberger, J. Peterson, T.S. Rao, J. Remacle, A.J. Schafer, T. Shibata, M.R. Stratton, J.G. Vockley, K. Watanabe, H. Yang, M.M. Yuen, B.M. Knoppers, M. Bobrow, A. Cambon-Thomsen, L.G. Dressler, S.O. Dyke, Y. Joly, K. Kato, K.L. Kennedy, P. Nicolas, M.J. Parker, E. Rial-Sebbag, C.M. Romeo-Casabona, K.M. Shaw, S. Wallace, G.L. Wiesner, N. Zeps, P. Lichter, A.V. Biankin, C. Chabannon, L. Chin, B. Clement, E. de Alava, F. Degos, M.L. Ferguson, P. Geary, D.N. Hayes, A.L. Johns, A. Kasprzyk, H. Nakagawa, R. Penny, M.A. Piris, R. Sarin, A. Scarpa, M. van de Vijver, P.A. Futreal, H. Aburatani, M. Bayes, D.D. Botwell, P.J. Campbell, X. Estivill, S.M. Grimmond, I. Gut, M. Hirst, C. Lopez-Otin, P. Majumder, M. Marra, J.D. McPherson, Z. Ning, X.S. Puente, Y. Ruan, H.G. Stunnenberg, H. Swerdlow, V.E. Velculescu, R.K. Wilson, H.H. Xue, L. Yang, P.T. Spellman, G.D. Bader, P.C. Boutros, P. Flicek, G. Getz, R. Guigo, G. Guo, D. Haussler, S. Heath, T.J. Hubbard, T. Jiang, S.M. Jones, Q. Li, N. Lopez-Bigas, R. Luo, L. Muthuswamy, B.F. Ouellette, J.V. Pearson, V. Quesada, B.J. Raphael, C. Sander, T.P. Speed, L.D. Stein, J.M. Stuart, J.W. Teague, Y. Totoki, T. Tsunoda, A. Valencia, D.A. Wheeler, H. Wu, S. Zhao, G. Zhou, M. Lathrop, G. Thomas, T. Yoshida, M. Axton, C. Gunter, L.J. Miller, J. Zhang, S.A. Haider, J. Wang, C.K. Yung, A. Cros, Y. Liang, S. Gnaneshan, J. Guberman, J. Hsu, D.R. Chalmers, K.W. Hasel, T.S. Kaan, W.W. Lowrance, T. Masui, L.L. Rodriguez, C. Vergely, D.D. Bowtell, N. Cloonan, A. deFazio, J.R. Eshleman, D. Etemadmoghadam, B.B. Gardiner, J.G. Kench, R.L. Sutherland, M.A. Tempero, N.J. Waddell, P.J. Wilson, S. Gallinger, M.S. Tsao, P.A. Shaw, G.M. Petersen, D. Mukhopadhyay, R.A. DePinho, S. Thayer, K. Shazand, T. Beck, M. Sam, L. Timms, V. Ballin, Y. Lu, J. Ji, X. Zhang, F. Chen, X. Hu, Q. Yang, G. Tian, L. Zhang, X. Xing, X. Li, Z. Zhu, Y. Yu, J. Yu, J. Tost, P. Brennan, I. Holcatova, D. Zaridze, A. Brazma, L. Egevard, E. Prokhortchouk, R.E. Banks, M. Uhlen, J. Viksna, F. Ponten, K. Skryabin, E. Birney, A. Borg, A.L. Borresen-Dale, C. Caldas, J.A. Foekens, S. Martin, J.S. Reis-Filho, A.L. Richardson, C. Sotiriou, G. Thoms, L. van't Veer, D. Birnbaum, H. Blanche, P. Boucher, S. Boyault, J.D. Masson-Jacquemier, I. Pauporte, X. Pivot, A. Vincent-Salomon, E. Tabone, C. Theillet, I. Treilleux, P. Bioulac-Sage, T. Decaens, D. Franco, M. Gut, D. Samuel, J. Zucman-Rossi, R. Eils, B. Brors, J.O. Korbel, A. Korshunov, P. Landgraf, H. Lehrach, S. Pfister, B. Radlwimmer, G. Reifenberger, M.D. Taylor, C. von Kalle, P.P. Majumder, P. Pederzoli, R.A. Lawlor, M. Delledonne, A. Bardelli, T. Gress, D. Klimstra, G. Zamboni, Y. Nakamura, S. Miyano, A. Fujimoto, E. Campo, S. de Sanjose, E. Montserrat, M. Gonzalez-Diaz, P. Jares, H. Himmelbauer, S. Bea, S. Aparicio, D.F. Easton, F.S. Collins, C.C. Compton, E.S. Lander, W. Burke, A.R. Green, S.R. Hamilton, O.P. Kallioniemi, T.J. Ley, E.T. Liu, B.J. Wainwright, International network of cancer genome projects. Nature 464, 993–998 (2010)
L. Ding, G. Getz, D.A. Wheeler, E.R. Mardis, M.D. McLellan, K. Cibulskis, C. Sougnez, H. Greulich, D.M. Muzny, M.B. Morgan, L. Fulton, R.S. Fulton, Q. Zhang, M.C. Wendl, M.S. Lawrence, D.E. Larson, K. Chen, D.J. Dooling, A. Sabo, A.C. Hawes, H. Shen, S.N. Jhangiani, L.R. Lewis, O. Hall, Y. Zhu, T. Mathew, Y. Ren, J. Yao, S.E. Scherer, K. Clerc, G.A. Metcalf, B. Ng, A. Milosavljevic, M.L. Gonzalez-Garay, J.R. Osborne, R. Meyer, X. Shi, Y. Tang, D.C. Koboldt, L. Lin, R. Abbott, T.L. Miner, C. Pohl, G. Fewell, C. Haipek, H. Schmidt, B.H. Dunford-Shore, A. Kraja, S.D. Crosby, C.S. Sawyer, T. Vickery, S. Sander, J. Robinson, W. Winckler, J. Baldwin, L.R. Chirieac, A. Dutt, T. Fennell, M. Hanna, B.E. Johnson, R.C. Onofrio, R.K. Thomas, G. Tonon, B.A. Weir, X. Zhao, L. Ziaugra, M.C. Zody, T. Giordano, M.B. Orringer, J.A. Roth, M.R. Spitz, I.I. Wistuba, B. Ozenberger, P.J. Good, A.C. Chang, D.G. Beer, M.A. Watson, M. Ladanyi, S. Broderick, A. Yoshizawa, W.D. Travis, W. Pao, M.A. Province, G.M. Weinstock, H.E. Varmus, S.B. Gabriel, E.S. Lander, R.A. Gibbs, M. Meyerson, R.K. Wilson, Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008)
B.A. Weir, M.S. Woo, G. Getz, S. Perner, L. Ding, R. Beroukhim, W.M. Lin, M.A. Province, A. Kraja, L.A. Johnson, K. Shah, M. Sato, R.K. Thomas, J.A. Barletta, I.B. Borecki, S. Broderick, A.C. Chang, D.Y. Chiang, L.R. Chirieac, J. Cho, Y. Fujii, A.F. Gazdar, T. Giordano, H. Greulich, M. Hanna, B.E. Johnson, M.G. Kris, A. Lash, L. Lin, N. Lindeman, E.R. Mardis, J.D. McPherson, J.D. Minna, M.B. Morgan, M. Nadel, M.B. Orringer, J.R. Osborne, B. Ozenberger, A.H. Ramos, J. Robinson, J.A. Roth, V. Rusch, H. Sasaki, F. Shepherd, C. Sougnez, M.R. Spitz, M.S. Tsao, D. Twomey, R.G. Verhaak, G.M. Weinstock, D.A. Wheeler, W. Winckler, A. Yoshizawa, S. Yu, M.F. Zakowski, Q. Zhang, D.G. Beer, I.I. Wistuba, M.A. Watson, L.A. Garraway, M. Ladanyi, W.D. Travis, W. Pao, M.A. Rubin, S.B. Gabriel, R.A. Gibbs, H.E. Varmus, R.K. Wilson, E.S. Lander, M. Meyerson, Characterizing the cancer genome in lung adenocarcinoma. Nature 450, 893–898 (2007)
M. Imielinski, A.H. Berger, P.S. Hammerman, B. Hernandez, T.J. Pugh, E. Hodis, J. Cho, J. Suh, M. Capelletti, A. Sivachenko, C. Sougnez, D. Auclair, M.S. Lawrence, P. Stojanov, K. Cibulskis, K. Choi, L. de Waal, T. Sharifnia, A. Brooks, H. Greulich, S. Banerji, T. Zander, D. Seidel, F. Leenders, S. Ansen, C. Ludwig, W. Engel-Riedel, E. Stoelben, J. Wolf, C. Goparju, K. Thompson, W. Winckler, D. Kwiatkowski, B.E. Johnson, P.A. Janne, V.A. Miller, W. Pao, W.D. Travis, H.I. Pass, S.B. Gabriel, E.S. Lander, R.K. Thomas, L.A. Garraway, G. Getz, M. Meyerson, Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 (2012)
Z. Chen, C.M. Fillmore, P.S. Hammerman, C.F. Kim, K.K. Wong, Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14, 535–546 (2014)
E.C. de Bruin, N. McGranahan, R. Mitter, M. Salm, D.C. Wedge, L. Yates, M. Jamal-Hanjani, S. Shafi, N. Murugaesu, A.J. Rowan, E. Gronroos, M.A. Muhammad, S. Horswell, M. Gerlinger, I. Varela, D. Jones, J. Marshall, T. Voet, P. Van Loo, D.M. Rassl, R.C. Rintoul, S.M. Janes, S.M. Lee, M. Forster, T. Ahmad, D. Lawrence, M. Falzon, A. Capitanio, T.T. Harkins, C.C. Lee, W. Tom, E. Teefe, S.C. Chen, S. Begum, A. Rabinowitz, B. Phillimore, B. Spencer-Dene, G. Stamp, Z. Szallasi, N. Matthews, A. Stewart, P. Campbell, C. Swanton, Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Sci 346, 251–256 (2014)
Cancer Genome Atlas Research Network, Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012)
M. Peifer, L. Fernandez-Cuesta, M.L. Sos, J. George, D. Seidel, L.H. Kasper, D. Plenker, F. Leenders, R. Sun, T. Zander, R. Menon, M. Koker, I. Dahmen, C. Muller, V. Di Cerbo, H.U. Schildhaus, J. Altmuller, I. Baessmann, C. Becker, B. de Wilde, J. Vandesompele, D. Bohm, S. Ansen, F. Gabler, I. Wilkening, S. Heynck, J.M. Heuckmann, X. Lu, S.L. Carter, K. Cibulskis, S. Banerji, G. Getz, K.S. Park, D. Rauh, C. Grutter, M. Fischer, L. Pasqualucci, G. Wright, Z. Wainer, P. Russell, I. Petersen, Y. Chen, E. Stoelben, C. Ludwig, P. Schnabel, H. Hoffmann, T. Muley, M. Brockmann, W. Engel-Riedel, L.A. Muscarella, V.M. Fazio, H. Groen, W. Timens, H. Sietsma, E. Thunnissen, E. Smit, D.A. Heideman, P.J. Snijders, F. Cappuzzo, C. Ligorio, S. Damiani, J. Field, S. Solberg, O.T. Brustugun, M. Lund-Iversen, J. Sanger, J.H. Clement, A. Soltermann, H. Moch, W. Weder, B. Solomon, J.C. Soria, P. Validire, B. Besse, E. Brambilla, C. Brambilla, S. Lantuejoul, P. Lorimier, P.M. Schneider, M. Hallek, W. Pao, M. Meyerson, J. Sage, J. Shendure, R. Schneider, R. Buttner, J. Wolf, P. Nurnberg, S. Perner, L.C. Heukamp, P.K. Brindle, S. Haas, R.K. Thomas, Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44, 1104–1110 (2012)
C.M. Rudin, S. Durinck, E.W. Stawiski, J.T. Poirier, Z. Modrusan, D.S. Shames, E.A. Bergbower, Y. Guan, J. Shin, J. Guillory, C.S. Rivers, C.K. Foo, D. Bhatt, J. Stinson, F. Gnad, P.M. Haverty, R. Gentleman, S. Chaudhuri, V. Janakiraman, B.S. Jaiswal, C. Parikh, W. Yuan, Z. Zhang, H. Koeppen, T.D. Wu, H.M. Stern, R.L. Yauch, K.E. Huffman, D.D. Paskulin, P.B. Illei, M. Varella-Garcia, A.F. Gazdar, F.J. de Sauvage, R. Bourgon, J.D. Minna, M.V. Brock, S. Seshagiri, Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44, 1111–1116 (2012)
J. George, J.S. Lim, S.J. Jang, Y. Cun, L. Ozretic, G. Kong, F. Leenders, X. Lu, L. Fernandez-Cuesta, G. Bosco, C. Muller, I. Dahmen, N.S. Jahchan, K.S. Park, D. Yang, A.N. Karnezis, D. Vaka, A. Torres, M.S. Wang, J.O. Korbel, R. Menon, S.M. Chun, D. Kim, M. Wilkerson, N. Hayes, D. Engelmann, B. Putzer, M. Bos, S. Michels, I. Vlasic, D. Seidel, B. Pinther, P. Schaub, C. Becker, J. Altmuller, J. Yokota, T. Kohno, R. Iwakawa, K. Tsuta, M. Noguchi, T. Muley, H. Hoffmann, P.A. Schnabel, I. Petersen, Y. Chen, A. Soltermann, V. Tischler, C.M. Choi, Y.H. Kim, P.P. Massion, Y. Zou, D. Jovanovic, M. Kontic, G.M. Wright, P.A. Russell, B. Solomon, I. Koch, M. Lindner, L.A. Muscarella, A. la Torre, J.K. Field, M. Jakopovic, J. Knezevic, E. Castanos-Velez, L. Roz, U. Pastorino, O.T. Brustugun, M. Lund-Iversen, E. Thunnissen, J. Kohler, M. Schuler, J. Botling, M. Sandelin, M. Sanchez-Cespedes, H.B. Salvesen, V. Achter, U. Lang, M. Bogus, P.M. Schneider, T. Zander, S. Ansen, M. Hallek, J. Wolf, M. Vingron, Y. Yatabe, W.D. Travis, P. Nurnberg, C. Reinhardt, S. Perner, L. Heukamp, R. Buttner, S.A. Haas, E. Brambilla, M. Peifer, J. Sage, R.K. Thomas, Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015)
E. Hodis, I.R. Watson, G.V. Kryukov, S.T. Arold, M. Imielinski, J.P. Theurillat, E. Nickerson, D. Auclair, L. Li, C. Place, D. Dicara, A.H. Ramos, M.S. Lawrence, K. Cibulskis, A. Sivachenko, D. Voet, G. Saksena, N. Stransky, R.C. Onofrio, W. Winckler, K. Ardlie, N. Wagle, J. Wargo, K. Chong, D.L. Morton, K. Stemke-Hale, G. Chen, M. Noble, M. Meyerson, J.E. Ladbury, M.A. Davies, J.E. Gershenwald, S.N. Wagner, D.S. Hoon, D. Schadendorf, E.S. Lander, S.B. Gabriel, G. Getz, L.A. Garraway, L. Chin, A landscape of driver mutations in melanoma. Cell 150, 251–263 (2012)
M.S. Lawrence, P. Stojanov, C.H. Mermel, J.T. Robinson, L.A. Garraway, T.R. Golub, M. Meyerson, S.B. Gabriel, E.S. Lander, G. Getz, Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014)
S. Horn, A. Figl, P.S. Rachakonda, C. Fischer, A. Sucker, A. Gast, S. Kadel, I. Moll, E. Nagore, K. Hemminki, D. Schadendorf, R. Kumar, TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013)
N.J. Fredriksson, L. Ny, J.A. Nilsson, E. Larsson, Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet 46, 1258–1263 (2014)
M. Collado, J. Gil, A. Efeyan, C. Guerra, A.J. Schuhmacher, M. Barradas, A. Benguria, A. Zaballos, J.M. Flores, M. Barbacid, D. Beach, M. Serrano, Tumour biology: Senescence in premalignant tumours. Nature 436, 642 (2005)
J. Bartkova, N. Rezaei, M. Liontos, P. Karakaidos, D. Kletsas, N. Issaeva, L.V. Vassiliou, E. Kolettas, K. Niforou, V.C. Zoumpourlis, M. Takaoka, H. Nakagawa, F. Tort, K. Fugger, F. Johansson, M. Sehested, C.L. Andersen, L. Dyrskjot, T. Orntoft, J. Lukas, C. Kittas, T. Helleday, T.D. Halazonetis, J. Bartek, V.G. Gorgoulis, Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 (2006)
M. Collado, M.A. Blasco, M. Serrano, Cellular senescence in cancer and aging. Cell 130, 223–233 (2007)
A. Efeyan, M. Murga, B. Martinez-Pastor, A. Ortega-Molina, R. Soria, M. Collado, O. Fernandez-Capetillo, M. Serrano, Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One 4, e5475 (2009)
M. Collado, M. Serrano, Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 10, 51–57 (2010)
P.A. Perez-Mancera, A.R. Young, M. Narita, Inside and out: the activities of senescence in cancer. Nat Rev Cancer 14, 547–558 (2014)
F. Rodier, J. Campisi, Four faces of cellular senescence. J Cell Biol 192, 547–556 (2011)
D. Dankort, E. Filenova, M. Collado, M. Serrano, K. Jones, M. McMahon, A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev 21, 379–384 (2007)
C. Michaloglou, L.C. Vredeveld, M.S. Soengas, C. Denoyelle, T. Kuilman, C.M. van der Horst, D.M. Majoor, J.W. Shay, W.J. Mooi, D.S. Peeper, BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 436, 720–724 (2005)
N. Dhomen, J.S. Reis-Filho, S. da Rocha Dias, R. Hayward, K. Savage, V. Delmas, L. Larue, C. Pritchard, R. Marais, Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15, 294–303 (2009)
M. Vergel, A. Carnero, Bypassing cellular senescence by genetic screening tools. Clin Transl Oncol 12, 410–417 (2010)
M. Braig, S. Lee, C. Loddenkemper, C. Rudolph, A.H. Peters, B. Schlegelberger, H. Stein, B. Dorken, T. Jenuwein, C.A. Schmitt, Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436, 660–665 (2005)
P.M. Voorhoeve, C. Le Sage, M. Schrier, A.J. Gillis, H. Stoop, R. Nagel, Y.-P. Liu, J. Van Duijse, J. Drost, A. Griekspoor, A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 124, 1169–1181 (2006)
J. Grillari, M. Hackl, R. Grillari-Voglauer, miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology 11, 501–506 (2010)
M. Gorospe, K. Abdelmohsen, MicroRegulators come of age in senescence. Trends Genet 27, 233–241 (2011)
E. Schraml, J. Grillari, From cellular senescence to age-associated diseases: the miRNA connection. Longev Healthspan 1, 10 (2012)
O. Bischof, R.I. Martinez-Zamudio, MicroRNAs and lncRNAs in senescence: A re-view. IUBMB Life 67, 255–267 (2015)
H. Toledano, The role of the heterochronic microRNA let-7 in the progression of aging. Exp Gerontol 48, 667–670 (2013)
V. Olive, I. Jiang, L. He, mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol 42, 1348–1354 (2010)
F. d’Adda di Fagagna, Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer 8, 512–522 (2008)
M. Fumagalli, F. d’Adda di Fagagna, SASPense and DDRama in cancer and ageing. Nat Cell Biol 11, 921–923 (2009)
J.T. Mendell, miRiad roles for the miR-17-92 cluster in development and disease. Cell 133, 217–222 (2008)
C.P. Concepcion, C. Bonetti, A. Ventura, The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J 18, 262–267 (2012)
L. Hong, M. Lai, M. Chen, C. Xie, R. Liao, Y.J. Kang, C. Xiao, W.Y. Hu, J. Han, P. Sun, The miR-17-92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res 70, 8547–8557 (2010)
V. Borgdorff, M.E. Lleonart, C.L. Bishop, D. Fessart, A.H. Bergin, M.G. Overhoff, D.H. Beach, Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1). Oncogene 29, 2262–2271 (2010)
G. Li, C. Luna, J. Qiu, D.L. Epstein, P. Gonzalez, Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev 130, 731–741 (2009)
Y. Liu, W. Qiang, X. Xu, R. Dong, A.M. Karst, Z. Liu, B. Kong, R.I. Drapkin, J.J. Wei, Role of miR-182 in response to oxidative stress in the cell fate of human fallopian tube epithelial cells. Oncotarget 6, 38983–38998 (2015)
P. Moskwa, F.M. Buffa, Y. Pan, R. Panchakshari, P. Gottipati, R.J. Muschel, J. Beech, R. Kulshrestha, K. Abdelmohsen, D.M. Weinstock, M. Gorospe, A.L. Harris, T. Helleday, D. Chowdhury, miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 41, 210–220 (2011)
S.M. Kooistra, L.C. Norgaard, M.J. Lees, C. Steinhauer, J.V. Johansen, K. Helin, A screen identifies the oncogenic micro-RNA miR-378a-5p as a negative regulator of oncogene-induced senescence. PLoS One 9, e91034 (2014)
O.C. Maes, H. Sarojini, E. Wang, Stepwise up-regulation of microRNA expression levels from replicating to reversible and irreversible growth arrest states in WI-38 human fibroblasts. J Cell Physiol 221, 109–119 (2009)
H. Tazawa, N. Tsuchiya, M. Izumiya, H. Nakagama, Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA 104, 15472–15477 (2007)
L.N. Bonifacio, M.B. Jarstfer, MiRNA profile associated with replicative senescence, extended cell culture, and ectopic telomerase expression in human foreskin fibroblasts. PLoS One 5, e12519 (2010)
N.R. Christoffersen, R. Shalgi, L.B. Frankel, E. Leucci, M. Lees, M. Klausen, Y. Pilpel, F.C. Nielsen, M. Oren, A.H. Lund, p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 17, 236–245 (2010)
D. Lodygin, V. Tarasov, A. Epanchintsev, C. Berking, T. Knyazeva, H. Korner, P. Knyazev, J. Diebold, H. Hermeking, Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle 7, 2591–2600 (2008)
D. Bhaumik, G.K. Scott, S. Schokrpur, C.K. Patil, A.V. Orjalo, F. Rodier, G.J. Lithgow, J. Campisi, MicroRNAs miR-146a/b negatively modulate the senescence-associated inflammatory mediators IL-6 and IL-8. Aging 1, 402–411 (2009)
D. Bhaumik, G.K. Scott, S. Schokrpur, C.K. Patil, J. Campisi, C.C. Benz, Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene 27, 5643–5647 (2008)
M. Vasa-Nicotera, H. Chen, P. Tucci, A.L. Yang, G. Saintigny, R. Menghini, C. Mahe, M. Agostini, R.A. Knight, G. Melino, M. Federici, miR-146a is modulated in human endothelial cell with aging. Atherosclerosis 217, 326–330 (2011)
R. Brosh, R. Shalgi, A. Liran, G. Landan, K. Korotayev, G.H. Nguyen, E. Enerly, H. Johnsen, Y. Buganim, H. Solomon, p53-repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol 4, 229 (2008)
G. Wan, R. Mathur, X. Hu, X. Zhang, X. Lu, miRNA response to DNA damage. Trends Biochem Sci 36, 478–484 (2011)
L. He, X. He, L.P. Lim, E. de Stanchina, Z. Xuan, Y. Liang, W. Xue, L. Zender, J. Magnus, D. Ridzon, A.L. Jackson, P.S. Linsley, C. Chen, S.W. Lowe, M.A. Cleary, G.J. Hannon, A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 (2007)
H. Hermeking, The miR-34 family in cancer and apoptosis. Cell Death Differ 17, 193–199 (2010)
C.D. Johnson, A. Esquela-Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. Wang, J. Shelton, J. Shingara, L. Chin, D. Brown, F.J. Slack, The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67, 7713–7722 (2007)
M. Benhamed, U. Herbig, T. Ye, A. Dejean, O. Bischof, Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol 14, 266–275 (2012)
A. Tzatsos, P. Paskaleva, S. Lymperi, G. Contino, S. Stoykova, Z. Chen, K.K. Wong, N. Bardeesy, Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem 286, 33061–33069 (2011)
R. Greussing, M. Hackl, P. Charoentong, A. Pauck, R. Monteforte, M. Cavinato, E. Hofer, M. Scheideler, M. Neuhaus, L. Micutkova, C. Mueck, Z. Trajanoski, J. Grillari, P. Jansen-Durr, Identification of microRNA-mRNA functional interactions in UVB-induced senescence of human diploid fibroblasts. BMC Genomics 14, 224 (2013)
A. Tzatsos, P. Paskaleva, F. Ferrari, V. Deshpande, S. Stoykova, G. Contino, K.K. Wong, F. Lan, P. Trojer, P.J. Park, N. Bardeesy, KDM2B promotes pancreatic cancer via Polycomb-dependent and -independent transcriptional programs. J Clin Invest 123, 727–739 (2013)
F. Kottakis, C. Polytarchou, P. Foltopoulou, I. Sanidas, S.C. Kampranis, P.N. Tsichlis, FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell 43, 285–298 (2011)
K. Lafferty-Whyte, C.J. Cairney, N.B. Jamieson, K.A. Oien, W.N. Keith, Pathway analysis of senescence-associated miRNA targets reveals common processes to different senescence induction mechanisms. Biochim Biophys Acta (BBA)-Mol Basis Dis 1792, 341–352 (2009)
C. Liu, K. Kelnar, A.V. Vlassov, D. Brown, J. Wang, D.G. Tang, Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res 72, 3393–3404 (2012)
R. Guo, J. Gu, Z. Zhang, Y. Wang, C. Gu, MicroRNA-410 functions as a tumor suppressor by targeting angiotensin II type 1 receptor in pancreatic cancer. IUBMB Life 67, 42–53 (2015)
Y. Goto, A. Kurozumi, H. Enokida, T. Ichikawa, N. Seki, Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol 22, 242–252 (2015)
J. Wang, Z. Li, Q. Ge, W. Wu, Q. Zhu, J. Luo, L. Chen, Characterization of microRNA transcriptome in tumor, adjacent, and normal tissues of lung squamous cell carcinoma. J Thorac Cardiovasc Surg 149, 1404–1414 e1404 (2015)
J. Ma, K. Mannoor, L. Gao, A. Tan, M.A. Guarnera, M. Zhan, A. Shetty, S.A. Stass, L. Xing, F. Jiang, Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol Oncol 8, 1208–1219 (2014)
R. Saab, Senescence and pre-malignancy: how do tumors progress? Semin Cancer Biol 21, 385–391 (2011)
M. Raica, A.M. Cimpean, D. Ribatti, Angiogenesis in pre-malignant conditions. Eur J Cancer 45, 1924–1934 (2009)
R. Paduch, The role of lymphangiogenesis and angiogenesis in tumor metastasis. Cell Oncol 39, 397–410 (2016)
S. Wang, E.N. Olson, AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev 19, 205–211 (2009)
A. Caporali, C. Emanueli, MicroRNA regulation in angiogenesis. Vasc Pharmacol 55, 79–86 (2011)
M.M. Santoro, S. Nicoli, miRNAs in endothelial cell signaling: the endomiRNAs. Exp Cell Res 319, 1324–1330 (2013)
N.M. Kane, A.J. Thrasher, G.D. Angelini, C. Emanueli, Concise review: MicroRNAs as modulators of stem cells and angiogenesis. Stem Cells 32, 1059–1066 (2014)
S.M. Weis, D.A. Cheresh, Tumor angiogenesis: molecular pathways and therapeutic targets. Nat Med 17, 1359–1370 (2011)
W. Wang, E. Zhang, C. Lin, MicroRNAs in tumor angiogenesis. Life Sci 136, 28–35 (2015)
J. Chou, P. Shahi, Z. Werb, microRNA-mediated regulation of the tumor microenvironment. Cell Cycle 12, 3262–3271 (2013)
H.I. Suzuki, A. Katsura, H. Matsuyama, K. Miyazono, MicroRNA regulons in tumor microenvironment. Oncogene 34, 3085–3094 (2015)
P.R. Kuninty, J. Schnittert, G. Storm, J. Prakash, MicroRNA targeting to modulate tumor microenvironment. Front Oncol 6, 3 (2016)
Z. Li, P. Chen, R. Su, Y. Li, C. Hu, Y. Wang, S. Arnovitz, M. He, S. Gurbuxani, Z. Zuo, A.G. Elkahloun, S. Li, H. Weng, H. Huang, M.B. Neilly, S. Wang, E.N. Olson, R.A. Larson, M.M. Le Beau, J. Zhang, X. Jiang, M. Wei, J. Jin, P.P. Liu, J. Chen, Overexpression and knockout of miR-126 both promote leukemogenesis. Blood 126, 2005–2015 (2015)
E.R. Lechman, B. Gentner, S.W. Ng, E.M. Schoof, P. van Galen, J.A. Kennedy, S. Nucera, F. Ciceri, K.B. Kaufmann, N. Takayama, S.M. Dobson, A. Trotman-Grant, G. Krivdova, J. Elzinga, A. Mitchell, B. Nilsson, K.G. Hermans, K. Eppert, R. Marke, R. Isserlin, V. Voisin, G.D. Bader, P.W. Zandstra, T.R. Golub, B.L. Ebert, J. Lu, M. Minden, J.C. Wang, L. Naldini, J.E. Dick, miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell 29, 602–606 (2016)
L. Poliseno, A. Tuccoli, L. Mariani, M. Evangelista, L. Citti, K. Woods, A. Mercatanti, S. Hammond, G. Rainaldi, MicroRNAs modulate the angiogenic properties of HUVECs. Blood 108, 3068–3071 (2006)
Z. Hua, Q. Lv, W. Ye, C.-K.A. Wong, G. Cai, D. Gu, Y. Ji, C. Zhao, J. Wang, B.B. Yang, MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One 1, e116 (2006)
C. Urbich, A. Kuehbacher, S. Dimmeler, Role of microRNAs in vascular diseases, inflammation and angiogenesis. Cardiovasc Res 9, 581–588 (2008)
M. Dews, A. Homayouni, D. Yu, D. Murphy, C. Sevignani, E. Wentzel, E.E. Furth, W.M. Lee, G.H. Enders, J.T. Mendell, Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38, 1060–1065 (2006)
K.J. Png, N. Halberg, M. Yoshida, S.F. Tavazoie, A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 481, 190–194 (2011)
P. Fasanaro, Y. D'Alessandra, V. Di Stefano, R. Melchionna, S. Romani, G. Pompilio, M.C. Capogrossi, F. Martelli, MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem 283, 15878–15883 (2008)
J.-g. Zhang, J.-j. Wang, F. Zhao, Q. Liu, K. Jiang, G.-h. Yang, MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta 411, 846–852 (2010)
L.-X. Yan, X.-F. Huang, Q. Shao, M.-Y. Huang, L. Deng, Q.-L. Wu, Y.-X. Zeng, J.-Y. Shao, MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14, 2348–2360 (2008)
I. Asangani, S. Rasheed, D. Nikolova, J. Leupold, N. Colburn, S. Post, H. Allgayer, MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 (2008)
L.Z. Liu, C. Li, Q. Chen, Y. Jing, R. Carpenter, Y. Jiang, H.F. Kung, L. Lai, B.H. Jiang, MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1alpha expression. PLoS One 6, e19139 (2011)
M.P. Boldin, K.D. Taganov, D.S. Rao, L. Yang, J.L. Zhao, M. Kalwani, Y. Garcia-Flores, M. Luong, A. Devrekanli, J. Xu, miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med 208, 1189–1201 (2011)
G. Ghosh, I.V. Subramanian, N. Adhikari, X. Zhang, H.P. Joshi, D. Basi, Y.S. Chandrashekhar, J.L. Hall, S. Roy, Y. Zeng, S. Ramakrishnan, Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest 120, 4141–4154 (2010)
Y. Zhang, P. Yang, T. Sun, D. Li, X. Xu, Y. Rui, C. Li, M. Chong, T. Ibrahim, L. Mercatali, D. Amadori, X. Lu, D. Xie, Q.J. Li and X.F. Wang, miR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol 15, 284–294 (2013)
D. Hanahan, L.M. Coussens, Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21, 309–322 (2012)
H.I. Suzuki, K. Yamagata, K. Sugimoto, T. Iwamoto, S. Kato, K. Miyazono, Modulation of microRNA processing by p53. Nature 460, 529–533 (2009)
X.L. Li, M.F. Jones, M. Subramanian, A. Lal, Mutant p53 exerts oncogenic effects through microRNAs and their target gene networks. FEBS Lett 588, 2610–2615 (2014)
M. Rokavec, H. Li, L. Jiang, H. Hermeking, The p53/miR-34 axis in development and disease. J Mol Cell Biol 6, 214–230 (2014)
M. Yamakuchi, C.J. Lowenstein, MiR-34, SIRT1, and p53: The feedback loop. Cell Cycle 8, 712–715 (2009)
A.L. Kasinski, F.J. Slack, miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res 72, 5576–5587 (2012)
F. Pichiorri, S.S. Suh, A. Rocci, L. De Luca, C. Taccioli, R. Santhanam, W. Zhou, D.M. Benson Jr., C. Hofmainster, H. Alder, M. Garofalo, G. Di Leva, S. Volinia, H.J. Lin, D. Perrotti, M. Kuehl, R.I. Aqeilan, A. Palumbo, C.M. Croce, Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 18, 367–381 (2010)
M.T. Le, C. Teh, N. Shyh-Chang, H. Xie, B. Zhou, V. Korzh, H.F. Lodish, B. Lim, MicroRNA-125b is a novel negative regulator of p53. Genes Dev 23, 862–876 (2009)
M.R. Junttila, G.I. Evan, p53--a Jack of all trades but master of none. Nat Rev Cancer 9, 821–829 (2009)
M.T. Le, N. Shyh-Chang, S.L. Khaw, L. Chin, C. Teh, J. Tay, E. O'Day, V. Korzh, H. Yang, A. Lal, J. Lieberman, H.F. Lodish, B. Lim, Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet 7, e1002242 (2011)
D.M. Burns, A. D'Ambrogio, S. Nottrott, J.D. Richter, CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473, 105–108 (2011)
M. Karin, NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol 1, a000141 (2009)
A. Oeckinghaus, M.S. Hayden, S. Ghosh, Crosstalk in NF-kappaB signaling pathways. Nat Immunol 12, 695–708 (2011)
N.D. Perkins, The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer 12, 121–132 (2012)
M.S. Hayden, S. Ghosh, NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26, 203–234 (2012)
K.W. McCool, S. Miyamoto, DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol Rev 246, 311–326 (2012)
K. Vazquez-Santillan, J. Melendez-Zajgla, L. Jimenez-Hernandez, G. Martínez-Ruiz, V. Maldonado, NF-κB signaling in cancer stem cells: a promising therapeutic target? Cell Oncol 38, 327–339 (2015)
D. Iliopoulos, S.A. Jaeger, H.A. Hirsch, M.L. Bulyk, K. Struhl, STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 39, 493–506 (2010)
M. Rokavec, M.G. Oner, H. Hermeking, lnflammation-induced epigenetic switches in cancer. Cell Mol Life Sci 73, 23–39 (2016)
X. Xue, W. Xia, H. Wenzhong, A modeled dynamic regulatory network of NF-kappaB and IL-6 mediated by miRNA. Biosystems 114, 214–218 (2013)
R. Zhou, S.P. O'Hara, X.M. Chen, MicroRNA regulation of innate immune responses in epithelial cells. Cell Mol Immunol 8, 371–379 (2011)
H.S. Cheng, M.S. Njock, N. Khyzha, L.T. Dang, J.E. Fish, Noncoding RNAs regulate NF-kappaB signaling to modulate blood vessel inflammation. Front Genet 5, 422 (2014)
L. Tong, Y. Yuan, S. Wu, Therapeutic microRNAs targeting the NF-kappa B signaling circuits of cancers. Adv Drug Deliv Rev 81, 1–15 (2015)
K. Bakirtzi, M. Hatziapostolou, I. Karagiannides, C. Polytarchou, S. Jaeger, D. Iliopoulos, C. Pothoulakis, Neurotensin signaling activates microRNAs-21 and -155 and Akt, promotes tumor growth in mice, and is increased in human colon tumors. Gastroenterology 141, 1749–1761 e1741 (2011)
C. Polytarchou, D. Iliopoulos, M. Hatziapostolou, F. Kottakis, I. Maroulakou, K. Struhl, P.N. Tsichlis, Akt2 regulates all Akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res 71, 4720–4731 (2011)
C. Polytarchou, D.W. Hommes, T. Palumbo, M. Hatziapostolou, M. Koutsioumpa, G. Koukos, A.E. van der Meulen-de Jong, A. Oikonomopoulos, W.K. van Deen, C. Vorvis, O.B. Serebrennikova, E. Birli, J. Choi, L. Chang, P.A. Anton, P.N. Tsichlis, C. Pothoulakis, H.W. Verspaget, D. Iliopoulos, MicroRNA214 Is Associated With Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology 149, 981–992 e911 (2015)
R. Zhou, G. Hu, A.Y. Gong, X.M. Chen, Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Res 38, 3222–3232 (2010)
A. Drakaki, M. Hatziapostolou, C. Polytarchou, C. Vorvis, G.A. Poultsides, J. Souglakos, V. Georgoulias, D. Iliopoulos, Functional microRNA high throughput screening reveals miR-9 as a central regulator of liver oncogenesis by affecting the PPARA-CDH1 pathway. BMC Cancer 15, 542 (2015)
R. Chen, A.B. Alvero, D.A. Silasi, M.G. Kelly, S. Fest, I. Visintin, A. Leiser, P.E. Schwartz, T. Rutherford, G. Mor, Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene 27, 4712–4723 (2008)
X.J. Kong, L.J. Duan, X.Q. Qian, D. Xu, H.L. Liu, Y.J. Zhu, J. Qi, Tumor-suppressive microRNA-497 targets IKKbeta to regulate NF-kappaB signaling pathway in human prostate cancer cells. Am J Cancer Res 5, 1795–1804 (2015)
P. Mechtler, R. Singhal, J.V. Kichina, J.E. Bard, M.J. Buck, E.S. Kandel, MicroRNA analysis suggests an additional level of feedback regulation in the NF-kappaB signaling cascade. Oncotarget 6, 17097–17106 (2015)
T. Li, M.J. Morgan, S. Choksi, Y. Zhang, Y.S. Kim, Z.G. Liu, MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 11, 799–805 (2010)
S. Masciarelli, G. Fontemaggi, S. Di Agostino, S. Donzelli, E. Carcarino, S. Strano, G. Blandino, Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 33, 1601–1608 (2014)
L. Santarpia, M. Nicoloso, G.A. Calin, MicroRNAs: a complex regulatory network drives the acquisition of malignant cell phenotype. Endocr Relat Cancer 17, F51–F75 (2010)
S.J. Hwang, H.J. Seol, Y.M. Park, K.H. Kim, M. Gorospe, D.H. Nam, H.H. Kim, MicroRNA-146a suppresses metastatic activity in brain metastasis. Mol Cells 34, 329–334 (2012)
E. Astarci, A.E. Erson-Bensan, S. Banerjee, Matrix metalloprotease 16 expression is downregulated by microRNA-146a in spontaneously differentiating Caco-2 cells. Develop Growth Differ 54, 216–226 (2012)
D. Sayed, M. Abdellatif, MicroRNAs in development and disease. Physiol Rev 91, 827–887 (2011)
D.R. Hurst, M.D. Edmonds, G.K. Scott, C.C. Benz, K.S. Vaidya, D.R. Welch, Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 69, 1279–1283 (2009)
S.L. Lin, A. Chiang, D. Chang, S.Y. Ying, Loss of mir-146a function in hormone-refractory prostate cancer. RNA 14, 417–424 (2008)
A.I. Garcia, M. Buisson, P. Bertrand, R. Rimokh, E. Rouleau, B.S. Lopez, R. Lidereau, I. Mikaelian, S. Mazoyer, Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med 3, 279–290 (2011)
M.A. Taylor, K. Sossey-Alaoui, C.L. Thompson, D. Danielpour, W.P. Schiemann, TGF-beta upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest 123, 150–163 (2013)
B. Wang, S.H. Hsu, S. Majumder, H. Kutay, W. Huang, S.T. Jacob, K. Ghoshal, TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene 29, 1787–1797 (2010)
J. Ji, T. Yamashita, X.W. Wang, Wnt/beta-catenin signaling activates microRNA-181 expression in hepatocellular carcinoma. Cell Biosci 1, 4 (2011)
R. Cuesta, A. Martinez-Sanchez, F. Gebauer, miR-181a regulates cap-dependent translation of p27(kip1) mRNA in myeloid cells. Mol Cell Biol 29, 2841–2851 (2009)
A. Bisso, M. Faleschini, F. Zampa, V. Capaci, J. De Santa, L. Santarpia, S. Piazza, V. Cappelletti, M. Daidone, R. Agami, G. Del Sal, Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle 12, 1679–1687 (2013)
F.J. Sheedy, E. Palsson-McDermott, E.J. Hennessy, C. Martin, J.J. O'Leary, Q. Ruan, D.S. Johnson, Y. Chen, L.A. O'Neill, Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol 11, 141–147 (2010)
R.T. Marquez, E. Wendlandt, C.S. Galle, K. Keck, A.P. McCaffrey, MicroRNA-21 is upregulated during the proliferative phase of liver regeneration, targets Pellino-1, and inhibits NF-kappaB signaling. Am J Physiol Gastrointest Liver Physiol 298, G535–G541 (2010)
A.M. Krichevsky, G. Gabriely, miR-21: a small multi-faceted RNA. J Cell Mol Med 13, 39–53 (2009)
X. Pan, Z.X. Wang, R. Wang, MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol Ther 10, 1224–1232 (2010)
R. Kumarswamy, I. Volkmann, T. Thum, Regulation and function of miRNA-21 in health and disease. RNA Biol 8, 706–713 (2011)
F. Talotta, A. Cimmino, M.R. Matarazzo, L. Casalino, G. De Vita, M. D'Esposito, R. Di Lauro, P. Verde, An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 28, 73–84 (2009)
P. Wang, C.F. Zhu, M.Z. Ma, G. Chen, M. Song, Z.L. Zeng, W.H. Lu, J. Yang, S. Wen, P.J. Chiao, Y. Hu, P. Huang, Micro-RNA-155 is induced by K-Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotarget 6, 21148–21158 (2015)
S. Costinean, N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, C.M. Croce, Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA 103, 7024–7029 (2006)
X.H. He, W. Zhu, P. Yuan, S. Jiang, D. Li, H.W. Zhang, M.F. Liu, miR-155 downregulates ErbB2 and suppresses ErbB2-induced malignant transformation of breast epithelial cells. Oncogene 35, 6015–6025 (2016)
R. Dinami, C. Ercolani, E. Petti, S. Piazza, Y. Ciani, R. Sestito, A. Sacconi, F. Biagioni, C. le Sage, R. Agami, R. Benetti, M. Mottolese, C. Schneider, G. Blandino, S. Schoeftner, miR-155 drives telomere fragility in human breast cancer by targeting TRF1. Cancer Res 74, 4145–4156 (2014)
P.M. Neilsen, J.E. Noll, S. Mattiske, C.P. Bracken, P.A. Gregory, R.B. Schulz, S.P. Lim, R. Kumar, R.J. Suetani, G.J. Goodall, D.F. Callen, Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 32, 2992–3000 (2013)
M. Subramanian, P. Francis, S. Bilke, X.L. Li, T. Hara, X. Lu, M.F. Jones, R.L. Walker, Y. Zhu, M. Pineda, C. Lee, L. Varanasi, Y. Yang, L.A. Martinez, J. Luo, S. Ambs, S. Sharma, L.M. Wakefield, P.S. Meltzer, A. Lal, A mutant p53/let-7i-axis-regulated gene network drives cell migration, invasion and metastasis. Oncogene 34, 1094–1104 (2015)
X. Cai, Y. Yin, N. Li, D. Zhu, J. Zhang, C.Y. Zhang, K. Zen, Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J Mol Cell Biol 4, 341–343 (2012)
A.K. Mitra, M. Zillhardt, Y. Hua, P. Tiwari, A.E. Murmann, M.E. Peter, E. Lengyel, MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov 2, 1100–1108 (2012)
J. Yang, R.A. Weinberg, Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 14, 818–829 (2008)
K. Polyak, R.A. Weinberg, Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 9, 265–273 (2009)
A. Hanisch, H.H. Sillje, E.A. Nigg, Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25, 5504–5515 (2006)
Z. Lu, Y. Li, A. Takwi, B. Li, J. Zhang, D.J. Conklin, K.H. Young, R. Martin, miR-301a as an NF-kappaB activator in pancreatic cancer cells. EMBO J 30, 57–67 (2011)
X. Ma, F. Yan, Q. Deng, F. Li, Z. Lu, M. Liu, L. Wang, D.J. Conklin, J. McCracken, S. Srivastava, A. Bhatnagar, Y. Li, Modulation of tumorigenesis by the pro-inflammatory microRNA miR-301a in mouse models of lung cancer and colorectal cancer. Cell Discov 1, 15005 (2015)
X. Xia, K. Zhang, G. Cen, T. Jiang, J. Cao, K. Huang, C. Huang, Q. Zhao, Z. Qiu, MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4. Oncotarget 6, 21046–21063 (2015)
Y. Lu, W. Gao, C. Zhang, S. Wen, H. Huangfu, J. Kang, B. Wang, Hsa-miR-301a-3p acts as an oncogene in laryngeal squamous cell carcinoma via target regulation of Smad4. J Cancer 6, 1260–1275 (2015)
X. Cui, C. Kong, Y. Zhu, Y. Zeng, Z. Zhang, X. Liu, B. Zhan, C. Piao, Z. Jiang, miR-130b, an onco-miRNA in bladder cancer, is directly regulated by NF-kappaB and sustains NF-kappaB activation by decreasing Cylindromatosis expression. Oncotarget 7, 48547–48561 (2016)
G. Zhu, Y. Wang, M. Mijiti, Z. Wang, P.F. Wu, D. Jiafu, Upregulation of miR-130b enhances stem cell-like phenotype in glioblastoma by inactivating the hippo signaling pathway. Biochem Biophys Res Commun 465, 194–199 (2015)
T. Yu, R. Cao, S. Li, M. Fu, L. Ren, W. Chen, H. Zhu, Q. Zhan, R. Shi, MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer 15, 29 (2015)
S. Ma, K.H. Tang, Y.P. Chan, T.K. Lee, P.S. Kwan, A. Castilho, I. Ng, K. Man, N. Wong, K.F. To, B.J. Zheng, P.B. Lai, C.M. Lo, K.W. Chan, X.Y. Guan, miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 7, 694–707 (2010)
P. Dong, M. Karaayvaz, N. Jia, M. Kaneuchi, J. Hamada, H. Watari, S. Sudo, J. Ju, N. Sakuragi, Mutant p53 gain-of-function induces epithelial-mesenchymal transition through modulation of the miR-130b-ZEB1 axis. Oncogene 32, 3286–3295 (2013)
B.L. Li, W. Lu, C. Lu, J.J. Qu, T.T. Yang, Q. Yan, X.P. Wan, CpG island hypermethylation-associated silencing of microRNAs promotes human endometrial cancer. Cancer Cell Int 13, 44 (2013)
C. Yang, J. Cai, Q. Wang, H. Tang, J. Cao, L. Wu, Z. Wang, Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol 124, 325–334 (2012)
Q. Chen, X. Zhao, H. Zhang, H. Yuan, M. Zhu, Q. Sun, X. Lai, Y. Wang, J. Huang, J. Yan, J. Yu, MiR-130b suppresses prostate cancer metastasis through down-regulation of MMP2. Mol Carcinog 54, 1292–1300 (2015)
S. Galardi, N. Mercatelli, M.G. Farace, S.A. Ciafre, NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res 39, 3892–3902 (2011)
S.A. Ciafre, S. Galardi, A. Mangiola, M. Ferracin, C.G. Liu, G. Sabatino, M. Negrini, G. Maira, C.M. Croce, M.G. Farace, Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334, 1351–1358 (2005)
S. Galardi, N. Mercatelli, E. Giorda, S. Massalini, G.V. Frajese, S.A. Ciafre, M.G. Farace, miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 282, 23716–23724 (2007)
S.W. Kim, K. Ramasamy, H. Bouamar, A.P. Lin, D. Jiang, R.C. Aguiar, MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci USA 109, 7865–7870 (2012)
A. Rodriguez, S. Griffiths-Jones, J.L. Ashurst, A. Bradley, Identification of mammalian microRNA host genes and transcription units. Genome Res 14, 1902–1910 (2004)
H. Yin, Y. Sun, X. Wang, J. Park, Y. Zhang, M. Li, J. Yin, Q. Liu, M. Wei, Progress on the relationship between miR-125 family and tumorigenesis. Exp Cell Res 339, 252–260 (2015)
Y.M. Sun, K.Y. Lin, Y.Q. Chen, Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol 6, 6 (2013)
C.E. Monk, G. Hutvagner, J.S. Arthur, Regulation of miRNA transcription in macrophages in response to Candida albicans. PLoS One 5, e13669 (2010)
M. Leotta, L. Biamonte, L. Raimondi, D. Ronchetti, M.T. Di Martino, C. Botta, E. Leone, M.R. Pitari, A. Neri, A. Giordano, P. Tagliaferri, P. Tassone, N. Amodio, A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol 229, 2106–2116 (2014)
W. Li, R. Duan, F. Kooy, S.L. Sherman, W. Zhou, P. Jin, Germline mutation of microRNA-125a is associated with breast cancer. J Med Genet 46, 358–360 (2009)
M. Bousquet, M.H. Harris, B. Zhou, H.F. Lodish, MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA 107, 21558–21563 (2010)
J. Liu, B. Guo, Z. Chen, N. Wang, M. Iacovino, J. Cheng, C. Roden, W. Pan, S. Khan, S. Chen, M. Kyba, R. Fan, S. Guo, J. Lu, miR-125b promotes MLL-AF9-driven murine acute myeloid leukemia involving a VEGFA-mediated non-cell-intrinsic mechanism. Blood 129, 1491–1502 (2017)
N. Xu, L. Zhang, F. Meisgen, M. Harada, J. Heilborn, B. Homey, D. Grander, M. Stahle, E. Sonkoly, A. Pivarcsi, MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem 287, 29899–29908 (2012)
Y. Guan, H. Yao, Z. Zheng, G. Qiu, K. Sun, MiR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int J Cancer 128, 2274–2283 (2011)
L. Huang, J. Luo, Q. Cai, Q. Pan, H. Zeng, Z. Guo, W. Dong, J. Huang, T. Lin, MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer 128, 1758–1769 (2011)
L. Liang, C.M. Wong, Q. Ying, D.N. Fan, S. Huang, J. Ding, J. Yao, M. Yan, J. Li, M. Yao, I.O. Ng, X. He, MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 52, 1731–1740 (2010)
M. Kappelmann, S. Kuphal, G. Meister, L. Vardimon, A.K. Bosserhoff, MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 32, 2984–2991 (2013)
L.H. Liu, H. Li, J.P. Li, H. Zhong, H.C. Zhang, J. Chen, T. Xiao, miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem Biophys Res Commun 416, 31–38 (2011)
G.K. Scott, A. Goga, D. Bhaumik, C.E. Berger, C.S. Sullivan, C.C. Benz, Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 282, 1479–1486 (2007)
S. Wang, J. Huang, H. Lyu, C.K. Lee, J. Tan, J. Wang, B. Liu, Functional cooperation of miR-125a, miR-125b, and miR-205 in entinostat-induced downregulation of erbB2/erbB3 and apoptosis in breast cancer cells. Cell Death Dis 4, e556 (2013)
Y. Wang, P. Tang, Y. Chen, J. Chen, R. Ma, L. Sun, Overexpression of microRNA-125b inhibits human acute myeloid leukemia cells invasion, proliferation and promotes cells apoptosis by targeting NF-kappaB signaling pathway. Biochem Biophys Res Commun 488, 60–66 (2017)
L. Yang, X. Zhang, Y. Ma, X. Zhao, B. Li, H. Wang, Ascites promotes cell migration through the repression of miR-125b in ovarian cancer. Oncotarget (2017). doi:10.18632/oncotarget.16846
R. Su, L. Dong, D. Zou, H. Zhao, Y. Ren, F. Li, P. Yi, L. Li, Y. Zhu, Y. Ma, J. Wang, F. Wang, J. Yu, microRNA-23a, −27a and −24 synergistically regulate JAK1/Stat3 cascade and serve as novel therapeutic targets in human acute erythroid leukemia. Oncogene 35, 6001–6014 (2016)
S. Hatzl, O. Geiger, M.K. Kuepper, V. Caraffini, T. Seime, T. Furlan, E. Nussbaumer, R. Wieser, M. Pichler, M. Scheideler, K. Nowek, M. Jongen-Lavrencic, F. Quehenberger, A. Wolfler, J. Troppmair, H. Sill, A. Zebisch, Increased expression of miR-23a mediates a loss of expression in the RAF kinase inhibitor protein RKIP. Cancer Res 76, 3644–3654 (2016)
T. Nguyen, A. Rich, R. Dahl, MiR-24 promotes the survival of hematopoietic cells. PLoS One 8, e55406 (2013)
K.A. Scheibner, B. Teaboldt, M.C. Hauer, X. Chen, S. Cherukuri, Y. Guo, S.M. Kelley, Z. Liu, M.R. Baer, S. Heimfeld, C.I. Civin, MiR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3theta. PLoS One 7, e50895 (2012)
S.U. Mertens-Talcott, S. Chintharlapalli, X. Li, S. Safe, The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 67, 11001–11011 (2007)
W. Tang, F. Yu, H. Yao, X. Cui, Y. Jiao, L. Lin, J. Chen, D. Yin, E. Song, Q. Liu, miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene 33, 2629–2638 (2014)
Y. Jing, Z. Han, S. Zhang, Y. Liu, L. Wei, Epithelial-mesenchymal transition in tumor microenvironment. Cell Biosci 1, 29 (2011)
C.L. Chaffer, R.A. Weinberg, A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011)
P. Mehlen, A. Puisieux, Metastasis: a question of life or death. Nat Rev Cancer 6, 449–458 (2006)
D.X. Nguyen, P.D. Bos, J. Massague, Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer 9, 274–284 (2009)
T. Brabletz, To differentiate or not--routes towards metastasis. Nat Rev Cancer 12, 425–436 (2012)
D.P. Tabassum, K. Polyak, Tumorigenesis: it takes a village. Nat Rev Cancer 15, 473–483 (2015)
R. Kalluri, EMT: when epithelial cells decide to become mesenchymal-like cells. J Clin Invest 119, 1417–1419 (2009)
R. Kalluri, R.A. Weinberg, The basics of epithelial-mesenchymal transition. J Clin Invest 119, 1420–1428 (2009)
T. Ni, X.Y. Li, N. Lu, T. An, Z.P. Liu, R. Fu, W.C. Lv, Y.W. Zhang, X.J. Xu, R. Grant Rowe, Y.S. Lin, A. Scherer, T. Feinberg, X.Q. Zheng, B.A. Chen, X.S. Liu, Q.L. Guo, Z.Q. Wu, S.J. Weiss, Snail1-dependent p53 repression regulates expansion and activity of tumour-initiating cells in breast cancer. Nat Cell Biol 18, 1221–1232 (2016)
C. Min, S.F. Eddy, D.H. Sherr, G.E. Sonenshein, NF-kappaB and epithelial to mesenchymal transition of cancer. J Cell Biochem 104, 733–744 (2008)
Y. Wu, J. Deng, P.G. Rychahou, S. Qiu, B.M. Evers, B.P. Zhou, Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 15, 416–428 (2009)
G. Storci, P. Sansone, S. Mari, G. D'Uva, S. Tavolari, T. Guarnieri, M. Taffurelli, C. Ceccarelli, D. Santini, P. Chieco, K.B. Marcu, M. Bonafe, TNFalpha up-regulates SLUG via the NF-kappaB/HIF1alpha axis, which imparts breast cancer cells with a stem cell-like phenotype. J Cell Physiol 225, 682–691 (2010)
L. Ma, R.A. Weinberg, MicroRNAs in malignant progression. Cell Cycle 7, 570–572 (2008)
S. Valastyan, R.A. Weinberg, MicroRNAs: Crucial multi-tasking components in the complex circuitry of tumor metastasis. Cell Cycle 8, 3506–3512 (2009)
J.L. Carstens, S. Lovisa, R. Kalluri, Microenvironment-dependent cues trigger miRNA-regulated feedback loop to facilitate the EMT/MET switch. J Clin Invest 124, 1458–1460 (2014)
P.A. Gregory, A.G. Bert, E.L. Paterson, S.C. Barry, A. Tsykin, G. Farshid, M.A. Vadas, Y. Khew-Goodall, G.J. Goodall, The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10, 593–601 (2008)
S.M. Park, A.B. Gaur, E. Lengyel, M.E. Peter, The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22, 894–907 (2008)
A. Cano, M.A. Nieto, Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition. Trends Cell Biol 18, 357–359 (2008)
D.L. Gibbons, W. Lin, C.J. Creighton, Z.H. Rizvi, P.A. Gregory, G.J. Goodall, N. Thilaganathan, L. Du, Y. Zhang, A. Pertsemlidis, Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev 23, 2140–2151 (2009)
M. Hahn, R. de Voer, N. Hoogerbrugge, M. Ligtenberg, R. Kuiper, A. Geurts van Kessel, The genetic heterogeneity of colorectal cancer predisposition-guidelines for gene discovery. Cell Oncol 39, 491–510 (2016)
E. Fessler, M. Jansen, E.M.F. De Sousa, L. Zhao, P.R. Prasetyanti, H. Rodermond, R. Kandimalla, J.F. Linnekamp, M. Franitza, S.R. van Hooff, J.H. de Jong, S.C. Oppeneer, C.J. van Noesel, E. Dekker, G. Stassi, X. Wang, J.P. Medema, L. Vermeulen, A multidimensional network approach reveals microRNAs as determinants of the mesenchymal colorectal cancer subtype. Oncogene 35, 6026–6037 (2016)
E.B. Rankin, A.J. Giaccia, Hypoxic control of metastasis. Science 352, 175–180 (2016)
A. Giannakakis, R. Sandaltzopoulos, J. Greshock, S. Liang, J. Huang, K. Hasegawa, C. Li, A. O'Brien-Jenkins, D. Katsaros, B.L. Weber, C. Simon, G. Coukos, L. Zhang, miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther 7, 255–264 (2008)
X. Huang, L. Ding, K.L. Bennewith, R.T. Tong, S.M. Welford, K.K. Ang, M. Story, Q.-T. Le, A.J. Giaccia, Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 35, 856–867 (2009)
T. van den Beucken, E. Koch, K. Chu, R. Rupaimoole, P. Prickaerts, M. Adriaens, J.W. Voncken, A.L. Harris, F.M. Buffa, S. Haider, M.H. Starmans, C.Q. Yao, M. Ivan, C. Ivan, C.V. Pecot, P.C. Boutros, A.K. Sood, M. Koritzinsky, B.G. Wouters, Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun 5, 5203 (2014)
Q. Qin, W. Furong, L. Baosheng, Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res 33, 50 (2014)
M. Beltran, I. Puig, C. Pena, J.M. Garcia, A.B. Alvarez, R. Pena, F. Bonilla, A.G. de Herreros, A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22, 756–769 (2008)
Y. Shimono, M. Zabala, R.W. Cho, N. Lobo, P. Dalerba, D. Qian, M. Diehn, H. Liu, S.P. Panula, E. Chiao, F.M. Dirbas, G. Somlo, R.A. Pera, K. Lao, M.F. Clarke, Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 (2009)
U. Wellner, J. Schubert, U.C. Burk, O. Schmalhofer, F. Zhu, A. Sonntag, B. Waldvogel, C. Vannier, D. Darling, A. zur Hausen, V.G. Brunton, J. Morton, O. Sansom, J. Schuler, M.P. Stemmler, C. Herzberger, U. Hopt, T. Keck, S. Brabletz, T. Brabletz, The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 11, 1487–1495 (2009)
D. Iliopoulos, M. Lindahl-Allen, C. Polytarchou, H.A. Hirsch, P.N. Tsichlis, K. Struhl, Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol Cell 39, 761–772 (2010)
H. Peinado, D. Olmeda, A. Cano, Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7, 415–428 (2007)
S. Brabletz, T. Brabletz, The ZEB/miR-200 feedback loop--a motor of cellular plasticity in development and cancer? EMBO Rep 11, 670–677 (2010)
N.H. Kim, H.S. Kim, X.Y. Li, I. Lee, H.S. Choi, S.E. Kang, S.Y. Cha, J.K. Ryu, D. Yoon, E.R. Fearon, R.G. Rowe, S. Lee, C.A. Maher, S.J. Weiss, J.I. Yook, A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol 195, 417–433 (2011)
H. Siemens, R. Jackstadt, S. Hunten, M. Kaller, A. Menssen, U. Gotz, H. Hermeking, miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 10, 4256–4271 (2011)
J.G. Gill, E.M. Langer, R.C. Lindsley, M. Cai, T.L. Murphy, M. Kyba, K.M. Murphy, Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells 29, 764–776 (2011)
Y.N. Liu, J.J. Yin, W. Abou-Kheir, P.G. Hynes, O.M. Casey, L. Fang, M. Yi, R.M. Stephens, V. Seng, H. Sheppard-Tillman, P. Martin, K. Kelly, MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene 32, 296–306 (2013)
R. Perdigao-Henriques, F. Petrocca, G. Altschuler, M.P. Thomas, M.T. Le, S.M. Tan, W. Hide, J. Lieberman, miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 35, 158–172 (2016)
S. Julien, I. Puig, E. Caretti, J. Bonaventure, L. Nelles, F. van Roy, C. Dargemont, A.G. de Herreros, A. Bellacosa, L. Larue, Activation of NF-kappaB by Akt upregulates snail expression and induces epithelium mesenchyme transition. Oncogene 26, 7445–7456 (2007)
Y. Yang, Y. Li, K. Wang, Y. Wang, W. Yin, L. Li, P38/NF-kappaB/snail pathway is involved in caffeic acid-induced inhibition of cancer stem cells-like properties and migratory capacity in malignant human keratinocyte. PLoS One 8, e58915 (2013)
M. Rokavec, M.G. Oner, H. Li, R. Jackstadt, L. Jiang, D. Lodygin, M. Kaller, D. Horst, P.K. Ziegler, S. Schwitalla, J. Slotta-Huspenina, F.G. Bader, F.R. Greten, H. Hermeking, IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest 124, 1853–1867 (2014)
U. Burk, J. Schubert, U. Wellner, O. Schmalhofer, E. Vincan, S. Spaderna, T. Brabletz, A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 9, 582–589 (2008)
C.J. Chang, C.H. Chao, W. Xia, J.Y. Yang, Y. Xiong, C.W. Li, W.H. Yu, S.K. Rehman, J.L. Hsu, H.H. Lee, M. Liu, C.T. Chen, D. Yu, M.C. Hung, p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 13, 317–323 (2011)
A. Puisieux, T. Brabletz, J. Caramel, Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol 16, 488–494 (2014)
P.A. Muller, K.H. Vousden, p53 mutations in cancer. Nat Cell Biol 15, 2–8 (2013)
Z. Zhang, X. Liu, B. Feng, N. Liu, Q. Wu, Y. Han, Y. Nie, K. Wu, Y. Shi, D. Fan, STIM1, a direct target of microRNA-185, promotes tumor metastasis and is associated with poor prognosis in colorectal cancer. Oncogene 34, 4808–4820 (2015)
Y. Takahashi, A.R. Forrest, E. Maeno, T. Hashimoto, C.O. Daub, J. Yasuda, MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One 4, e6677 (2009)
M. Liu, N. Lang, X. Chen, Q. Tang, S. Liu, J. Huang, Y. Zheng, F. Bi, miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett 301, 151–160 (2011)
J.S. Imam, K. Buddavarapu, J.S. Lee-Chang, S. Ganapathy, C. Camosy, Y. Chen, M.K. Rao, MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene 29, 4971–4979 (2010)
R. Wang, S. Tian, H.B. Wang, D.P. Chu, J.L. Cao, H.F. Xia, X. Ma, MiR-185 is involved in human breast carcinogenesis by targeting Vegfa. FEBS Lett 588, 4438–4447 (2014)
P. Yuan, X.H. He, Y.F. Rong, J. Cao, Y. Li, Y. Hu, Y. Liu, D. Li, W. Lou, M.F. Liu, KRAS-NFkappaB-YY1-miR-489 signaling axis controls pancreatic cancer metastasis. Cancer Res 77, 100–111 (2016)
J. Li, H. Wu, W. Li, L. Yin, S. Guo, X. Xu, Y. Ouyang, Z. Zhao, S. Liu, Y. Tian, Z. Tian, J. Ju, B. Ni, H. Wang, Downregulated miR-506 expression facilitates pancreatic cancer progression and chemoresistance via SPHK1/Akt/NF-kappaB signaling. Oncogene 35, 5501–5514 (2016)
S.F. Tavazoie, C. Alarcon, T. Oskarsson, D. Padua, Q. Wang, P.D. Bos, W.L. Gerald, J. Massague, Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008)
T. Yokobori, S. Suzuki, N. Tanaka, T. Inose, M. Sohda, A. Sano, M. Sakai, M. Nakajima, T. Miyazaki, H. Kato, MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci 104, 48–54 (2013)
X. Peng, W. Guo, T. Liu, X. Wang, X.a. Tu, D. Xiong, S. Chen, Y. Lai, H. Du, G. Chen, Identification of miRs-143 and-145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PloS One 6, e20341 (2011)
N. Pencheva, S.F. Tavazoie, Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol 15, 546–554 (2013)
N. Kosaka, H. Iguchi, Y. Yoshioka, K. Hagiwara, F. Takeshita, T. Ochiya, Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem 287, 1397–1405 (2012)
A. Lujambio, G.A. Calin, A. Villanueva, S. Ropero, M. Sanchez-Cespedes, D. Blanco, L.M. Montuenga, S. Rossi, M.S. Nicoloso, W.J. Faller, W.M. Gallagher, S.A. Eccles, C.M. Croce, M. Esteller, A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA 105, 13556–13561 (2008)
X. Sun, J. Liu, C. Xu, S.C. Tang, H. Ren, The insights of let-7 miRNAs in oncogenesis and stem cell potency. J Cell Mol Med 20, 1779–1788 (2016)
N. Nadiminty, R. Tummala, W. Lou, Y. Zhu, X.-B. Shi, J.X. Zou, H. Chen, J. Zhang, X. Chen, J. Luo, MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS One 7, e32832 (2012)
B. Zhao, H. Han, J. Chen, Z. Zhang, S. Li, F. Fang, Q. Zheng, Y. Ma, J. Zhang, N. Wu, MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett 342, 43–51 (2014)
A. Esquela-Kerscher, P. Trang, J.F. Wiggins, L. Patrawala, A. Cheng, L. Ford, J.B. Weidhaas, D. Brown, A.G. Bader, F.J. Slack, The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 7, 759–764 (2008)
Y.S. Lee, A. Dutta, The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21, 1025–1030 (2007)
C. Mayr, M.T. Hemann, D.P. Bartel, Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315, 1576–1579 (2007)
S. Valastyan, F. Reinhardt, N. Benaich, D. Calogrias, A.M. Szasz, Z.C. Wang, J.E. Brock, A.L. Richardson, R.A. Weinberg, A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137, 1032–1046 (2009)
M. Yamagishi, K. Nakano, A. Miyake, T. Yamochi, Y. Kagami, A. Tsutsumi, Y. Matsuda, A. Sato-Otsubo, S. Muto, A. Utsunomiya, K. Yamaguchi, K. Uchimaru, S. Ogawa, T. Watanabe, Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer Cell 21, 121–135 (2012)
I. Uribesalgo, C. Ballare, L. Di Croce, Polycomb regulates NF-kappaB signaling in cancer through miRNA. Cancer Cell 21, 5–7 (2012)
Q. Huang, K. Gumireddy, M. Schrier, C. Le Sage, R. Nagel, S. Nair, D.A. Egan, A. Li, G. Huang, A.J. Klein-Szanto, The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 10, 202–210 (2008)
C. Liu, K. Kelnar, B. Liu, X. Chen, T. Calhoun-Davis, H. Li, L. Patrawala, H. Yan, C. Jeter, S. Honorio, The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17, 211–215 (2011)
S. Knoll, K. Furst, B. Kowtharapu, U. Schmitz, S. Marquardt, O. Wolkenhauer, H. Martin, B.M. Putzer, E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep 15, 1315–1329 (2014)
V. Kulda, M. Pesta, O. Topolcan, V. Liska, V. Treska, A. Sutnar, K. Rupert, M. Ludvikova, V. Babuska, L. Holubec, Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 200, 154–160 (2010)
S. Zhu, H. Wu, F. Wu, D. Nie, S. Sheng, Y.-Y. Mo, MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18, 350–359 (2008)
L. Ma, J. Young, H. Prabhala, E. Pan, P. Mestdagh, D. Muth, J. Teruya-Feldstein, F. Reinhardt, T.T. Onder, S. Valastyan, F. Westermann, F. Speleman, J. Vandesompele, R.A. Weinberg, miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12, 247–256 (2010)
G. Martello, A. Rosato, F. Ferrari, A. Manfrin, M. Cordenonsi, S. Dupont, E. Enzo, V. Guzzardo, M. Rondina, T. Spruce, A.R. Parenti, M.G. Daidone, S. Bicciato, S. Piccolo, A MicroRNA targeting dicer for metastasis control. Cell 141, 1195–1207 (2010)
L. Ma, J. Teruya-Feldstein, R.A. Weinberg, Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–688 (2007)
C.W. Li, W. Xia, L. Huo, S.O. Lim, Y. Wu, J.L. Hsu, C.H. Chao, H. Yamaguchi, N.K. Yang, Q. Ding, Y. Wang, Y.J. Lai, A.M. LaBaff, T.J. Wu, B.R. Lin, M.H. Yang, G.N. Hortobagyi, M.C. Hung, Epithelial-mesenchymal transition induced by TNF-alpha requires NF-kappaB-mediated transcriptional upregulation of Twist1. Cancer Res 72, 1290–1300 (2012)
M. Al-Hajj, M.S. Wicha, A. Benito-Hernandez, S.J. Morrison, M.F. Clarke, Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100, 3983–3988 (2003)
R. Chhabra, N. Saini, MicroRNAs in cancer stem cells: current status and future directions. Tumour Biol 35, 8395–8405 (2014)
M. Garofalo, C.M. Croce, Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev 81, 53–61 (2015)
E. Anastasiadou, F.J. Slack, Cancer. Malicious exosomes. Science 346, 1459–1460 (2014)
M.A. Antonyak, R.A. Cerione, Microvesicles as mediators of intercellular communication in cancer. Methods Mol Biol 1165, 147–173 (2014)
J. Skog, T. Wurdinger, S. van Rijn, D.H. Meijer, L. Gainche, M. Sena-Esteves, W.T. Curry Jr., B.S. Carter, A.M. Krichevsky, X.O. Breakefield, Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10, 1470–1476 (2008)
K. Al-Nedawi, B. Meehan, J. Micallef, V. Lhotak, L. May, A. Guha, J. Rak, Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10, 619–624 (2008)
M.T. Le, P. Hamar, C. Guo, E. Basar, R. Perdigao-Henriques, L. Balaj, J. Lieberman, miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest 124, 5109–5128 (2014)
M. Fabbri, A. Paone, F. Calore, R. Galli, E. Gaudio, R. Santhanam, F. Lovat, P. Fadda, C. Mao, G.J. Nuovo, N. Zanesi, M. Crawford, G.H. Ozer, D. Wernicke, H. Alder, M.A. Caligiuri, P. Nana-Sinkam, D. Perrotti, C.M. Croce, MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA 109, E2110–E2116 (2012)
M. Fabbri, TLRs as miRNA receptors. Cancer Res 72, 6333–6337 (2012)
J.R. Chevillet, Q. Kang, I.K. Ruf, H.A. Briggs, L.N. Vojtech, S.M. Hughes, H.H. Cheng, J.D. Arroyo, E.K. Meredith, E.N. Gallichotte, E.L. Pogosova-Agadjanyan, C. Morrissey, D.L. Stirewalt, F. Hladik, E.Y. Yu, C.S. Higano, M. Tewari, Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci USA 111, 14888–14893 (2014)
G. Di Leva, C.M. Croce, miRNA profiling of cancer. Curr Opin Genet Dev 23, 3–11 (2013)
H. Ling, M. Fabbri, G.A. Calin, MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12, 847–865 (2013)
D. Kim, Y.M. Sung, J. Park, S. Kim, J. Kim, H. Ha, J.Y. Bae, D. Baek, General rules for functional microRNA targeting. Nat Genet 48, 1517–1526 (2016)
A.G. Bader, miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 3, 120 (2012)
Acknowledgements
This research has, in part, been co-financed by the European Union (European Social Fund - ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) - Research Funding Program: THALES (CancerTFs, MIS 379435), Investing in knowledge society through the European Social Fund to E.K. (Grand number 80799), in part, by the Broad Medical Research Program at the Chron’s & Colitis Foundation of America and the Pancreatic Cancer UK-Research Innovation Fund (RIF2016_A08), by a research grant in Biomedical Sciences from FONDATION SANTÉ to E.R. and E.K., and a State (IKY) fellowship of Excellence for Postdoctoral Research in Greece - SIEMENS programme to G.M. and E.K.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Markopoulos, G.S., Roupakia, E., Tokamani, M. et al. A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol. 40, 303–339 (2017). https://doi.org/10.1007/s13402-017-0341-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-017-0341-9