Abstract
The technological goal of hydrogen/deuterium exchange-mass spectrometry (HDX-MS) is to determine backbone amide hydrogen exchange rates. The most critical challenge to achieve this goal is obtaining the deuterium incorporation in single-amide resolution, and gas-phase fragmentation may provide a universal solution. The gas-phase fragmentation may generate the daughter ions which differ by a single amino acid and the difference in deuterium incorporations in the two analogous ions can yield the deuterium incorporation at the sub-localized site. Following the pioneering works by Jørgensen and Rand, several papers utilized the electron transfer dissociation (ETD) to determine the location of deuterium in single-amide resolution. This paper demonstrates further advancement of the strategy by determining backbone amide hydrogen exchange rates, instead of just determining deuterium incorporation at a single time point, in combination with a wide time window monitoring. A method to evaluate the effects of scrambling and to determine the exchange rates from partially scrambled HDX-ETD-MS data is described. All parent ions for ETD fragmentation were regio-selectively scrambled: The deuterium in some regions of a peptide ion was scrambled while that in the other regions was not scrambled. The method determined 31 backbone amide hydrogen exchange rates of cytochrome c in the non-scrambled regions. Good fragmentation of a parent ion, a low degree of scrambling, and a low number of exchangeable hydrogens in the preceding side chain are the important factors to determine the exchange rate. The exchange rates determined by the HDX-MS are in good agreement with those determined by NMR.

ᅟ
Similar content being viewed by others
Introduction
To determine the exchange rates of all backbone amide hydrogens of an analyte protein in single-amide resolution is the primary goal of hydrogen/deuterium exchange-mass spectrometry (HDX-MS) as a technology [1, 2]. Two technical challenges must be met to achieve the goal: detection of deuterium incorporation in a wide time window and localization of deuterium incorporation at a single amino acid resolution [3].
For the first challenge, it is critical to monitor from the fastest exchanging amide hydrogen to the slowest one in an analyte protein. The fastest amide hydrogen starts exchanging around 0.01 s at pH 7 and room temperature, while it may take several years for the slowest one to exchange at the same condition [4]. This challenge was met by varying pH and temperature for cytochrome c [1, 2]. Since backbone amide hydrogen exchange reactions are primarily base-catalyzed reactions near neutral pH, increasing one pH unit of the exchange reaction accelerates the intrinsic exchange rates tenfold and decreasing one pH unit of the reaction decelerates the intrinsic exchange rates to one-tenth. Similarly, lowering the reaction temperature from 23 to 0 °C decreases the intrinsic exchange rates 0.088-fold [5]. Cytochrome c does not change its dynamic properties in a wide pH range, and thus it was possible to cover a wide time window (0.013–1000,000 s at 23 °C at pD 7.4) by varying both pH and temperature. This approach may not be applicable for an analyte protein which is unstable or changes its dynamic properties at desired experimental conditions.
The second challenge to obtain single-amide resolution can be achieved by subtracting the deuterium incorporations in two analogous peptides or in two analogous daughter ions. Relative non-specificity of acid proteases [1, 6,7,8] and utility of new protease(s) [3, 9,10,11,12] can generate a pair of analogous peptides in bottom up HDX-MS. For example, subtraction of the deuterium incorporations in peptides 1–10 and 1–9 gives the deuterium incorporation at the amide hydrogen of residue 10. Similarly, gas-phase fragmentation may provide a pair of analogous daughter ions. For example, subtraction of the deuterium incorporations in the two analogous ions c3+ and c4+ gives the deuterium incorporation at the amide hydrogen of the fifth residue (region III in Fig. 1).
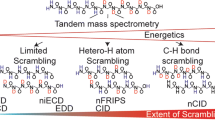
A potential issue to obtain single-amide resolution by gas-phase fragmentation is intramolecular hydrogen/deuterium exchange (scrambling) [13,14,15,16,17,18,19], because it obscures all localized information within the ion. The initial efforts to explore this possibility using collision-induced dissociation (CID) faded due to the scrambling [20,21,22,23,24,25,26,27,28,29,30]. Recently, electron capture dissociation (ECD) and electron transfer dissociation (ETD) showed better potential for sub-localization of deuterium without scrambling. Jørgensen’s synthetic peptide for the detection of scrambling [13] was a breakthrough to apply gas-phase fragmentation for HDX-MS [14,15,16]. Following their efforts, several papers used ECD/ETD for the sub-localization of deuterium in bottom-up HDX-MS studies [17,18,19, 31,32,33,34] as well as in top down HDX-MS studies [35,36,37,38,39].
More ECD/ETD fragmentation data for HDX-MS with various peptide sequences need to be accumulated to establish the effects of functional groups and sequence on the degree of scrambling. Applicability and limitation of gas-phase fragmentation for HDX-MS are not completely understood. We cannot predict the degree of scrambling from the MS parameters and the sequence of a peptide. Although the MS parameters which affect the degree of scrambling may be known [13, 14, 32, 34], scrambling cannot be eliminated completely in some case and the degree of scrambling can be site-specific and peptide specific [40].
This paper describes the application of ETD for the sub-localization of deuterium to determine the backbone amide hydrogen exchange rates in cytochrome c. To obtain the exchange rates, a wide range of exchange time (0.013–1000,000 s at 23°C at pD 7.4) was monitored by varying both pH and temperature. The approach successfully determined the exchange rates of 31 residues, whereas incomplete fragmentation and partial scrambling prevented the sub-localization of deuterium and the determination of exchange rates for the other residues.
Simulation
Hypothetical Peptide
Prior to analyzing the ETD data, the deuterium buildup curve in each segment of a hypothetical peptide, Ala-Leu-Arg-Glu-Lys, was simulated with three different scrambling percentages (Fig. 1). The purpose of this simulation is to understand and visualize what to expect when hydrogen and deuterium are partially or completely scrambled during ETD analysis. For this simulation, the following assumptions were made.
-
(A)
ETD fragmentation of +3 charge state of a hypothetical model peptide is performed to generate a series of c and z ions at +1 charge state (Fig. 1).
-
(B)
The peptide retains deuterium at backbone amides of Arg, Glu, and Lys prior to the MS analysis, analogous to a standard bottom-up HDX-MS experiment with LC separation.
-
(C)
The three backbone amide hydrogens have different exchange rates: Arg-NH, 10−2 (s−1); Glu-NH, 10−4 (s−1); and Lys-NH, 10−6 (s−1).
-
(D)
The exchange experiment is performed in 100% D2O, and there is no back exchange during the downstream processing for simpler presentation.
-
(E)
Scrambling of hydrogen and deuterium occurs uniformly among all exchangeable hydrogen positions in the whole molecule for simpler analysis.
Deuterium Distribution with 0% Scrambling
When the hypothetical peptide is deuterated, subtraction of deuterium incorporations in two analogous daughter ions can determine the deuterium incorporation in a segment of the peptide (Fig. 1). For example, the subtraction of c3 from c4 can yield the total deuterium incorporation in backbone amide of Lys and side chain COOH of Glu (region III in Fig. 1). In the absence of scrambling, all deuterium in the region III resides in the Lys-NH, because Glu-COOH cannot retain deuterium after LC separation (assumption B). In this case, the original sigmoidal deuterium buildup curve for each backbone amide is clearly observable and the sum of deuterium incorporation in the three central regions (I, II, and III) matches with the deuterium incorporation in the parent peptide ion (Fig. 2a).
Simulation of deuterium incorporation in each segment of the model peptide, Ala-Leu-Arg-Glu-Lys, with various degrees of hydrogen/deuterium scrambling. Black solid line deuterium buildup curve of the parent ion. Blue dash line deuterium buildup curve of the region I (c2–c1 or z4–z3). Red solid line deuterium buildup curve of the region II (c3–c2 or z3–z2). Green dot line deuterium buildup curve of the region III (c4–c3 or z2–z1)
Deuterium Distribution with 100% Scrambling
All local information within the molecule is lost under complete scrambling conditions (Fig. 2c). The deuterium buildup curve of each region has the same shape (with an attenuated slope) as that of the parent ion. From this type of data, the exchange rate of each backbone amide cannot be determined. In this case, the sum of deuterium incorporations in the three central regions is approximately one half of the deuterium incorporation in the parent ion (Fig. 2c), because the other half of deuterium resides at N- and C-terminal regions in the hypothetical peptide.
Deuterium Distribution with 50% Scrambling
Some of the local information may still be retained when a half of hydrogen and deuterium is scrambled (Fig. 2b). In this case, the number of exchangeable hydrogen atoms in the preceding side chain has a big influence on the possibility of determining a backbone amide hydrogen exchange rate.
Determining the exchange rate for the backbone amide of Arg is probably feasible using the experimental deuterium buildup curve in the region I even with 50% scrambling (blue dash in Fig. 2b). While 50% of the original deuterium attached at the backbone amide of Arg is lost for scrambling, the remaining 50% stays at the site. Also, because there is no exchangeable hydrogen in the preceding Leu side chain, scrambling adds only one-seventeenth of deuterium (there are 17 exchangeable hydrogens in the +3 parent ion) from the other two backbone amides to this region. The deuterium gained from the other two sites provides a subtle slope from 103 to 107 s (up to two-seventeenths of deuterium). As a total, the deuterium buildup curve of the region I with 50% scrambling (blue dash in Fig. 2b) is very close to the 50% attenuated original deuterium buildup curve of the backbone amide of Arg (blue dash in Fig. 2a). Although the region may lose a significant amount of deuterium in a case of partial scrambling, the exchange rate may still be determined because the preceding residue does not have exchangeable hydrogens in the side chain.
Determination of the exchange rate for the backbone amide of Glu using the experimental deuterium incorporation in the region II may be difficult with 50% scrambling (red solid in Fig. 2b). Because there are four exchangeable hydrogens (in the neutral state) in the preceding Arg side chain, the region II attracts five-seventeenths of deuterium available upon scrambling. The gained deuterium adds gentle slopes around 102 and 106 s obscuring the remaining 50% of the original sigmoidal curve for backbone amide of Glu around 104 s.
Determining the exchange rate for Lys-NH using the experimental deuterium incorporation in the region III (green dot in Fig. 2b) should be slightly more difficult than the Arg-NH yet easier than Glu-NH. While the region III keeps 50% of the original deuterium at Lys-NH, it lures two-seventeenths of all scrambled deuterium due to the presence of one exchangeable hydrogen in the preceding Glu side chain. The deuterium buildup curve for the region III has a weak slope up to 104 s from the scrambling and a sharper slope around 106 s from the original sigmoidal curve.
Experimental
Materials
All reagents were obtained from Sigma-Aldrich (St. Louis, MO). Cytochrome c is from equine heart (product number, C7752).
pH
The actual pD (pDcorr) value is slightly higher than the readings of pH meter (pDread) in D2O (pDcorr = pDread + 0.4) [41].
On-Exchange Experiment for HDX-MS
On-exchange reaction was initiated by mixing 2 μL of 1 mg/mL cytochrome c in H2O with 18 μL of deuterated buffer (20 mM citrate at pDcorr 5.4 or 6.4; 20 mM phosphate, pDcorr 7.4 or 8.4; 20 mM serine pDcorr 9.4 or 10.4). The reaction mixture was incubated for 15, 50, 150, or 500 s at 0 °C or 30, 100, 300, or 1000 s 23 °C. The on-exchanged solution was quenched by the addition of chilled 30 μL of 1.6 M guanidine hydrochloride (GuHCl) in 0.8% aqueous formic acid and immediately analyzed.
Fully Deuterated Experiment for HDX-MS
The fully deuterated sample was prepared by incubating the mixture of 2 μL of 1 mg/mL cytochrome c with 18 μL of D2O at 60 °C for 3 h. The sample was then quenched identically to an on-exchanged solution.
General Protein Process for HDX-MS
HDX-MS analysis was carried out fully automated system described previously [2, 42].
MS Analysis
Mass spectrometric analyses were carried out using an LTQ™ Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) with the capillary temperature at 275 °C and resolution 15,000. For MS/MS, normalized ETD collision energy was set at 35%, activation Q was 0.250, and Activation time was 300 ms. Other MS/MS acquisition parameters are listed in Table 1.
Data Analysis
HDExaminer version 2.1 (Sierra Analytics, Modesto, CA) was used to extract centroid values from the MS and MS/MS raw data files. Then Excel was used to process and present the data.
Evaluating the Effects of Scrambling and Calculating the Backbone Amide Hydrogen Exchange Rates
Based on the simulation results, two curve fittings were calculated to evaluate the presence or absence of significant effects by scrambling at a sub-localized segment i (e.g., c4–c3 in Fig. 1): One assuming 0% scrambling, Χ0%, i2, and the other assuming 100% scrambling, Χ100%, i2.
The Χ0%, i2, the deviation from a pseudo-first order kinetic model, should fit well for a deuterium buildup curve in the absence of scrambling (like Fig. 2a). D0% (i, t) is calculated deuterium incorporation in the segment i at time point t with assuming 0% scrambling. Dobs (i, t) is the observed deuterium incorporation in the segment i at time point t. Dmax, i is the maximum deuterium incorporation, and k i is the exchange rate of the backbone amide in the segment i. Here Dmax, i and k i are optimized to minimize Χ0%, i2. In a few cases where a segment contains two backbone amides (e.g., c4–c2 in Fig. 1), two sets of Dmax, i and k i are optimized to minimize Χ0%, i2.
The Χ100%2, the deviation from an attenuated deuterium build curve of the parent ion, should fit well for a deuterium buildup curve with complete scrambling (like Fig. 2c). D100% (i, t) is calculated deuterium incorporation in the segment i at time point t with assuming 100% scrambling. Dobs (parent, t) is the observed deuterium incorporation in the parent ion at time point t. Datt is the attenuation factor for the segment i. Here Datt is optimized to minimize Χ100%, i2.
If the minimized Χ0%, i2 is smaller than the minimized Χ100%, i2, the exchange rate for the backbone amide in the segment i is determined. In such a situation, the non-scrambling model can fit the experimental data better than the complete scrambling model and the effect of scrambling in the segment i is considered small enough to determine the exchange rate. The ki which gives the minimum Χ0%, i2 is the exchange rate for the backbone amide in the segment i. If the minimized Χ0%, i2 is larger than the minimized Χ100%, i2, the effect of scrambling in the segment i is too large to determine the exchange rate.
Results
Evaluation of Scrambling Using Fully Deuterated Samples
All four +3 parent ions generated the corresponding (M–NH3) ions in the ETD spectra, and all of them indicated partial scrambling (Table 2). Prior to determining the exchange rates of cytochrome c backbone amides, the degree of scrambling in each parent peptide ion was evaluated using the results from fully deuterated samples. Rand et al. proposed to use the deuterium loss in NH3 by comparing the deuterium incorporation in a parent ion and that in the corresponding (M–NH3) ion to quickly evaluate the degree of scrambling [15]. Whereas four +3 peptide ions and eight +2 ions were used as parent ions in the current study, only the +3 parent ions produced the corresponding (M–NH3) ions. The scrambling percentages were estimated between 20% and 33% for those +3 ions from the analysis of the deuterium loss in NH3, assuming the scrambling is uniform within each parent peptide ion.
None of +2 parent peptide ions produced (M–NH3) ions and many +2 parent ions produced (M–19) ions which correspond to the loss of H3O+ (Table 2). The (M–H3O+) species are likely to be produced via CID mechanism not via ETD, because the charge states are only one less than the parent ions while losing H3O+ [43]. Interestingly significantly more deuterium was lost by losing H3O+ (presumably via CID) than NH3 via ETD mechanism.
The analysis of deuterium incorporation in c and z ions generated from fully deuterated samples also indicated partial scrambling for all parent peptide ions (Fig. 3 and Supporting Information Fig. S1). Since the assessment of scrambling by the deuterium loss in NH3 did not provide information for any of +2 parent ions, the deuterium incorporation in c and z ions of fully deuterated samples was also monitored. In this assessment, the observed deuterium incorporation in each of c and z ions was compared with the calculated values with 0% scrambling and 100% scrambling. To calculate the deuterium incorporation in each daughter ion with 0% scrambling, it was assumed that the deuterium incorporated in the parent ion is equally distributed among all backbone amides except the one in the second residue (◊ in Fig. 3). The calculated deuterium incorporation with 0% scrambling may not be accurate, because the deuterium incorporation at each backbone amide may not be equal due to the various back exchange properties [5]. To calculate the deuterium incorporation in each daughter ion with 100% scrambling, it was assumed that the deuterium incorporated in the parent ion is equally distributed among all exchangeable hydrogen positions in the ion (○ in Fig. 3). In many cases, an observed deuterium incorporation (X in Fig. 3) falls between the two calculated values with 0% scrambling and 100% scrambling (◊ and ○ in Fig. 3).
Deuterium incorporation in c and z ions of peptide 67–82 (+3) in full-deuterated samples. X is observed deuterium incorporation.  is calculated deuterium incorporation with 0% scrambling.
is calculated deuterium incorporation with 0% scrambling.  is calculated deuterium incorporation with 100% scrambling. The largest z ion (z16 in this case) corresponds to the (M–NH3) ion
is calculated deuterium incorporation with 100% scrambling. The largest z ion (z16 in this case) corresponds to the (M–NH3) ion
Determination of Backbone Amide Hydrogen Exchange Rates in Peptide 83–96
Regio-selective scrambling was observed in ETD fragmentation of cytochrome c peptide 83–96 (Figs. 4 and 5; Supporting Information Fig. S9). While the hydrogens and deuteriums in the central part of the peptide are heavily scrambled, those near N- and C-terminals show little enough scrambling to determine the exchange rates. The sub-localized segment with I85-NH, K86-NH, I95-NH, or A96-NH has a smaller Χ0%, i2 value than the corresponding Χ100%, i2 value, meeting the criterion of small effects by scrambling. On the other hand, all other segments have a larger Χ0%, i2 values than the Χ100%, i2 values.
Potential daughter ions by ETD fragmentation of parent peptide 83–96 (MH22+) and the exchange rate determination of each backbone amide hydrogen.  is the backbone amide whose exchange rate can be determined by ETD sub-localization.
is the backbone amide whose exchange rate can be determined by ETD sub-localization.  is the backbone amide whose exchange rate cannot be determined by ETD sub-localization due to scrambling
is the backbone amide whose exchange rate cannot be determined by ETD sub-localization due to scrambling
The deuterium buildup curve for the segment including I85-NH showed a very clean sigmoid shape expected for data with minimal scrambling (Fig. 5b). The deuterium buildup curve for the segment including I85-NH was obtained by subtracting the deuterium incorporation in z12 from that in z13 (Fig. 4). The exchange rate of I85-NH determined by this method, 1.4 × 10−3 s−1, is in good agreement with the value determined by NMR, 1.1 × 10−3 s−1 (Table 3) [4]. The deuterium incorporations in fully deuterated experiments were 0.70 (Fig. 5b), which is close to the average deuterium incorporation expected at each backbone amide without scrambling, 0.67 (= 8.0 / 12; 8.0 is deuterium incorporated in the parent ion and 12 is the number of backbone amides which can retain deuterium during digestion and LC separation). The value is significantly higher than what expected for 100% scrambling, 0.25 (= 8.0 / 32; 32 is the number of exchangeable hydrogens in the parent ion), suggesting that this site did not lose much deuterium via scrambling.
The segment including K86-NH showed the deuteration level flat at around 0.34 throughout the time course except for the very first time point, indicating that K86-NH has a very fast exchange rate (Fig. 5d). The deuterium buildup curve for the segment was obtained by subtracting the deuterium incorporation in z11 from that in z12 (Fig. 4). This flatness favors a non-scrambling model of a sigmoidal curve (Χ0%, i2) with a very fast exchange rate over a scrambling model of gradual increase (Χ100%, i2). If the deuterium incorporation in this segment is the result of scrambling, it should have gradual increase as seen in Fig. 5a. This segment, however, is not free from scrambling. The deuterium incorporation in the fully deuterated sample was low at around 0.36, suggesting that approximately a half of deuterium from K86-NH might have been lost via scrambling.
The segment including I95-NH showed little deuterium incorporation even at the longest exchange time point, indicating that I95-NH has a very slow exchange rate (Fig. 5f). The deuterium buildup curve for I95-NH was obtained by subtracting the deuterium incorporation in c11 from that in c12 (Fig. 4). There are two possible reasons for the low-deuterium incorporation during the exchange experiments; lack of deuterium incorporation at the amide position during the experimental time window employed or loss of deuterium due to a high degree of scrambling. In this case, the low-deuterium incorporation cannot be a result of scrambling, because this segment retains a significant amount of deuterium in fully deuterated experiments (0.70). If the low-deuterium incorporation at the segment is a result of scrambling, the deuterium incorporation in fully deuterated experiments should also be low. There must be a sigmoidal increase of deuterium incorporation after the longest time point. The deuterium buildup curve observed for the segment including I95-NH (Fig. 5f) would be reminiscent of the curve simulated for the region III of the hypothetical peptide with partial scrambling (green dot line in Fig. 2b). The exchange rate determined for this residue (7.1 × 10−7 s−1) is probably faster than the actual rate, because the transition of the sigmoid curve was not observed within the time window employed. Determination of the accurate exchange rate for this residue requires longer exchange time points. Nonetheless, these results are consistent with the NMR results which showed that I95-NH is one of the slowest exchanging sites and a part of the very stable C-terminal helix in cytochrome c [4].
The backbone amide hydrogen exchange rates of residues between K87 and L94 cannot be determined due to scrambling (Fig. 4). Deuterium incorporations in the segments including these amides gradually increase in similar ways as that in the parent ion (Fig. 5a, c and e; Supporting Information Fig. S9). None of these segments show a sigmoidal curve like the segment including I85-NH (Fig. 5b), an even high-deuteration level throughout the time window like the segment including K86-NH (Fig. 5d) or an even low-deuterium incorporation like the segment including I95-NH (Fig. 5f). The deuterium incorporations in these segments in fully deuterated experiments are also close to what expected for total scrambling. For example, the observed deuterium incorporations at the segments including K88-NH and E92-NH in fully deuterated samples were 0.73 and 1.19, respectively. The calculated values with 100% scrambling are 0.75 and 1.25, respectively (Fig. 5c and e). The segment including E92-NH accumulates a large amount of deuterium due to the presence of guanidino group in the preceding R91 side chain upon scrambling.
Determination of Backbone Amide Hydrogen Exchange Rates of Cytochrome c
Overall, the HDX-MS sub-localization method with ETD determined 31 out of 100 backbone amide hydrogen exchange rates of cytochrome c (Table 3). Previously, the HDX-MS sub-localization method using analogous peptides determined 15 backbone amide hydrogen exchange rates (Fig. 6) [2]. Total 38 backbone amide hydrogen exchange rates were determined by HDX-MS with the exchange rates of eight residues being determined by both methods. Out of the 31 exchange rates determined by ETD method, only ten of them were also determined by HDX-NMR. The current method could determine fast exchange rates which NMR method could not monitor (Fig. 6).
The backbone amide hydrogen exchange rates determined by subtraction of two analogous ETD ions generally agreed well with those determined by HDX-NMR (Fig. 7). Most of the exchange rates determined were within tenfold of those determined by the HDX-NMR method. The exchange rates deviated most were those for the residues I95, A96, and L98, for which HDX-ETD-MS method could not determine the exchange rates accurately due to the lack of longer exchange time points (arrows in Figs. 6 and 7).
Comparison of backbone amide hydrogen exchange rates determined by HDX-NMR and HDX-MS; comparison of the value determined by HDX-NMR [4] versus that determined with analogous peptides [2]; and
comparison of the value determined by HDX-NMR [4] versus that determined with analogous peptides [2]; and , comparison of the value determined by HDX-NMR versus that determined with analogous ETD ions. The exchange rates determined with analogous ETD ions for I95, A96, and L98 are likely to be even slower (see main text and Table 3)
, comparison of the value determined by HDX-NMR versus that determined with analogous ETD ions. The exchange rates determined with analogous ETD ions for I95, A96, and L98 are likely to be even slower (see main text and Table 3)
The backbone amide hydrogen exchange rates determined by subtraction of two analogous ETD ions were consistent within the data set. For example, the exchange rate of K60 can be determined by the subtraction of c11 ion from c12 ion or z5 from z6 of parent ion 48–64 (+3). The exchange rates determined by the two subtractions were within 20% the difference (1.1 × 10−4 and 0.9 × 10−4, respectively; Table S1 in Supporting Information). There are three pairs of such subtractions, and the differences of the exchange rates were within twofold in all cases. Also, the exchange rate of K79 was determined by two different parent ions, c14 ion–c13 ion of 65–80 (+3) and z5 ion-z4 ion of 67–82 (+3). In this case, the difference was about fourfold (8.2 × 10−2 and 2.3 × 10−2, respectively; Table S1 in Supporting Information).
Discussion
The Factors which Affect the Determination of Backbone Amide Hydrogen Exchange Rates
The feasibility of determining exchange rate from two analogous daughter ions generated in ETD fragmentation depends on a few factors; (i) the presence and quality of daughter ions (from fragmentation of a parent ion), (ii) the degree of scrambling, and (iii) the number of exchangeable hydrogens in the preceding side chain. Table 4 summarizes the success of cytochorme c exchange rate determination by ETD sub-localization.
ETD fragmentation needs to produce a series of c and z ions at high quality [18]. CID can identify more peptides than ETD in general when an analyte protein does not have labile posttranslational modifications [44], meaning that CID can generate a nicer series of daughter ions than ETD. In the current study, parent peptides 1–21 and 22–36 produced no/little c and z ions of high enough quality and the fragmentation of many parent ions did not produce a series of sufficiently good quality daughter ions to generate a complete sub-localization (= in Table 4). Also, subtraction of the deuterium incorporations in two analogous daughter ions adds the errors from both ions in the same manner as subtraction of the deuterium incorporations in two analogous peptides. Because the quality (signal-to-noise ratio) of an ETD daughter ion is usually not as good as that of a peptide ion, the quality issue adds an extra challenge to ETD sub-localization.
Low degree of scrambling is required to determine exchange rate. In the current study, 32 backbone amide hydrogen exchange rates were determined (O in Table 4; Lys79 was determined twice) while 40 exchange rates were unable to determined due to scrambling (X in Table 4; 44% success rate). The deuterium buildup curves fit better with the scrambling model (Χ100%, i2) than the non-scrambling model (Χ0%, i2) for these 40 segments (see Experimental), even though the c or z ions necessary to determine the exchange rate have high enough quality (Supporting Information Figs. S3–S10). Scrambling introduces deuterium from other sites to the segment of interest to obscure the original sigmoidal curve.
The lower the number of exchangeable hydrogens in the preceding side chain, the higher the chance to determine a backbone amide hydrogen exchange rate. Upon sub-localization of deuterium using c and z-ion series, the minimum observable segment is a combination of a backbone amide, the preceding α-carbon, and the preceding side chain (Fig. 1). Since this minimum segment reports the sum of deuterium incorporation in all exchangeable hydrogens, any deuterium incorporation in the preceding side chain obscures the deuterium incorporation at the backbone amide. The absence of exchangeable hydrogen in the preceding side chain minimizes the effects of deuterium incorporations at other sites of the parent peptide through scrambling. In this study, the exchange rates of 13 out of 23 residues following amino acids without exchangeable side chain hydrogens were determined (57% success rate). On the other hand, none of the exchange rates were determined for the three residues following Arg (four exchangeable hydrogens in neutral state; 0% success rate) and only three exchange rates were determined out of the 13 residues following Lys (two exchangeable hydrogens in neutral state; 23% success rate).
Regio-selective Scrambling
All eight parent ions used for the analysis showed regio-selective scrambling [40]: There were some residues whose exchange rates were determined and others whose exchange rates were not determined due to scrambling in each parent ion (Table 4). This is consistent with the observation that all parent ions in the fully deuterated sample were partially scrambled (Table 2; Fig. 3; and Supporting Information Fig. S1). One trend is that scrambling is more pronounced near the regions where two or more Lys and Arg residues locate in proximity in each parent peptide ion (Pink in Table 4). It is not clear, however, what mechanisms are operative for scrambling.
Scrambling is primarily caused by a series of paired intramolecular proton transfer and deuteron transfer reactions. Most scrambling occurs prior to the generation of radical species during ETD fragmentation; considering all suggested parameters to suppress scrambling is in ionization and isolation steps, such as capillary temperature and isolation width [14, 32]. Then, intramolecular hydrogen/deuterium exchange reactions which result in scrambling are pairs of intramolecular proton transfer and deuteron transfer reactions within a positively charged peptide ion [45]. For an intramolecular proton/deuteron transfer reaction to occur, a proton/deuteron donor and a proton/deuteron acceptor are required to be in proximity via charge solvation or salt bridge formation within the ion [46].
The most important proton/deuteron transfer reactions for scrambling are those involved with backbone amide groups, because they are the only groups which retain deuterium after bottom-up HDX-MS experiments prior to scrambling [5]. From here, we speculate possible mechanism of intramolecular proton/deuteron transfer reactions involving backbone amide groups.
The intramolecular proton/deuteron transfer reaction via charge solvation involving ammonium group is the most probable pathway to move the deuterons from backbone amide groups (Fig. 8a). Amino and guanidino groups are two of the most basic groups in the peptides. Because the total number of the two functional groups is larger than the number of charges in each peptide ion in the current study, all charges should reside on the amino or guanidino groups. Therefore, when a peptide ion takes a certain conformation involving charge solvation, the charge group is either ammonium or guanidinium group [46]. The investigations by Lias et al. showed that difference in the proton affinities of two bases involved should be 20 kcal/mol or less for proton transfer reaction to occur [47, 48]. If this is the case, proton transfer reaction does not occur from guanidinium to amide, because the proton affinity of guanidine (235.7 kcal/mol [49]) is too large compared with that of amide (212.4 kcal/mol for N-methylacetamide [49]). On the other hand, the proton affinity of amine (214.9 kcal/mol for methylamine [49]) is small enough to allow the reaction. The mechanism of this reaction is homologous to that of acid-catalyzed amide hydrogen/deuterium exchange reaction in solution (Fig. 8a) [50, 51].
The intramolecular proton/deuteron transfer reaction via salt bridge is another pathway which cannot be completely excluded to move the deuterons from backbone amides (Fig. 8b) [52, 53]. Two functional groups, one acidic and one basic, may form a salt bridge. Amino and guanidino groups are two of the most basic groups in the peptides and are the best candidates for the proton/deuteron acceptor of the salt bridge. There is at least one neutral amino group in each peptide ion in the current study, as the total number of the amino and guanidino groups is larger than the number of charges in each peptide ion (All guanidino groups should carry a charge because of the higher proton affinity). The transition state of this reaction is analogous to that of base-catalyzed backbone amide hydrogen/deuterium exchange reaction in solution [50, 51] and is stabilized by ammonium group in this case (Fig. 8b). Although intermolecular proton/deuteron exchange reaction between carboxylic acid (ΔH for deprotonation of acetic acid, 348.7 kcal/mol [49]) and amino group via salt bridge formation is observed with help from the nearby functional group [52] that between amide (ΔH for deprotonation of N-methylacetamide, 361.9 kcal/mol [49]) and amino group may be too costly in gas phase [46, 54, 55].
To predict more precise regio-selectivity of scrambling, the gas-phase conformation of each parent peptide ion may need to be investigated. Many papers describe gas-phase intermolecumar hydrogen/deuterium exchange reactions of various peptides with deuterating reagents, such as ND3, D2O, CD3OD, and CD3COOD [46,47,48, 52, 53, 56, 57]. These papers rationalize the reactivity of the intermolecular hydrogen/deuterium exchange reactions using peptide conformations in the gas-phase and relay mechanisms [46, 52, 53, 57]. The conformation of a peptide is equally or more important for intramolecular hydrogen/deuterium exchange reaction than for intermolecular reaction. The conformation of peptide determines the alignment of all functional groups involved in the intramolecular reaction and thus should have a strong influence on the reactivity.
The Implication of the Current Study
Common practices among the most publications which utilize ETD for deuterium sub-localization include the following: (i) using one set of the MS parameters which were optimized to minimize the scrambling for a model peptide and (ii) applying ETD to sub-localize deuterium for single (or a very few) time point not for the entire time course.
A caution must be exercised when ETD is used for sub-localization of deuterium: it is critical to check the degree of scrambling strictly for each parent peptide ion (related to (i)). It is practical to use one set of MS parameters for all peptide ions in one HDX-MS study to sub-localize the deuterium by ETD, instead of optimizing the parameters for each peptide ion. However, one should be aware that the degree of scrambling may vary from one peptide ion to another and thus no scrambling in the model peptide ion does not warrant no scrambling in the peptide ions of interest, because scrambling may be regio-selective and thus peptide-selective [40]. The easiest method to check the degree of scrambling is monitoring the deuterium loss in ammonia proposed by Rand et al. [15]. In the paper, they showed the deuterium loss of 0.0 to 0.2 in ammonia (corresponding to 0 to 29% scrambling) depending on peptide ions using one set of optimized MS parameters. The high end of the scrambling percent observed in the Rand paper (25–29%) is close to the values observed in the current study (20–33% in Table 2). The current study showed that even 20% scrambling estimated by the deuterium loss in ammonia method results in scrambled deuterium buildup curves in some residues due to regio-selective scrambling (peptide ion 67–82 (+3) in Tables 2 and 4). This suggests that the deuterium loss in ammonia should be “very” close to zero to ensure the absence of scrambling.
Application of ETD sub-localization to a wide time window, instead of a single time point, presents an opportunity to differentiate non-scrambled regions from scrambled regions (related to (ii)). The deuterium buildup curve of each sub-localized segment should have a distinctive sigmoidal curve in the absence of scrambling (Fig. 2a). On the other hand, the deuterium buildup curve of each segment should have a very similar shape as that of the parent ion with a different slope upon complete scrambling (Fig. 2c). Therefore, the shape of each deuterium buildup curve, which is not attainable by a single time point experiment, can tell the effects of scrambling in the segment. In the current study, a criterion to decide whether the effects of scrambling are small enough to determine the exchange rate in a segment is described for a time course experiment. According to the criterion, all eight parent ions analyzed had both segments with low enough scrambling to determine the exchange rates and segments with too much scrambling to determine the exchange rates (Table 4). This criterion also provides a method to obtain backbone amide hydrogen exchange rates from partially scrambled ETD data.
Abbreviations
- CID:
-
Collision-induced dissociation
- ECD:
-
Electron capture dissociation
- ETD:
-
Electron transfer dissociation
- GuHCl:
-
Guanidine hydrochloride
- HDX:
-
Hydrogen/deuterium exchange
- LC:
-
Liquid chromatography
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- TFA:
-
Trifluoroacetic acid
References
Coales, S.J., E, S.Y., Lee, J.E., Ma, A., Morrow, J.A., Hamuro, Y.: Expansion of time window for mass spectrometric measurement of amide hydrogen/deuterium exchange reactions. Rapid Commun. Mass Spectrom. 24, 3585–3592 (2010)
Hamuro, Y.: Determination of equine cytochrome c backbone amide hydrogen/deuterium exchange rates by mass spectrometry using a wider time window and isotope envelope. J. Am. Soc. Mass Spectrom. 28, 486–497 (2017)
Mayne, L., Kan, Z.Y., Chetty, P.S., Ricciuti, A., Walters, B.T., Englander, S.W.: Many overlapping peptides for protein hydrogen exchange experiments by the fragment separation-mass spectrometry method. J. Am. Soc. Mass Spectrom. 22, 1898–1905 (2011)
Milne, J.S., Mayne, L., Roder, H., Wand, A.J., Englander, S.W.: Determinants of protein hydrogen exchange studied in equine cytochrome c. Protein Sci. 7, 739–745 (1998)
Bai, Y., Milne, J.S., Mayne, L.C., Englander, S.W.: Primary structure effects on peptide group hydrogen exchange. Proteins Struct. Funct. Genet. 17, 75–86 (1993)
Anand, G.S., Hughes, C.A., Jones, J.M., Taylor, S.S., Komives, E.A.: Amide H/2H exchange reveals communication between the cAMP and catalytic subunit-binding sites in the RIα subunit of protein kinase A. J. Mol. Biol. 323, 377–386 (2002)
Hamuro, Y., Burns, L.L., Canaves, J.M., Hoffman, R.C., Taylor, S.S., Woods Jr., V.L.: Domain organization of D-AKAP2 revealed by enhanced deuterium exchange-mass spectrometry (DXMS). J. Mol. Biol. 321, 703–714 (2002)
Hamuro, Y., Coales, S.J., Molnar, K.S., Tuske, S.J., Morrow, J.A.: Specificity of immobilized porcine pepsin in H/D exchange compatible conditions. Rapid Commun. Mass Spectrom. 22, 1041–1046 (2008)
Zhang, H.M., Kazazic, S., Schaub, T.M., Tipton, J.D., Emmett, M.R., Marshall, A.G.: Enhanced digestion efficiency, peptide ionization efficiency, and sequence resolution for protein hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 80, 9034–9041 (2008)
Rey, M., Yang, M., Burns, K.M., Yu, Y., Lees-Miller, S.P., Schriemer, D.C.: Nepenthesin from monkey cups for hydrogen/deuterium exchange mass spectrometry. Mol. Cell. Proteomics. 12, 464–472 (2013)
Kadek, A., Mrazek, H., Halada, P., Rey, M., Schriemer, D.C., Man, P.: Aspartic protease nepenthesin-1 as a tool for digestion in hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 86, 4287–4294 (2014)
Yang, M., Hoeppner, M., Rey, M., Kadek, A., Man, P., Schriemer, D.C.: Recombinant Nepenthesin II for hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 87, 6681–6687 (2015)
Rand, K.D., Jørgensen, T.J.: Development of a peptide probe for the occurrence of hydrogen (1H/2H) scrambling upon gas-phase fragmentation. Anal. Chem. 79, 8686–8693 (2007)
Rand, K.D., Adams, C.M., Zubarev, R.A., Joørgensen, T.J.: Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 130, 1341–1349 (2008)
Rand, K.D., Zehl, M., Jensen, O.N., Jørgensen, T.J.: Loss of ammonia during electron-transfer dissociation of deuterated peptides as an inherent gauge of gas-phase hydrogen scrambling. Anal. Chem. 82, 9755–9762 (2010)
Rand, K.D., Zehl, M., Jørgensen, T.J.: Measuring the hydrogen/deuterium exchange of proteins at high spatial resolution by mass spectrometry: overcoming gas-phase hydrogen/deuterium scrambling. Acc. Chem. Res. 47, 3018–3027 (2014)
Rand, K.D., Pringle, S.D., Morris, M., Brown, J.M.: Site-specific analysis of gas-phase hydrogen/deuterium exchange of peptides and proteins by electron transfer dissociation. Anal. Chem. 84, 1931–1940 (2012)
Seger, S.T., Breinholt, J., Faber, J.H., Andersen, M.D., Wiberg, C., Schjodt, C.B., Rand, K.D.: Probing the conformational and functional consequences of disulfide bond engineering in growth hormone by hydrogen-deuterium exchange mass spectrometry coupled to electron transfer dissociation. Anal. Chem. 87, 5973–5980 (2015)
Leurs, U., Beck, H., Bonnington, L., Lindner, I., Pol, E., Rand, K.: Mapping the interactions of selective biochemical probes of antibody conformation by hydrogen–deuterium exchange mass spectrometry. Chembiochem. 18, 1016–1021 (2017)
Deng, Y., Pan, H., Smith, D.L.: Selective isotope labeling demonstrates that hydrogen exchange at individual peptide amide linkages can be determined by collision-induced dissociation mass spectrometry. J. Am. Chem. Soc. 121, 1966–1967 (1999)
Kim, M.Y., Maier, C.S., Reed, D.J., Deinzer, M.L.: Site-specific amide hydrogen/deuterium exchange in E. coli thioredoxins measured by electrospray ionization mass spectrometry. J. Am. Chem. Soc. 123, 9860–9866 (2001)
Akashi, S., Naito, Y., Takio, K.: Observation of hydrogen-deuterium exchange of ubiquitin by direct analysis of electrospray capillary-skimmer dissociation with Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 71, 4974–4980 (1999)
Eyles, S.J., Speir, P., Kruppa, G., Gierasch, L.M., Kaltashov, I.A.: Protein conformational stability probed by Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Chem. Soc. 122, 495–500 (2000)
Demmers, J.A.A., Haverkamp, J., Heck, A.J.R., Koeppe-II, R.E., Killian, J.A.: Electrospray ionization mass spectrometry as a tool to analyze hydrogen/deuterium exchange kinetics of transmembrane peptides in lipid bilayers. Proc. Natl. Acad. Sci. U. S. A. 97, 3189–3194 (2000)
Johnson, R.S., Krylov, D., Walsh, K.A.: Proton mobility within electrosprayed peptide ions. J. Mass Spectrom. 30, 386–387 (1995)
Demmers, J.A.A., Rijkers, D.T.S., Haverkamp, J., Killian, J.A., Heck, A.J.R.: Factors affecting gas-phase deuterium scrambling in peptide ions and their implications for protein structure determination. J. Am. Chem. Soc. 124, 11191–11198 (2002)
Jørgensen, T.J., Gardsvoll, H., Ploug, M., Roepstorff, P.: Intramolecular migration of amide hydrogens in protonated peptides upon collisional activation. J. Am. Chem. Soc. 127, 2785–2793 (2005)
Bulleigh, K., Howard, A., Do, T., Wu, Q., Anbalagan, V., Stipdonk, M.V.: Investigation of intramolecular proton migration in a series of model, metal-cationized tripeptides using in situ generation of an isotope label. Rapid Commun. Mass Spectrom. 20, 227–232 (2006)
Ferguson, P.L., Pan, J., Wilson, D.J., Dempsey, B., Lajoie, G., Shilton, B., Konermann, L.: Hydrogen/deuterium scrambling during quadrupole time-of-flight MS/MS analysis of a zinc-binding protein domain. Anal. Chem. 79, 153–160 (2007)
Hamuro, Y., Tomasso, J.C., Coales, S.J.: A simple test to detect hydrogen/deuterium scrambling during gas-phase peptide fragmentation. Anal. Chem. 80, 6785–6790 (2008)
Huang, R.Y., Garai, K., Frieden, C., Gross, M.L.: Hydrogen/deuterium exchange and electron-transfer dissociation mass spectrometry determine the interface and dynamics of apolipoprotein E oligomerization. Biochemist. 50, 9273–9282 (2011)
Landgraf, R.R., Chalmers, M.J., Griffin, P.R.: Automated hydrogen/deuterium exchange electron transfer dissociation high resolution mass spectrometry measured at single-amide resolution. J. Am. Soc. Mass Spectrom. 23, 301–309 (2012)
Donohoe, G.C., Arndt, J.R., Valentine, S.J.: Online deuterium hydrogen exchange and protein digestion coupled with ion mobility spectrometry and tandem mass spectrometry. Anal. Chem. 87, 5247–5254 (2015)
Masson, G.R., Maslen, S.L., Williams, R.L.: Analysis of phosphoinositide 3-kinase inhibitors by bottom-up electron-transfer dissociation hydrogen/deuterium exchange mass spectrometry. Biochem. J. 474, 1867–1877 (2017)
Pan, J., Han, J., Borchers, C.H., Konermann, L.: Electron capture dissociation of electrosprayed protein ions for spatially resolved hydrogen exchange measurements. J. Am. Chem. Soc. 130, 11574–11575 (2008)
Abzalimov, R.R., Kaplan, D.A., Easterling, M.L., Kaltashov, I.A.: Protein conformations can be probed in top-down HDX MS experiments utilizing electron transfer dissociation of protein ions without hydrogen scrambling. J. Am. Soc. Mass Spectrom. 20, 1514–1517 (2009)
Pan, J., Jun Han, J., Borchers, C.H., Konermann, L.: Characterizing short-lived protein Foldin intermediates by top-down hydrogen exchange mass spectrometry. Anal. Chem. 82, 8591–8597 (2010)
Sterling, H.J., Williams, E.R.: Real-time hydrogen/deuterium exchange kinetics via supercharged electrospray ionization tandem mass spectrometry. Anal. Chem. 82, 9050–9057 (2010)
Pan, J., Heath, B.L., Jockusch, R.A., Konermann, L.: Structural interrogation of electrosprayed peptide ions by gas-phase H/D exchange and electron capture dissociation mass spectrometry. Anal. Chem. 84, 373–378 (2012)
Hamuro, Y.: Regio-selective intramolecular hydrogen/deuterium exchange in gas-phase electron transfer dissociation. J. Am. Soc. Mass Spectrom. 28, 971–977 (2017)
Glasoe, P.K., Long, F.A.: Use of glass electrodes to measure acidities in deuterium oxide. J. Phys. Chem. 64, 188–189 (1960)
Hamuro, Y., Coales, S.J., Southern, M.R., Nemeth-Cawley, J.F., Stranz, D.D., Griffin, P.R.: Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J. Biomol. Tech. 14, 171–182 (2003)
Yulin Huang, Y., Pasa-Tolic, L., Guan, S., Marshall, A.G.: Collision-induced dissociation for mass spectrometric analysis of biopolymers: high-resolution Fourier transform ion cyclotron resonance MS4. Anal. Chem. 66, 4385–4389 (1994)
Molina, H., Matthiesen, R., Kandasamy, K., Pandey, A.: Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal. Chem. 80, 4825–4835 (2008)
Eigen, M.: Proton transfer, acid-base catalysis, and enzymatic hydrolysis. Angew. Chem. Int. Ed. 3, 1–19 (1964)
Wyttenbach, T., Paizs, B., Barran, P., Breci, L., Liu, D., Suhai, S., Wysocki, V.H., Bowers, M.T.: The effect of the initial water of hydration on the energetics, structures, and H/D exchange mechanism of a family of pentapeptides: an experimental and theoretical study. J. Am. Chem. Soc. 125, 13768–13775 (2003)
Ausloos, P., Lias, S.G.: Thermoneutral isotope exchange reactions of cations in the gas phase. J. Am. Chem. Soc. 103, 3641–3647 (1981)
Lias, S.G.: Thermoneutral isotope exchange reactions in proton-bound complexes of water with organic molecules: correlations with energetics of formation of the corresponding association ions. J. Phys. Chem. 88, 4401–4407 (1984)
Hunter, E.P., Lias, S.G.: In Mallard, W. G., Linstrom, P. J. (eds.) Nist Standard Reference Database Number, p. 69. National Institute of Standards and Technology (http://webbook.nist.gov), Gaithersburg (2008)
Perrin, C.L., Lollo, C.P.: Mechanisms of NH proton exchange in amides and proteins: solvent effects and solvent accessibility. J. Am. Chem. Soc. 106, 2154–2151 (1984)
Perrin, C.L.: Proton exchange in amides: surprises from simple systems. Acc. Chem. Res. 22, 268–275 (1989)
Campbell, S., Rodgers, M.T., Marzluff, E.M., Beauchamp, J.L.: Deuterium exchange reactions as a probe of biomolecule structure. Fundamental studies of gas phase WD exchange reactions of protonated glycine oligomers with D2O, CD3OD, CD3CO2D, and ND3. J. Am. Chem. Soc. 117, 12840–12854 (1995)
Wyttenbach, T., Bowers, M.T.: Gas phase conformations of biological molecules: the hydrogen/deuterium exchange mechanism. J. Am. Soc. Mass Spectrom. 10, 9–14 (1999)
Locke, M.J., McIver, R.T.: Effect of solvation on the acid/base properties of glycine. J. Am. Chem. Soc. 105, 4226–4232 (1983)
Chapo, C.J., Paul, J.B., Provencal, R.A., Roth, K., Saykally, R.J.: Is arginine Zwitterionic or neutral in the gas phase? Results from IR cavity ringdown spectroscopy. J. Am. Chem. Soc. 120, 12956–12957 (1998)
Mao, D., Douglas, D.J.: H/D exchange of gas phase bradykinin ions in a linear quadrupole ion trap. J. Am. Soc. Mass Spectrom. 14, 85–94 (2003)
Huang, Y., Marini, J.A., McLean, J.A., Tichy, S.E., Russell, D.H.: A mechanistic study of the H/D exchange reactions of protonated arginine and arginine-containing di- and tripeptides. J. Am. Soc. Mass Spectrom. 20, 2049–2057 (2009)
Acknowledgments
The author thanks Stephen J. Coales, Jessica E. Lee, and Anita Ma for their technical support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 490 kb)
Rights and permissions
About this article
Cite this article
Hamuro, Y., E, S. Determination of Backbone Amide Hydrogen Exchange Rates of Cytochrome c Using Partially Scrambled Electron Transfer Dissociation Data. J. Am. Soc. Mass Spectrom. 29, 989–1001 (2018). https://doi.org/10.1007/s13361-018-1892-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-018-1892-3


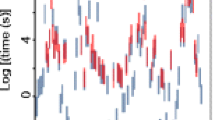






 determined by HDX-NMR [
determined by HDX-NMR [ determined by HDX-MS with analogous peptides [
determined by HDX-MS with analogous peptides [ determined by HDX-MS with analogous ETD ions
determined by HDX-MS with analogous ETD ions
