Abstract
The economic value of xylanolytic enzymes is derived from their use in a variety of industrial processes, which necessitates a cost-effective manufacturing procedure. In the current study, forty bacterial isolates were isolated from water samples and investigate their efficacy to producing xylanase enzyme. The most potent bacterial isolate was identified by sequencing and amplifications of 16Sr RNA gene as Bacillus haynesii strain K6. The impacts of various culture conditions on the productivity of xylane were examined. Data showed that the highest xylanase production was achieved at pH 7, in presence of 3 g/L xylan, 5 g/L peptone, and incubated at 40 °C for 24 h. The Box-Behnken model was used to find the best parameters for the relevant variables, and the results revealed an increase in xylanase production with values of 35.02 U/mL. The maximum precipitation of xylanase from the optimized culture was attained by ammonium sulfate (60%) followed by purification using dialysis and sephadex G100 column chromatography. The purified xylanase had a 12-fold enrichment, with a specific activity of 84 U/mg and a molecular weight approximately 439 KDa determined by thin-layer chromatography (TLC)/mass spectrometry. The amino acid analysis of the purified xylanase enzyme revealed the presence of 15 amino acids, with the highest concentrations of 1940 and 1520 mg/L for proline and cysteine, respectively. Finally, the physical properties of wastepaper pulp were improved after treatment with xylanase enzyme. The whiteness and softness of xylanase-treated wastepaper were improved with percentages of 34.6% and 16.2%, respectively. Therefore, we recommend the use of xylanase enzyme in the bleaching process as it is a biologically synthetic material, safe, and suitable for industrial use, and it reduces the use of harmful chemicals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Xylan is the second most prevalent polysaccharide in nature, accounting for around one-third of all renewable organic carbon on the earth. It is a key component of hemicellulose in plants [1]. Hydrolysis of β-1, 4 glycosidic linkages among xylopranosyl units of xylan and releasing xylose and xylooligosaccharides is achievable with xylanase. Xylanases and related de-branching enzymes are prepared by various microorganisms (bacteria, actinomycetes, and fungi), and are responsible for hemicellulose hydrolysis [2, 3]. In addition to their usage in food, agro-fiber, and the paper and pulp sectors, where the enzymes aid to lessen ecological impact, xylanases and the organisms that develop them are being exploited in pollution control to breakdown xylan to sustainable chemicals and fuels [2, 4]. Thermotolerant bacteria are microorganisms that can withstand high temperatures. The thermotolerant bacteria found in geothermal heat zones survive at temperatures of 45 °C or higher. At these temperatures, the proteins in these microorganisms are at their most active. As a result, it is advantageous in a variety of biotechnological applications[5]. Large-scale culture of fungi and actinomycetes is generally challenging due to sluggish creation times for very viscous polymers and limited oxygen transmission [6,7,8]. There are types of bacteria where Bacillus sp. is used more extensively than other bacteria in industrial fermentation since they secrete most of their enzymes [9, 10]. Due to their excellent specificity, gentle reactions, little substrate loss, and end product formation, microbial xylanases are the ideal catalysts for xylan breakdown.
Microorganism-derived xylanases might be useful in the feed, food, wastepaper, and various agricultural sectors [11, 12]. Xylanase has been shown to save energy and improve yield and pulp properties in biomechanical pulping processes, through pre-treatment of pulps before bleaching in the pulp and paper industry [13, 14]. Recently, ink from effluent generated from paper and pulp industries can be remove by using xylanase and laccase enzymes [15]. In recycling of wastepaper occurs deinking process which involves dislodgement of ink particles from paper [16]. Conventionally, large amount of chlorine, sodium carbonate, chlorine-based derivatives, sodium silicate, hydrogen peroxide, sodium hydroxide, and chelating agents have been used which resulted in hazardous effluent disposal problem [17]. As a result, the goal of this paper was to identify thermotolerant bacteria from hot springs and their ability to produce xylanase. Additionally, improved xylanase production was achieved using the Box-Behnken Design, which was defined by a number of characteristics. Finally, xylanase was added to the wastepaper pulp to bio-bleaching.
2 Materials and methods
2.1 Materials
Nutrient agar plate, Logule’s iodine solution, dialysis bag, Sephadex G-200 (Sigma Chemicals, Cairo, Egypt), Xylan powder, 3, 5 dinitrosallicylic acid, NaOH, sodium potassium tartrate, and ammonium sulfate (El-Noor Company for Chemicals, in Qasr Al-Aini, Cairo, Egypt).
2.2 Sampling and bacterial isolation
In the current study, water samples were collected from Ain-Helwan Spring, Helwan, Cairo, Egypt (GPS N 29° 51′, E 31° 19′) in sterilized falcon tubes. The pH (6–7) and temperature (35–40 °C) of the collected water samples were measured upon collection. The collected water samples were transferred directly to laboratory for bacterial isolation. Approximately, 200 µL of each sample was spread over the surface of nutrient agar plates (contained g/L beef extract 3.0, peptone 5.0, NaCl 0.5, agar 20.0, and distilled H2O 1 L; pH = 7) and incubated at 45 °C for 24 h. The different bacterial colonies that appeared at the end of incubation period were picked up and re-inoculated onto new plate to check the purity. The purified bacterial isolates were inoculated on nutrient agar slants and preserved at −4 °C for further study.
2.3 Screening for xylanase producing bacteria
The efficacy of the purified bacterial isolates to secretion of xylanase enzymes was screened on mineral salt media (contained g/L NaNO3, 5; KH2PO4, 1; K2HPO4, 2, MgSO4.7H2O, 0.5; KCl, 0.1; CaCl2, 0.01; FeSO4.7H2O, 0.02; agar, 17, distilled H2O, 1 L; pH = 7) supplemented with 2 g, w/v xylan powder, and sterilized at 120 °C (1.5 psi) for 20 min [18], pouring the sterilized media onto Petri-dishes under aseptic conditions, left to solidify, inoculated with bacterial strains, and incubated for 24 h at 45 °C. At the end of incubation period, the inoculated plates were flooded with Logule’s iodine solution, and the results were recorded as a diameter of clear zone around bacterial growth which indicate the successful xylanase production.
2.4 Identification of the bacterial isolates
The most potent bacterial isolate (high xylanase enzyme producers) was subjected to morphological, physiological, and biochemical identification according to standard keys [19]. The molecular identification by amplification and sequencing of 16S rRNA gene was accomplished to confirm the traditional identification. The 16S rDNA gene was amplified using universal primers of 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) [20]. The obtained sequence was compared with those deposited in GeneBank database by ClustalX 1.8 software package (http://www-igbmc.u-strasbg.fr/BioInfo/clustalx). The phylogenetic tree was drawn by the neighbor-joining method using MEGA (Version 6.1) software. The confidence level of each branch (1000 repeats) was tested by bootstrap analysis.
2.5 Production of crude xylanase enzyme
The production of xylanase enzyme was achieved by mineral salt broth media supplemented with xylan (2 g/L, w/v). The prepared broth media were inoculated by bacterial culture (1 %, v/v) and incubated for 24 h at 45 °C under 200 rpm shaking condition. Before assay, the inoculated media were centrifuged at 5000 rpm for 10 min at 4 °C to obtain the supernatant (crude enzyme) which used to xylanase enzyme assay [17].
2.6 Quantitative assay for xylanase activity and protein concentration
The quantitative activity of xylanase enzyme was achieved by detecting the amount of reducing sugars from xylan substrate using the dinitrosalcylic acid (DNS) method [21]. The DNS assay was carried out as follows: 0.2 mL of culture filtrate was mixed with 1 mL of 1 %, w/v xylan (prepared in phosphate buffer, pH 7) in a test tube, and incubated at 45 °C for 30 min. At the end of incubation period, 1 mL of DNS reagent was added, and the mixture was subjected to boiling at 100 °C for 5 min. After cooling, the color intensity was measured at 540 nm. The amount of reducing sugars released was measured using glucose as xylanase activity standard. One unit of xylanase enzyme activity was defined as the amount of enzyme that released 1µmol of reducing sugar per minute. The total soluble protein was determined by Bradford method [22]. This procedure depends upon using the protein reagent Coomassie Brilliant Blue (CBB) G-250 prepared by dissolving the CBB G-250 in 50mL of 95% ethanol. A total of 100mL of o-phosphoric acid was added later and the whole reagent was diluted to 200 mL to make 5× concentrated dye, where the CBB G-250 bind to protein and the color of reaction changes from light green to blue. The absorbance was read off a spectrophotometer at wavelength of 595 nm.
2.7 Optimization production parameters
Different environmental conditions were investigated to detect the best condition for xylanase enzyme by the most potent bacterial strain. To achieve this goal, the mineral salt broth media supplemented with xylene were prepared, autoclaved, and inoculated by 2 % v/v of overnight bacterial culture. The analyzed environmental conditions were incubation period (6, 12, 18, 24, and 30 h), different temperatures (30, 35, 40, 45, and 50 °C), different substrate concentration (1, 2, 3, and 4 g/L, w/v), different pH values (6, 7, 8, 9, and 10), different inoculums size (1, 2, 3, and 4 %, v/v), and different nitrogen sources (ammonium nitrate, ammonium chloride, ammonium hydrogen orthophosphate, beef extract, urea, and peptone). Each experiment was achieved in triplicate. At the end of incubation period, 2 mL of inoculated media was withdrawn and centrifuged at 5000 rpm for 10 min at 4 °C to collect a clear supernatant which was used after that to detect enzyme activity using DNS method [23].
2.8 Optimization with Box-Behnken design
The Box-Behnken design (BBD) was used for major factors determining which influence on xylanase production. The factors and their levels are found in Table 1 used in BBD for xylanase production. Where these factors of basal medium were six numerical factors, the factors include temperature, pH, inoculum size, substrate concentration (xylan), incubation period, and different nitrogen source (peptone). Fifty-four experimental trials comprised as the experimental design, where all runs involve three levels (−1, 0, +1) as shown in Table 1. In the BBD, three levels were used for maximum xylanase production determining what was obtained at −1 or +1 of the variables by comparing them with the experimental results obtained from 0 point values. According to this rule R=n+1, a 54 run experiment was generated, where R is the run numbers and n is the number of variables. The Box-Behnken experimental design was achieved based on the following first-order model:
where Y represents the response (xylanase activity U/mL), β0 is the model intercept, Bi is the linear coefficient, Xi is the level of independent variable, and k is the number of involved variables.
2.9 Extraction and purification
The crude enzyme (supernatant collected from inoculated media after centrifugation) was mixed with ammonium sulfate (60 %) which added drop-wisely under stirring conditions till saturation [24]. The previous mixture was stored in cold conditions for 3 h followed by centrifugation at 10,000 rpm for 30 min and the residue was collected. The collected final product was dissolved in phosphate buffer (pH 7.0) to form turbid suspension which subjected to centrifugation for 10 min at 10,000 rpm to form clear supernatant which was undergone to more purification by dialysis bag and column chromatography.
2.10 Partially purification with dialysis process
The crude extract from ammonium sulfate step was put into dialysis bag against distilled water for 3 h, followed by transfer to phosphate buffer at pH 7. The obtained xylanase was concentrated by putting dialysis bag against sucrose crystals and kept in the refrigerator at 4 °C for further purification.
2.11 Completely purification with sephadex G-100 chromatography
By using phosphate buffer, a sephadex G-100 column (1.5× 50 cm) was equilibrated. After purification with dialysis bag, the concentrated xylanase fraction was loaded onto the column and chromatographed by using phosphate buffer subsequently. At a flow rate, 5 mL/h fractions of 5 mL were collected and the active fractions pooled [24].
2.12 Detection of molecular weight by TLC/CMS
In this study, the molecular weight of the purified enzyme was detected by an Advion compact mass spectrometer (CMS) provided with TLC reader. First, make adjustment parameters with mode of fragmentation: typical. Its mass ranges from 100 to 1200, with mass type (ESI). NB standard sulphadiazine (M. Wt = 250) was injected to assure the quality of analysis.
2.13 Identification of amino acids
Standard preparation: where the stock solution contains 18 amino acids (aspartic acid, threonine, serine, glutamic acid, proline, glycine, alanine, cystine, valine, methionine, isoleucine, leucine, tyrosine, phenylalanine, histadine, lysine, ammonia, arginine), all amino acid concentration are 2.5 µMol/mL, except cystine 1.25 µMol/mL, then dilute 60 µL in a 1.5-mL vial with sample dilution buffer then filtered using a 0.22-µm syringe filter then 100 µL was injected. Sample preparation: 1gm of sample was mixed with 5mL hexane. The mixture was allowed to macerate for 24 h. Then, the mixture was filtered on Whatman No. 1 filter paper and the residue was transferred into a test tube where it was incubated in an oven with 10 mL 6N HCl for 24h at 110°C. After the incubation, the sample was filtered on Whatman No. 1 filter paper, evaporated on rotary evaporator, and dissolved completely in 20-mL dilution buffer, filtered using 0.22 µm syringe filter, and 100 µL was injected. Xylanase amino acids were identified by Sykam Amino Acid Analyzer (Sykam GmbH, Germany) equipped with Solvent Delivery System S 2100 (quaternary pump with flow range 0.01 to 10.00 mL/min and maximum pressure up to 400 bar), Autosampler S 5200, Amino Acid Reaction Module S4300 (with built-in dual filter photometer between 440 and 570 nm with constant signal output and signal summary option), and Refrigerated Reagent Organizer S 4130.
2.14 Application of xylanase on wastepaper
Whatman filter paper was treated with 3 mL of xylanase enzyme and incubated under optimal conditions at pH 7, temperature 45 °C, for 1.5 h. A similar type of Whitman filter paper treated with distilled water instead of enzyme was running as a control. After incubation, the treated samples and control investigated by testing machines instruments (TMI) device for softness measurement, and Techno bright device for measuring brightness and darkness [25].
2.15 Statistical analysis
All results presented in this study are the means of three independent replicates. Data were subjected to analysis of variance one-way (ANOVA) by a statistical package Minitab v19. The mean difference comparison between the treatments was analyzed by the Tukey HSD at p < 0.05.
3 Results and discussion
3.1 Isolation and identification of the most xylanase producer bacterial isolate
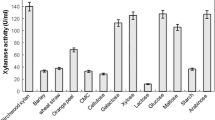
Totally, 40 bacterial isolates were obtained from collected water samples and showed high efficacy to grown at 45 °C. Among these isolates, 21 (52 %) bacterial isolates have the efficacy to produce xylanase enzyme detected by rapid qualitative agar plate after flooding with Logule’s iodine solution (Table 2). The bacterial isolate designated as K6 was selected based on high producing of xylanase enzyme and kept for further study. Compatible with our study, out of 257 isolates obtained from soil samples, 112 isolates represented by 44 % of total bacterial isolates showed xylanase enzyme activity detected by agar plate method. Among 112 isolates, 19 isolates were selected based on highest clear zone formed around their growth [26]. Also, out of 6 isolates, one isolate designated as Bact-1 was xylanase producer detected by agar plate methods [27]. In the current study, the isolate K6 was subjected to morphological, physiological, and biochemical identification. Data showed that the bacterial strain was Gram-positive, bacilli, spore former, facultative anaerobic, the growth was inhibited at 20 °C, and weakly growth was observed at 55–60 °C. The bacterial strain has the efficacy to fermenting different sugars including glucose, galactose, fructose, xylose, arabinose, maltose, mannose, and starch forming acid and gas, whereas lack the potentiality to ferment mannitol and lactose. The physiological tests including methyl red, Vogues-Proskauer, citrate utilization, and nitrate reduction were positive. Based on the above results, the bacterial strain was identified as Bacillus haynesii [19]. The traditional identification was confirmed by amplification and sequencing of 16S rRNA gene. Data showed that the strain K6 has a similarity percentage of 99.8 % with bacterial strain Bacillus haynesii (accession number NR157609). The obtained sequence showed that the bacterial strain K6 was clustered in the group of B. haynesii. Therefore, the most potent bacterial strain in the current study was identified as B. haynesii strain K6 (Fig. 1). The obtained sequence in the current study was registered in GenBank under accession number of OM469329. Xylanases are synthesized by different microbial strains such as bacteria, fungi, actinomycetes, yeasts, and algae. The production of xylanase enzymes using different microbial strains showed varying biochemical characteristics which enables it to integrate into various biotechnological and industrial applications [28]. Although the fungal xylanases have taken more attention because of their high activity, the restrictions that faced the large scale limited their productions and this paved the way for bacterial xylanase production [29]. Various bacterial species such as Bacillus, Micrococcus, Paenibacillus, Rhodothermus, Microbacterium, Arthrobacter, Staphylococcus, Anoxybacillus, and Pseudoxanthomonas have been reported as xylanase producers [24, 29, 30]. Bacillus are considered the most bacterial strains for xylanase production. Various Bacillus spp. such as Bacillus stearothermophilus, Bacillus circulans, Bacillus halodurans, Bacillus subtilis, Bacillus amyloliquefaciens, Bacillus safensis, Bacillus altitudinis, and Bacillus pumilus have been showed xylanolytic activity [23, 29, 31, 32]. To the best of our knowledge, this is the first report for production of xylanase using thermotolerant B. haynesii.
3.2 Evaluation of different xylanase production parameters
Although choosing the right strain is important for a successful industrial enzyme factory, it will not ensure maximal enzyme output until the production process is tuned properly. The kind of fermentation technique employed, the temperature, the pH of the production medium, the nutrition supply provided, and the length of fermentation are all aspects that determine the adequacy of a fermentation process. For reaching the optimum amount of enzyme production, proper exploration and adjustment of these factors are critical [33]. In the current study, the maximum xylanase production using Bacillus haynesii was achieved at pH 7.0 (Fig. 2A). The ambient pH has a significant impact on ionization and the transport of nutritional components across the cell membrane; hence, enzyme synthesis is largely reliant on it. As a result, their accessibility to the bacteria cell and enzymatic activities is reduced, which has an impact on cell proliferation and product creation. As shown, the enzyme activity at different pH values was statistically not significant. These findings contrast with those of Limkar et al. [34], who demonstrated that Bacillus spp. produces the most xylanase at pH 8.5. Bacterial xylanases have a pH that is neutral or alkaline, making them suitable for a wide range of industrial applications. The optimal temperature for xylanase synthesis varies per organism [35]. At different temperatures, such as 30, 35, 40, 45, and 50 °C, the effect of temperature on production of xylanase was investigated. The greatest production of xylanase by B. haynesii strain K6 was found at 40 °C with productivity of 11.17 U/mL (Fig. 2B). Temperatures above or below 40 °C signify a decrease in xylanase production; the most powerful strain’s xylanase enzyme is a thermotolerant enzyme. Similarly, the maximum xylanase productivity was attained at temperature of 35 °C by Bacillus subtilis and 40 °C by Bacillus megaterium [36]. For enhancing xylanase production using B. haynesii K6, different inoculum sizes (1–4 %) were tested as shown in Fig. 2C. The maximum xylanase output occurred when the inoculum size was 1 %, and the enzyme production decreased as the inoculum size increased. Bacillus subtilis and Bacillus megaterium exhibited highest xylanase activity at inoculum size of 2 % and 1.5 %, respectively [36]. The findings demonstrated that when the inoculum size was increased by more than 1%, the rate of xylanase enzyme production decreased. Also, the highest xylanase synthesis was accomplished at 2 % inoculum size of Bacillus mojavensis in submerged fermentation [37]. As shown in Figure (2D), the maximum xylanase productivity using B. haynesii strain was 18.63 U/mL and was achieved by adding 3 g/L xylan in fermentative media. Substrate concentrations below and above this value showed decreasing gradually as compared to the optimal one. Incompatible with the current study, the highest xylanase activity (7.3 to 8.4 U/mL) by Bacillus pumilus was attained in broth media supplemented with 0.5 to 1 % xylan, and the amount of xylanase activity decreased as the substrate concentration increased or decreased [38]. The effect of incubation period was studied on the xylanase production as displayed in Figure (2E). Data analysis showed that the maximum xylanase activity (26.27 U/mL) from Bacillus haynesii strain K6 was achieved after 24 h of incubation. The xylanase activity decreased when the incubation time became longer or less than 24 h. The obtained data completely agreed with those reported by Rosli, et al., who reported that the maximum xylanase activity (10.86 U/mL) by Bacillus tequilensis was achieved after 24 h of incubation period [39]. In the current study, the efficacy of extra-inorganic and organic nitrogen source on xylanase activity was investigated. Analysis of variance showed that the maximum xylanase activity (29.57 U/mL) was achieved in presence of peptone, whereas the enzyme activity was decreased in presence of other nitrogen sources (Fig. 2F). In a similar study, the xylanolytic activity of Bacillus sp. was the highest value (7.896 U/mL) in the presence of beef extract in fermentative media [38].
Evaluation of different xylanase production variables: (A) pH, (B) temperature, (C) inoculum size, (D) substrate concentration, (E) incubation period, and (F) different nitrogen sources. Different letters between columns denote that mean values are significantly different (p ≤ 0.05) by Tukey test, means ± SD (n = 4)
3.3 The optimal conditions with Box-Behnken design
In this study, the BBD of response surface methodology for studying the effect of six factors with six levels affecting on xylanase production was investigated. The six factors were temperature, pH, peptone, inoculum size, xylan, and incubation period. As shown in Table 3 and Fig. 3, the xylanase activity ranged from 17.63 to 35.02 U/mL in all runs. Run No. 17 provided the maximum xylanase activity with values of 35.02 U/mL, where these factors were 40 °C, pH 7, 5 g/L peptone as a best concentration of the best nitrogen source, 1 % inoculum size, 3 g/L xylan concentration, and 24 h of incubation period. Naz et al. [40] reported that the maximum xylanase activity was 175.6945 U/mL in presence of 3 % corncob concentration, 0.05 % peptone, and 0.5 % of KH2PO4. The constructed experimental model is highly significant and accurately portrays the real connection between the impacts of factors and enzyme response linked to xylanase activity, according to the findings of ANOVA obtained from the current investigation. The expected vs. actual plots and normal residual plots of xylanase activity of the xylanolytic bacterial strain B. haynesii showed more stability in the residual plot (Fig. 4). The distribution of response variable from several experimental circumstances revealed that most of the components contributed equally. The likelihood graphs also showed that the expected and real xylanase activities were quite comparable.
3.4 Purification of xylanase enzyme
Under optimum broth-state fermentation conditions, xylanase enzyme was produced using B. haynesii strain. At the end of experiment, different concentration of (NH4)2SO4 (10–80 %) was used to precipitate the proteins. The obtained data showed that the precipitation of protein was increased by increasing the (NH4)2SO4 concentration, whereas the enzyme activity increased up to 60 % (NH4)2SO4. The crude enzyme precipitated with saturation of 60 % (NH4)2SO4 and showed highest enzyme activity (58.62 U/mL) as compared with those precipitated with different (NH4)2SO4 concentrations (Fig. 5). The obtained data are compatible with those reported by Kapilan [41], who investigate the efficacy of different concentration of (NH4)2SO4 (10–70 %) in protein precipitation and xylanase enzyme activity. Who reported that the protein precipitation was increased by increasing the concentration of (NH4)2SO4, whereas the highest xylanase enzyme activity (33.16 U/mg protein) was increased up to 50 % (NH4)2SO4. In the current study, the xylanase enzyme was subjected to dialysis against sucrose and the result showed that the specific activity after dialysis was 27.9 U/mg as shown in (Table 4). The concentrated enzyme from dialysis process was loaded on sephadex G-100 column and receives 10 fractions, each one containing 5 mL of purified xylanase enzyme and estimated the xylanase activity, protein content, and specific enzyme activity in each fraction (Table 5). Data showed that the highest enzyme activity was attained in fraction number 6 with enzyme activity, protein content, and the specific enzyme activity values of 93.23 U/mL, 0.48 mg/mL, and 204.65 U/mg, respectively (Table 5). According to the obtained data, the xylanase enzyme activity was varied due to purification steps. The obtained finding was in agreement with those reported that the specific activities of xylanase enzyme synthesized by Paenibacillus macquariensis were 3.2 U/mg after precipitation with 30 to 60 % (NH4)2SO4, 14.8 U/mg after DEAE-cellulose chromatography purification, and 25.2 U/mg after passage through sephadex G-100 chromatography [42].
3.5 Molecular weight and amino acid content of xylanase
In this study, the molecular weight of xylanase enzyme produced by B. haynesii strain K6 was 439 KDa (Fig. 6A). The molecular weight of xylanase enzyme synthesized by Thermoanaerobacterium sp. was 350 KDa [43], whereas those synthesized by Paenibacillus macerans have a molecular weight of 205 KDa [44]. Other study reported that the molecular weight of xylanase enzyme produced by B. subtilis was 340 KDa [45]. The amino acid sequencing of xylanase enzyme produced by B. haynesii strain K6 is represented in Fig. 6B. Data analysis showed the xylanase enzyme under study containing 15 amino acids which were aspartic acid followed by threonine, serine, glutamic, proline, glycine, cystine, valine, methionine, isoleucine, leucine, tyrosine, phenyalanine, histidine, and lysine. The results showed that the highest values were 1940 and 1520 mg/L for proline and cysteine, respectively, while the lowest value for valine, tyrosine, and histidine was 40 mg/L. Similarly, Dutta et al. [46] found that the amino acid composition of xylanase enzyme synthesized by different bacterial species were theronine (9.5 %), followed by glycine (8.8 %), alanine (8.2 %), serine (7.9 %), and aspartic acid (6.54 %).
3.6 Bio-bleaching of wastepaper by xylanase enzyme
The xylanase enzymes used in bio-bleaching process should be characterized by active alkaline pH, high temperature, and do not contain a cellulolytic enzyme to avoid cellulose fiber degradation [47]. The researchers were interested in using xylanase enzyme in the bleaching process since it helps to reduce the consumption of chlorine compounds [48, 49]. Xylanase produced by Bacillus pumilus ASH5 was used in bio-bleaching of kraft pulp to improve its whiteness and brightness and hence reduce the usage of chlorine and chlorine dioxide with percentages of 20 % and 10 %, respectively [50]. In the current study, the thermotolerant xylanase enzyme produced by B. haynesii K6 was used to improve the properties of wastepaper. Data showed that the whiteness of wastepaper was enhanced after treatment with xylanase enzyme as compared with untreated sample (control). The whiteness of untreated samples was 48 % and improved after treatment with xylanase up to 64.5%. Interestingly, the darkness of papers after treatment with xylanase tend to highly decrease (96.4%) as compared to control (86.9%) (Table 6). It is clear from the obtained results of bio-bleaching of wastepaper with xylanase enzyme that the physical properties of the treated paper are improved compared with untreated (Table 6). Similarly, the xylanase enzyme demonstrated a remarkable brightness of up to 13 % for the paper pulp using the bio-bleaching approach in a prior study [31]. Xylanase enzymes are helping in breakdown of xylan that linked with lignin and cellulose of the pulp and hence improve the separation of these components that enhance fiber wall swelling and improve extraction of lignin from the treated pulp [51]. The treatment of cellulosic fibers with xylanase enzyme is a useful tool to increase paper strength through breakdown of xylan and lignin removal [52]. Therefore, the pretreatment of paper pulp with xylanase enzyme has been more effective, high selective, eco-friendly, reduces the use of hazard chemicals, and non-toxic method for bio-bleaching [28].
4 Conclusion
In this study, it was discovered that an efficient xylanase-producing B. haynesii strain capable of manufacturing extracellular xylanase could be developed. B. haynesii strain has ability to produce xylanase after incubation period 24 h, incubation temperature 40 °C; the concentration of xylan used in this study was 3 g/L, pH was 7, and the best nitrogen source used was peptone with concentration 5 g/L, where the optimal conditions subjected to statistical analysis by Tukey HSD at p<0.05. The BBD was utilized to find the best conditions for producing huge amounts of the xylanase enzyme. The results of this investigation can be used to improve settings for future fermenter scale-up experiments and to determine the economization of the xylanase enzyme manufacturing process. Xylanase was characterized by amino acid analyzer and TLC mass. The xylanase enzyme has a molecular weight of 439 KDa and contains 15 amino acids in various quantities. The use of xylanase on wastepaper had positive results, with a brightness ratio of up to 16 % improving the qualities of the treated wastepaper. This suggests that xylanase is appropriate for industrial usage, particularly in the bleaching process, because it lowers the use of toxic chemicals and is economically safe.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
References
Wierzbicki MP, Maloney V, Mizrachi E, Myburg AA (2019) Xylan in the middle: understanding xylan biosynthesis and its metabolic dependencies toward improving wood fiber for industrial processing. Front Plant Sci 10:176. https://doi.org/10.3389/fpls.2019.00176
Walia A, Guleria S, Mehta P, Chauhan A, Parkash J (2017) Microbial xylanases and their industrial application in pulp and paper biobleaching: a review. 3 Biotech 7(1):11. https://doi.org/10.1007/s13205-016-0584-6
Fouda A, Eid AM, Elsaied A, El-Belely EF, Barghoth MG, Azab E, Gobouri AA, Hassan SE (2021) Plant growth-promoting endophytic bacterial community inhabiting the leaves of Pulicaria incisa (Lam.) DC inherent to arid regions. Plants (Basel, Switzerland) 10 (1). https://doi.org/10.3390/plants10010076
Chaudhary R, Kuthiala T, Singh G, Rarotra S, Kaur A, Arya SK, Kumar P (2021) Current status of xylanase for biofuel production: a review on classification and characterization. Biomass Conversion and Biorefinery:1-19
Yamini C, Sharmila G, Muthukumaran C, Pavithran K, Manojkumar N (2022) Proteomic perspectives on thermotolerant microbes: an updated review. Mol Biol Rep 49(1):629–646. https://doi.org/10.1007/s11033-021-06805-z
Ahmed NE, Salem SS, Hashem AH (2021) Statistical optimization, partial purification, and characterization of phytase produced from Talaromyces purpureogenus NSA20 using potato peel waste and its application in dyes de-colorization. Biointerface Research in Applied Chemistry 12(4):4417–4431. https://doi.org/10.33263/BRIAC124.44174431
Hendy Mahmoud, Hashem Amr, Sultan Mahmoud, El-Ghamery Abbas, Abdelraof Mohamed (2021) L-methionine γ-lyase from Thermo-tolerant fungi: Isolation, Identification of the potent producers, and Statistical Optimization of Production via Response surface methodology. Egyptian Journal of Chemistry 0(0):0–0. https://doi.org/10.21608/ejchem.2021.79178.4555
Selim Mohamed T., Salem Salem S., Mohamed Asem A., El-Gamal Mamdouh S., Awad Mohamed F., Fouda Amr (2021) Biological Treatment of Real Textile Effluent Using Aspergillus flavus and Fusarium oxysporium and Their Consortium along with the Evaluation of Their Phytotoxicity. Journal of Fungi 7(3):193. https://doi.org/10.3390/jof7030193
Parab P, Khandeparker R (2021) Xylanolytic enzyme consortia from Bacillus sp. NIORKP76 for improved biobleaching of kraft pulp. Bioprocess Biosyst Eng 44(12):2513–2524. https://doi.org/10.1007/s00449-021-02623-6
Contesini FJ, Melo RRd, Sato HH (2018) An overview of Bacillus proteases: from production to application. Crit Rev Biotechnol 38(3):321–334
Motta F, Andrade C, Santana M (2013) A review of xylanase production by the fermentation of xylan: classification, characterization and applications. Sustain Degrad Lignocellulosic Biomass-techniques, Appl Commercialization 1:251–276
Khalil AMA, Hassan SE, Alsharif SM, Eid AM, Ewais EE, Azab E, Gobouri AA, Elkelish A, Fouda A (2021) Isolation and characterization of fungal endophytes isolated from medicinal plant Ephedra pachyclada as plant growth-promoting. Biomolecules 11 (2). https://doi.org/10.3390/biom11020140
Przybysz Buzała K, Przybysz P, Kalinowska H, Derkowska M (2016) Effect of cellulases and xylanases on refining process and kraft pulp properties. PLoS One 11(8):e0161575
Tao W, Guo L, Meng A, Wang L, Ren H, Zhai H (2019) Effects of xylanase pretreatment on the quality of refiner mechanical mulberry branch fibers. Advances in Polymer Technology 2019
Gupta GK, Dixit M, Kapoor RK, Shukla P (2021) Xylanolytic enzymes in pulp and paper industry: new technologies and perspectives. Molecular biotechnology:1-14
Hasanin Mohamed Sayed, Hashem Amr Hosny, Abd El-Sayed Essam S., El-Saied Houssni (2020) Green ecofriendly bio-deinking of mixed office waste paper using various enzymes from Rhizopus microsporus AH3: efficiency and characteristics. Cellulose 27(8):4443–4453. https://doi.org/10.1007/s10570-020-03071-3
Maity C, Ghosh K, Halder SK, Jana A, Adak A, Das Mohapatra PK, Pati BR, Mondal KC (2012) Xylanase isozymes from the newly isolated Bacillus sp. CKBx1D and optimization of its deinking potentiality. Appl Biochem Biotechnol 167(5):1208–1219. https://doi.org/10.1007/s12010-012-9556-4
Samanta A, Kolte AP, Senani S, Sridhar M, Jayapal N (2011) A simple and efficient diffusion technique for assay of endo β-1, 4-xylanase activity. Braz J Microbiol 42:1349–1353
Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer K-H, Whitman WB (2011) Bergey’s manual of systematic bacteriology: Volume 3: The Firmicutes, vol 3. Springer Science & Business Media,
Abdel-Hamid MS, Fouda A, El-Ela HKA, El-Ghamry AA, Hassan SE (2021) Plant growth-promoting properties of bacterial endophytes isolated from roots of Thymus vulgaris L. and investigate their role as biofertilizers to enhance the essential oil contents. Biomolecular concepts 12(1):175–196. https://doi.org/10.1515/bmc-2021-0019
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Kumar SS, Abrar A, Jai G, Raj MD (2019) Partial purification and characterization of xylan degrading alkaline xylanase from Bacillus subtilis’. Res J Chem Environ 23 (9)
Yadav P, Maharjan J, Korpole S, Prasad GS, Sahni G, Bhattarai T, Sreerama L (2018) Production, purification, and characterization of thermostable alkaline xylanase from Anoxybacillus kamchatkensis NASTPD13. Front Bioeng Biotechnol 6:65
Sridevi A, Narasimha G, Devi PS (2019) Production of xylanase by Penicillium sp. and its biobleaching efficiency in paper and pulp industry. Int J Pharm Sci Res 10:1307–1311
Rodrigues IDSV, e Silva CGS, da Silva RS, Dolabella SS, Fernandes MF, Fernandes RPM (2019) Screening of bacterial extracellular xylanase producers with potential for cellulose pulp biobleaching. Acta Scientiarum Biol Sci 41:42101
Chitchaowana N, Klawech W, Sutthimusik S, Lertworapreecha M (2016) Characterization and optimization of xylanase producing strain of Bacillus subtilis isolated from the cabbage looper (Trichoplusia ni (Hübner)) intestine. Malaysian J Microbiol:239-244
Bhardwaj N, Kumar B, Verma P (2019) A detailed overview of xylanases: an emerging biomolecule for current and future prospective. Biores Bioprocess 6(1):40. https://doi.org/10.1186/s40643-019-0276-2
Chakdar H, Kumar M, Pandiyan K, Singh A, Nanjappan K, Kashyap PL, Srivastava AK (2016) Bacterial xylanases: biology to biotechnology. 3 Biotech 6(2):1–15
Chapla D, Patel H, Madamwar D, Shah A (2012) Assessment of a thermostable xylanase from Paenibacillus sp ASCD2 for application in prebleaching of eucalyptus kraft pulp. Waste and Biomass Valorization 3(3):269–274
Gupta V, Garg S, Capalash N, Gupta N, Sharma P (2015) Production of thermo-alkali-stable laccase and xylanase by co-culturing of Bacillus sp and B halodurans for biobleaching of kraft pulp and deinking of waste paper. Bioprocess Biosyst Eng 38(5):947–956
Thite VS, Nerurkar AS, Baxi NN (2020) Optimization of concurrent production of xylanolytic and pectinolytic enzymes by Bacillus safensis M35 and Bacillus altitudinis J208 using agro-industrial biomass through Response Surface Methodology. Sci Rep 10(1):3824. https://doi.org/10.1038/s41598-020-60760-6
Tarafdar A, Sirohi R, Gaur VK, Kumar S, Sharma P, Varjani S, Pandey HO, Sindhu R, Madhavan A, Rajasekharan R (2021) Engineering interventions in enzyme production: lab to industrial scale. Biores Technol 326:124771
Limkar MB, Pawar SV, Rathod VK (2019) Statistical optimization of xylanase and alkaline protease co-production by Bacillus spp using Box-Behnken Design under submerged fermentation using wheat bran as a substrate. Biocatal Agric Biotechnol 17:455–464. https://doi.org/10.1016/j.bcab.2018.12.008
Alokika SB (2019) Production, characteristics, and biotechnological applications of microbial xylanases. Appl Microbiol Biotechnol 103(21):8763–8784. https://doi.org/10.1007/s00253-019-10108-6
Irfan M, Asghar U, Nadeem M, Nelofer R, Syed Q (2016) Optimization of process parameters for xylanase production by Bacillus sp in submerged fermentation. J Radiat Res Appl Sci 9(2):139–147. https://doi.org/10.1016/j.jrras.2015.10.008
Akhavan Sepahy A, Ghazi S, Akhavan Sepahy M (2011) Cost-effective production and optimization of alkaline xylanase by indigenous Bacillus mojavensis AG137 fermented on agricultural waste. Enzyme Research 2011
Lawrence R, Kumar Y, Singh AK, Singh S (2013) Production and optimization of xylanase by thermophilic Bacillus sp isolated from soil. J Pure Appl Microbiol 9(2):1117–1128
Rosli SNA, Man RC, Masngut N (2019) Screening of culture conditions for production of xylanase from landfill soil bacteria. Indonesian J Chem 19(2):470–478
Naz S, Irfan M, Farooq MU (2017) Xylanase production from Bacillus subtilis in submerged fermentation using Box-Behnken design. Pakistan J Biotechnol 14(2):151–156
Kapilan R (2015) Purification of xylanase from Bacillus subtilis BS166. J Sci 5(7):511–515
Sharma M, Mehta S, Kumar A (2013) Purification and characterization of alkaline xylanase secreted from Paenibacillus macquariensis.
Shao W, Deblois S, Wiegel J (1995) A high-molecular-weight, cell-associated xylanase isolated from exponentially growing Thermoanaerobacterium sp strain JW/SL-YS485. Appl Environ Microbiol 61(3):937–940
Dheeran P, Nandhagopal N, Kumar S, Jaiswal YK, Adhikari DK (2012) A novel thermostable xylanase of Paenibacillus macerans IIPSP3 isolated from the termite gut. J Ind Microbiol Biotechnol 39(6):851–860
Sá-Pereira P, Costa-Ferreira M, Aires-Barros MR (2002) Enzymatic properties of a neutral endo-1,3(4)-β-xylanase Xyl II from Bacillus subtilis. J Biotechnol 94(3):265–275. https://doi.org/10.1016/S0168-1656(01)00436-9
Dutta B, Banerjee A, Chakraborty P, Bandopadhyay R (2018) In silico studies on bacterial xylanase enzyme: structural and functional insight. J Gene Eng Biotechnol 16(2):749–756
Walia A, Mehta P, Guleria S, Chauhan A, Shirkot C (2015) Molecular cloning and sequencing of alkalophilic Cellulosimicrobium cellulans CKMX1 xylanase gene isolated from mushroom compost and characterization of the gene product. Braz Arch Biol Technol 58:913–922
Nagar S, Gupta VK (2021) Hyper production and eco-friendly bleaching of kraft pulp by xylanase from Bacillus pumilus SV-205 using agro waste material. Waste and Biomass Valorization 12(7):4019–4031
Yang S, Yang B, Duan C, Fuller DA, Wang X, Chowdhury SP, Stavik J, Zhang H, Ni Y (2019) Applications of enzymatic technologies to the production of high-quality dissolving pulp: a review. Biores Technol 281:440–448
Battan B, Sharma J, Dhiman SS, Kuhad RC (2007) Enhanced production of cellulase-free thermostable xylanase by Bacillus pumilus ASH and its potential application in paper industry. Enzyme Microb Technol 41(6–7):733–739
Thomas L, Sindhu R, Binod P, Pandey A (2015) Production of an alkaline xylanase from recombinant Kluyveromyces lactis (KY1) by submerged fermentation and its application in bio-bleaching. Biochem Eng J 102:24–30
Lin X, Wu Z, Zhang C, Liu S, Nie S (2018) Enzymatic pulping of lignocellulosic biomass. Ind Crops Prod 120:16–24
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bakry, M.M., Salem, S.S., Atta, H.M. et al. Xylanase from thermotolerant Bacillus haynesii strain, synthesis, characterization, optimization using Box-Behnken Design, and biobleaching activity. Biomass Conv. Bioref. 14, 9779–9792 (2024). https://doi.org/10.1007/s13399-022-03043-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-022-03043-6