Abstract
Hydrogen deuterium exchange mass spectrometry (HDX-MS) is a well established method for the measurement of solution-phase deuterium incorporation into proteins, which can provide insight into protein conformational mobility. However, most HDX measurements are constrained to regions of the protein where pepsin proteolysis allows detection at peptide resolution. Recently, single-amide resolution deuterium incorporation has been achieved by limiting gas-phase scrambling in the mass spectrometer. This was accomplished by employing a combination of soft ionization and desolvation conditions coupled with the radical-driven fragmentation technique electron transfer dissociation (ETD). Here, a hybrid LTQ-Orbitrap XL is systematically evaluated for its utility in providing single-amide deuterium incorporation for differential HDX analysis of a nuclear receptor upon binding small molecule ligands. We are able to show that instrumental parameters can be optimized to minimize scrambling and can be incorporated into an established and fully automated HDX platform making differential single-amide HDX possible for bottom-up analysis of complex systems. We have applied this system to determine differential single amide resolution HDX data for the peroxizome proliferator activated receptor bound with two ligands of interest.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Amide hydrogen/deuterium exchange mass spectrometry (HDX-MS) has become a recognized method to study protein structure and dynamics [1–5]. Recently, several studies using differential HDX have provided valuable insight into protein–ligand interactions [6–9]. Conformational changes due to global unfolding and refolding as well as those induced by ligand binding causes variations in amide exposure to surrounding solvent, and these variations can be measured by the rate at which amide hydrogens exchange with solvent deuterium. Typical HDX experiments involve incubating the protein in a deuterated environment for a predefined period of time immediately followed by the addition of a quench solution (low pH, low temperature) containing denaturants to slow further on-exchange of deuterium and minimize back-exchange with nondeuterated solvent. The protein is then digested with acid-stable proteases, desalted, and analyzed by reversed phase liquid chromatography electrospray ionization mass spectrometry. Deuterium uptake is determined by measuring the increase in the intensity weighted centroid m/z values for each peptide ion isotopic distribution. To probe protein–ligand interactions, this procedure is performed on both the free protein and ligand bound protein. Differences in the deuterium uptake between the experiments reveals regions of the protein that exhibit differential exchange kinetics as a result of ligand binding.
While this technique provides valuable insight, the measurements are constrained to regions of the protein where digestion with pepsin allows detection of a proteolytic peptide. In addition, the measured HDX data are the average of all amides within the peptide; therefore the resolution of the HDX information obtained is equal to the size of the peptide (however, excluding the first two amino acids). Although NMR can provide single-amide HDX data, the upper protein size is limited compared with HDX MS and significantly more sample is required. Recently, single-amide deuterium incorporation measurements with mass spectrometry have been applied to model peptides systems [10] and to the apo form of β2-microglobulin [11]. Single-amide resolution was achieved by following the deuterium uptake of sequential fragment ions of a peptide. However, such measurements are only achievable by minimizing the degree of gas-phase intramolecular scrambling of the deuterium label across all amides and side chains within the peptide. Scrambling occurs as a result of the ion reaching the internal energy threshold that mobilizes protons before fragmentation occurs. To reduce scrambling, instrumental parameters involved in ion desolvation and transfer into the mass analyzer must be optimized to limit the amount of energy imparted to the precursor ion. The low energy, radical-driven fragmentation techniques electron capture dissociation (ECD) and electron transfer dissociation (ETD) are also employed to avoid excess vibrational energy associated with collision-induced dissociation (CID) [12–15]. Single amide HDX data has also been determined from top-down MS/MS experiments, where the intact protein is fragmented [16, 17]. However, this top-down workflow is unsuited to large proteins and protein–protein complexes due to the difficulty in generating intact protein ESI MS data for large (>50 kDa) proteins. For example, our recent VDR:RXR:SRC:DNA protein complex was in excess of 130 kDa and contained three proteins, two small molecule ligands, and DNA [18]. Such complexes are amenable to the bottom up workflow described here, but are beyond the current capabilities of top-down HDX or NMR methods.
While the measurement of hydrogen exchange at individual backbone amides can provide insight into protein folding and dynamics, it can also provide site specific information into protein-ligand interactions. To demonstrate the utility of this method, we have applied bottom up differential HDX-ETD using a high resolution mass spectrometer (HRMS) towards the analysis of peroxisome proliferator-activated receptor γ (PPARγ) in the presence and absence of small molecule ligands. PPARγ belongs to the nuclear receptor superfamily of ligand-dependent transcription factors, and when in complex with its coreceptor retinoid X receptor α (RXRα) controls genes involved in energy, carbohydrate, and lipid metabolism [19, 20], making PPARγ a highly desirable drug target for the treatment of type II diabetes. The drug class known as the glitazones or thiazolidinediones (TZDs) have shown potent and robust antidiabetic efficacy and improved insulin sensitivity in diabetic patients [21, 22]. Unfortunately, the use of TZDs have been associated with weight gain, plasma volume expansion, and increased incidences of cardiovascular events [19, 23]. Due to these clinical observations, emphasis has shifted to the development of selective PPARγ modulators (SPPARγMs) that exhibit comparable insulin sensitization efficacy as the TZDs but with decreased adverse side effects [19, 24, 25]. A recent study of PPARγ by HDX showed that subtle structural differences among PPARγ ligands can produce significantly different outcomes in terms of receptor conformational dynamics, ligand binding mode, and magnitude of agonist activity [26]. However, like all differential HDX studies performed to date, those HDX data were limited in resolution to the size of the proteolytic peptides (minus the first two amino acids). Here we re-examined two of the ligands from the 2007 study, specifically the full agonist and TZD rosiglitazone and the partial agonist MRL24. By utilizing differential HDX-ETD-HRMS, we were able to sublocalize deuterium incorporation to individual backbone amides and more clearly define the interaction of these ligands within the ligand binding pocket (LBP) of the receptor. This study represents the application of single-amide resolution HDX to provide site-specific information on protein–ligand interactions that were not previously available from NMR HDX data.
2 Experimental
2.1 Materials and Reagents
PPARG ligand binding domain was cloned, expressed, and purified as previously described [26]. Rosiglitazone was purchased from Sigma-Aldrich and MRL24 was synthesized as previously described [26].
2.2 Monitoring Gas-Phase Scrambling
To optimize the instrument parameters and to monitor gas-phase scrambling, we employed the model peptide HHHHHHIIKIIK described by the Jorgensen group (American Peptide Company, Sunnyvale, CA, USA) [12 ]. Complete deuteration was achieved by incubation of the peptide in deuterated buffer (20 mM NaH2PO4, pH 2.6) overnight at 4 °C. HDX off-exchange was initiated by a 50-fold dilution into direct infusion buffer (50% MeOH, 0.5 M CH3COOH, pH 2.6) and directly infused at regular time intervals in a precooled syringe. Full-scan and ETD MS/MS spectra were acquired on an LTQ Orbitrap XL ETD (Thermo Scientific, Bremen, Germany). The ion source and ion transfer optics were optimized for proper desolvation of the analyte ion as well as isolation width in the ion trap to minimize gas-phase scrambling (full details below).
2.3 Hydrogen/Deuterium Exchange
Differential, solution phase HDX experiments were performed with a fully automated system using a LEAP Technologies Twin HTS PAL liquid handling robot interfaced to an LTQ Orbitrap XL ETD mass spectrometer (Thermo Scientific, Bremen, Germany) [27]. Each exchange reaction was initiated by incubating 4 μL of protein (10 μM, with or without 100 μM ligand) with 16 μL of D2O protein buffer for a predetermined time (10, 30, 60, 300, 900, and 3600 in a randomized order) at 4 °C. The exchange reaction was quenched by mixing the protein solution with 30 μL of 3 M Urea, 1% TFA at 1 °C. The mixture was passed across an in-house packed pepsin column (1 mm × 20 mm) at 50 uL/min and digested peptides were captured onto a 1 mm × 10 mm C8 trap column (Thermo Scientific) and desalted (total time for digestion and desalting was 2.5 min). Peptides were then separated across a 1 mm × 50 mm C18 column (5 μm Hypersil Gold, Thermo Scientific) with linear gradient of 5%–50% CH3CN, 0.3% formic acid, over 5 min. Protein digestion and peptide separation were performed within a thermal chamber (Mécour) held at 15 °C and 1 °C, respectively, to promote more efficient digestion and to reduce D/H back exchange.
2.4 ESI-MS Parameters
Electrospray ionization parameters were set as the following to minimize the internal energy of the ion before precursor selection: sheath gas 30 au, auxiliary gas 20 au, sweep gas 20 au, spray voltage 4.0 kV, capillary temperature 50 °C, capillary voltage 45 V, tube lens 120 V. ETD optimized settings resulted in a 2- to 3-fold decrease in signal intensity, however our standard protein stock solution concentration (10 uM) was sufficient to generate all data shown here. Mass spectrometric analyses were acquired with a measured resolving power of 100,000 at m/z 400. Three replicates were performed for each ion-exchange time point. ETD MS/MS of the peptide, spanning residues 279–287, was performed by isolating the [IRIFQGCQF + 2H]2+ precursor ion at m/z 556.288 from retention times 5.1–5.2 min with an isolation width of 12 u to prevent sideband activation during precursor ion selection. Mixing time of the analyte cations and fluoranthene radical anions was set at 100 ms.
2.5 Calculating the Deuterium Content of Single Backbone Amides
The intensity weighted average m/z value (centroid) of each c-ion isotopic envelope was calculated with MagTran software. The deuterium content was then manually determined by measuring the difference of the centroid values of sequential fragment ions from the labeled peptide and comparing it to the corresponding mass difference of the unlabeled peptide and then multiplying by the charge state:
where D is the deuterium content, n is the residue number, l is the ligand bound protein, a is the apo protein, and z is the charge of the ion.
3 Results and Discussion
3.1 HDX-MS of PPARγ
Our current bottom-up HDX platform provides a rapid, robust and quantitative assessment of amide HDX at peptide resolution [18, 27, 28]. In such on-exchange experiments, labile hydrogen atoms exchange with deuterons from the surrounding solvent where highly exposed sites exchange rapidly, while little to no exchange occurs at sites that are shielded from the solvent or involved in hydrogen bonding networks. The typical work flow continually acquires full scan spectra for each peptide eluting off the analytical column over the 5 min gradient. The on exchange time points for apo and ligand bound protein were acquired in an interlaced, randomized, order at 10, 30, 60, 300, 900, and 3600 s in triplicate.
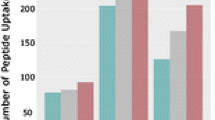
The molecular basis for ligand-mediated activation of nuclear receptors is often described by the “helix 12” or AF-2 (activation function-2) conformational model. This model suggests a switch between an active and inactive conformation or stabilization of the conformational mobility of the C-terminal helix, helix 12, of the ligand binding domain (LBD) [29]. However, a recent HDX study of the PPARγ ligand binding domain (LBD) revealed that both full and partial agonists can activate the receptor in a helix 12 independent manner. The study demonstrated that increased stabilization of helix 3 or the β sheet region of the PPARγ LBP could compensate for lack of interaction with helix 12. Figure 1a depicts the three-dimensional representation of the PPARγ LBD with a peptide in helix 3 (IRIFQGCQF) highlighted in blue that has shown HDX stabilization upon binding of rosiglitazone and MRL24. The peptide is shown in more detail in Figure 1b. In agreement with published data, rosiglitazone and MRL24 show a reduction in HDX of helix 3 when bound to PPARγ compared with the apo-receptor (Figure 1c) [26].
(a) Three-dimensional representation of the PPARγ ligand binding domain. The area highlighted in blue is the peptide IRIFQGCQF in helix 3. (b) Structural details of helix 3 peptide. (c) Build up curves illustrating the differences in deuterium incorporation upon binding of rosiglitazone and MRL24 to PPARγ
3.2 Bottom up HDX-ETD-HRMS
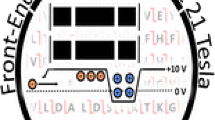
To improve HDX resolution to the single amide level, the instrument tune method was set to isolate the peptide of interest, IRIFQGCQF2+, eluting from 5.1 to 5.2 min and fragment it using ETD (Figure 2). The resulting c3–c8 product ions were followed for all time points in triplicate for the apo form of PPARγ as well as liganded PPARγ exposed to saturating amounts of MRL-24 and rosglitazone. The c1 and c2 ions were below the S/N threshold and were not followed. On average, a resolving power of 100,000 allowed three MS/MS scans to be acquired across the chromatographic peak of the peptide. At lower resolving power, isobaric c- and z-ions were unable to be baseline resolved (data not shown).
3.3 Gauging Gas-Phase Scrambling
Instrumental parameters were first optimized on a model peptide system HHHHHHIIKIIK [12] for proof of concept that scrambling does occur but can be minimized. Parameters found most pertinent to gas-phase scrambling were the temperature of the heated capillary and the isolation width in the ion trap for precursor selection. An example of varying degrees of scrambling based on heated capillary temperature can be seen in Supplemental Figure 1. Varying isolation widths were also investigated and it was concluded that isolation widths below 10 u induced a high level of scrambling (data not shown). For a thorough study of instrument parameters affecting hydrogen scrambling see reference [13]. Gauging the extent of gas-phase scrambling by monitoring sequential fragment ions over several experimental and instrumental conditions can be labor intensive and time consuming. Rand et al. have established a more routine method for reporting the degree of gas-phase scrambling by monitoring the loss of ammonia from the N-terminus of the peptide [30]. This ion is frequently observed in ETD product ion spectra and results from the loss of NH3 from the charged-reduced species of the peptide. Figure 3 depicts the IRIFQGCQF2+ product ion spectra for the singly charged reduced species and the corresponding NH3-deficient ion for the apo form of PPARγ and the MRL24 and rosiglitazone bound forms after 3600 s exposure to heavy water. Similar isotopic distributions between the charged reduced species and the NH3-deficient ions indicated that no significant depletion of deuterium occurred. The average deuterium loss observed for the apo, MRL24, and rosiglitazone bound forms of the protein were −0.1 ± 0.05, –0.02 ± 0.02, and −0.1 ± 0.01, respectively. The theoretically calculated deuterium depletion assuming 100% scrambling [30] were 0.15 ± 0.0, 0.04 ± 0.0, and 0.12 ± 0.0, respectively, which are considerably higher than the observed values.
3.4 Measuring Single-Amide Deuterium Content
The c-series ions generated by ETD fragmentation of the peptide IRIFQGCQF2+ were followed over seven on-exchange time points. An example of sequential c-ions for the unlabeled and 60 s on-exchange apo, MRL24 bound and rosiglitazone bound peptide is shown in Figure 4. Obvious shifts in the isotopic distribution of the c 4–c 8 ions for the apo sample compared with the unlabeled sample indicate a high degree of deuterium incorporation. More subtle changes in the isotopic envelopes of the MRL24 and rosiglitazone samples indicate deuterium incorporation but to a lesser extent. The residue specific deuterium content was calculated by comparing the difference in centroid values of sequential ions of the unlabeled peptide with the corresponding difference in centroid values for the labeled samples and multiplying by the charge state (see Calculating the Deuterium Content of Single Backbone Amides in the Methods section). This calculation was performed for each c-ion over all seven on-exchange time points. The quantitative data is depicted graphically in Figure 5. Note that because ETD fragments originate from the cleavage of the N–Cα backbone, the amide of residue n is contained in the c n-1 ion. No deuterium incorporation was observed in the c 3 fragment ion for the apo and liganded receptor across all time points. The residue corresponding to the c 3 ion is F282, indicating that the amide at that position is fully protected to exchange. A notable increase in deuterium content is observed for the c 4 ion again for the apo and liganded receptor across all time points, signifying an exchangeable amide at this amino acid. A nearly linear increase in deuterium content is observed across the remaining c-ions for the apo sample, suggesting these amides are equally protected. The deuterium content for the c 5 and c 6 ions of the MRL24 and rosiglitazone samples is equal to that of the c 4 ions, which implies the amides at residues G284 and C285 are protected to exchange. Another increase in deuterium content is observed for the c 7 ion, while the deuterium content of the c 8 ion remains constant in comparison, again indicating a site of exchange at Q286 and protection at F287 for the MRL24 and rosiglitazone samples. Due to the difference in deuterium content of the apo sample compared with the MRL24 and rosiglitazone samples, the protection observed for the ligand bound samples can be attributed to stabilization of this region of the receptor induced by the individual ligands themselves. This difference is mapped onto the three-dimensional structure in Figure 6. Figure 6a and c show the protection observed across the entire peptide with the lighter blue representing less protection to exchange. Figure 6b and d show the protection observed at the single-amide level. Black indicates no difference in HDX kinetics between the apo and ligand bound form of the receptor. A color change is indicative of a residue showing protection of the ligand bound receptor in comparison to the apo protein, again with lighter (cooler) colors indicating more protection to exchange. While the same residues are identified as exchanging or protected for the ligand bound samples, a notable difference in total deuterium content is observed. Supplemental Figure 2 shows the structural details of helix 3 of PPARγ bound to MRL24 and rosiglitazone as well as the superimposed images. MRL24 lies close to the helix 3 and makes more hydrophobic contacts than rosiglitazone, affording a higher degree of protection to exchange.
Isotopic patterns of the c 3–c 8 product ions with no deuterium labeling, 60 s on-exchange of apo PPARγ and 60 s on-exchange of the MRL24 and rosiglitazone bound PPARγ. These ETD spectra were acquired with the same “low scrambling” instrument parameters used to generate the data shown in Supplemental Figure 1
Differential HDX of the peptide IRIFQGCQF overlaid on the three-dimensional structure (PDB 2PRG). Protection observed at the peptide level for PPARγ bound to (a) MRL24 and (b) rosiglitazone. Protection observed at the single-amide level for PPARγ bound to (b) MRL24 and (d) rosiglitazone. Helices 10, 11, and 12 have been removed for a clearer view of helix 3
4 Conclusions
HDX-MS has emerged as a rapid and sensitive approach for characterization of protein dynamics and protein–ligand interactions [8, 31, 32]. Our laboratory and others have successfully applied the use of HDX to the mechanistic analysis of nuclear receptor activation [7, 8, 26, 33–37]. HDX analysis, particularly when combined with X-ray crystallography, can provide insights into protein structure function and into ligand activation of receptors.
Previously, it has been demonstrated that single amide resolution HDX can be obtained either by subtracting overlapping peptic peptides that differ by one amino acid generated in the typical bottom up HDX approach or by top down ETD or ECD. Here we demonstrate single-amide deuterium incorporation analysis applied to a fully automated bottom up HDX-ETD-MS system. Differential HDX kinetics were detected comparing apo PPARγ with receptor bound to rosiglitazone and MRL24. More importantly, single-amide HDX revealed a difference in magnitude of protection between the full agonist rosiglitazone and the partial agonist MRL24. Thus, HDX at single-amide resolution gave structural insight from an important region of the ligand binding pocket of PPARγ. This illustrates the utility of bottom-up HDX to provide site specific protein–ligand information on complex systems that has previously not been achievable by other HDX methods.
References
Bai, Y., Milne, J.S., Mayne, L., Englander, S.W.: Primary structure effects on peptide group hydrogen exchange. Proteins 17, 75–86 (1993)
Zhang, Z., Smith, D.L.: Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993)
Chamberlain, A.K., Marqusee, S.: Touring the landscapes: partially folded proteins examined by hydrogen exchange. Structure 5, 859–863 (1997)
Englander, S.W., Mayne, L., Bai, Y., Sosnick, T.R.: Hydrogen exchange: the modern legacy of Linderstrom-Lang. Protein Sci. 6, 1101–1109 (1997)
Engen, J.R., Smith, D.L.: Investigating protein structure and dynamics by hydrogen exchange MS. Anal Chem. 73, 256A–265A (2001)
Hamuro, Y., Zawadzki, K.M., Kim, J.S., Stranz, D.D., Taylor, S.S., Woods Jr., V.L.: Dynamics of cAPK type IIβ activation revealed by enhanced amide H/2H exchange mass spectrometry (DXMS). J. Mol. Biol. 327, 1065–1076 (2003)
Yan, X., Broderick, D., Leid, M.E., Schimerlik, M.I., Deinzer, M.L.: Dynamics and ligand-induced solvent accessibility changes in human retinoid X receptor homodimer determined by hydrogen deuterium exchange and mass spectrometry. Biochemistry 43, 909–917 (2004)
Hamuro, Y., Coales, S.J., Morrow, J.A., Molnar, K.S., Tuske, S.J., Southern, M.R., Griffin, P.R.: Hydrogen/deuterium-exchange (H/D-Ex) of PPARgamma LBD in the presence of various modulators. Protein Sci. 15, 1883–1892 (2006)
Zhang, J., Chalmers, M.J., Stayrook, K.R., Burris, L.L., Garcia-Ordonez, R.D., Pascal, B.D., Burris, T.P., Dodge, J.A., Griffin, P.R.: Hydrogen/deuterium exchange reveals distinct agonist/partial agonist receptor dynamics within vitamin D receptor/retinoid X receptor heterodimer. Structure 18, 1332–1341 (2010)
Zehl, M., Rand, K.D., Jensen, O.N., Jorgensen, T.J.: Electron transfer dissociation facilitates the measurement of deuterium incorporation into selectively labeled peptides with single residue resolution. J. Am. Chem. Soc. 130, 17453–17459 (2008)
Rand, K.D., Zehl, M., Jensen, O.N., Jorgensen, T.J.: Protein hydrogen exchange measured at single-residue resolution by electron transfer dissociation mass spectrometry. Anal. Chem. 81, 5577–5584 (2009)
Rand, K.D., Jorgensen, T.J.: Development of a peptide probe for the occurrence of hydrogen (1H/2H) scrambling upon gas-phase fragmentation. Anal. Chem. 79, 8686–8693 (2007)
Rand, K.D., Adams, C.M., Zubarev, R.A., Jorgensen, T.J.: Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 130, 1341–1349 (2008)
Rand, K.D.L., Amon, F.W.S., Jorgensen, T.J.D.: Investigation of amide hydrogen back-exchange in Asp and His repeats measured by hydrogen (1H/2H) exchange mass spectrometry. Int. J. Mass Spectrom. 302, 110–115 (2011)
Charlebois, J.P., Patrie, S.M., Kelleher, N.L.: Electron capture dissociation and 13 C,15N depletion for deuterium localization in intact proteins after solution-phase exchange. Anal. Chem. 75, 3263–3266 (2003)
Kaltashov, I.A., Bobst, C.E., Abzalimov, R.R.: H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: is there a need for a top-down approach? Anal. Chem. 81, 7892–7899 (2009)
Konermann, L., Pan, J.X., Liu, Y.H.: Hydrogen exchange mass spectrometry for studying protein structure and dynamics. Chem. Soc. Rev. 40, 1224–1234 (2011)
Zhang, J., Chalmers, M.J., Stayrook, K.R., Burris, L.L., Wang, Y., Busby, S.A., Pascal, B.D., Garcia-Ordonez, R.D., Bruning, J.B., Istrate, M.A., Kojetin, D.J., Dodge, J.A., Burris, T.P., Griffin, P.R.: DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat. Struct. Mol. Biol. 18, 556–563 (2011)
Berger, J., Wagner, J.A.: Physiological and therapeutic roles of peroxisome proliferator-activated receptors. Diabetes Technol. Ther. 4, 163–174 (2002)
Chawla, A., Repa, J.J., Evans, R.M., Mangelsdorf, D.J.: Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866–1870 (2001)
Day, C.: Thiazolidinediones: a new class of antidiabetic drugs. Diabet. Med. 16, 179–192 (1999)
Pearson, S.L., Cawthorne, M.A., Clapham, J.C., Dunmore, S.J., Holmes, S.D., Moore, G.B., Smith, S.A., Tadayyon, M.: The thiazolidinedione insulin sensitiser, BRL 49653, increases the expression of PPAR-gamma and aP2 in adipose tissue of high-fat-fed rats. Biochem. Biophys. Res. Commun. 229, 752–757 (1996)
Nissen, S.E., Wolski, K.: Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. N. Engl. J. Med. 356, 2457–2471 (2007)
Rangwala, S.M., Lazar, M.A.: The dawn of the SPPARMs? Sci STKE. 2002, pe9 (2002)
Rocchi, S., Picard, F., Vamecq, J., Gelman, L., Potier, N., Zeyer, D., Dubuquoy, L., Bac, P., Champy, M.F., Plunket, K.D., Leesnitzer, L.M., Blanchard, S.G., Desreumaux, P., Moras, D., Renaud, J.P., Auwerx, J.: A unique PPARgamma ligand with potent insulin-sensitizing yet weak adipogenic activity. Mol. Cell. 8, 737–747 (2001)
Bruning, J.B., Chalmers, M.J., Prasad, S., Busby, S.A., Kamenecka, T.M., He, Y., Nettles, K.W., Griffin, P.R.: Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure 15, 1258–1271 (2007)
Chalmers, M.J., Busby, S.A., Pascal, B.D., He, Y., Hendrickson, C.L., Marshall, A.G., Griffin, P.R.: Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 78, 1005–1014 (2006)
Griffin, P.R., Chalmers, M.J., Pascal, B.D., Willis, S., Zhang, J., Iturria, S.J., Dodge, J.A.: Methods for the analysis of high precision differential hydrogen-deuterium exchange data. Int. J. Mass Spectrom. 302, 59–68 (2011)
Heldring, N., Pike, A., Andersson, S., Matthews, J., Cheng, G., Hartman, J., Tujague, M., Ström, A., Treuter, E., Warner, M., Gustafsson, J.-Å.: Estrogen receptors: how do they signal and what are their targets. Physiol. Rev. 87, 905–931 (2007)
Rand, K.D., Zehl, M., Jensen, O.N., Jorgensen, T.J.: Loss of ammonia during electron-transfer dissociation of deuterated peptides as an inherent gauge of gas-phase hydrogen scrambling. Anal. Chem. 82, 9755–9762 (2010)
Iacob, R.E., Pene-Dumitrescu, T., Zhang, J., Gray, N.S., Smithgall, T.E., Engen, J.R.: Conformational disturbance in Abl kinase upon mutation and deregulation. Proc. Natl. Acad. Sci. U.S.A. 106, 1386–1391 (2009)
Hsu, Y.H., Burke, J.E., Li, S., Woods Jr., V.L., Dennis, E.A.: Localizing the membrane binding region of Group VIA Ca2 + −independent phospholipase A2 using peptide amide hydrogen/deuterium exchange mass spectrometry. J. Biol. Chem. 284, 23652–23661 (2009)
Hamuro, Y., Coales, S.J., Southern, M.R., Nemeth-Cawley, J.F., Stranz, D.D., Griffin, P.R.: Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J. Biomol. Tech. 14, 171–182 (2003)
Yan, X., Perez, E., Leid, M., Schimerlik, M.I., de Lera, A.R., Deinzer, M.L.: Deuterium exchange and mass spectrometry reveal the interaction differences of two synthetic modulators of RXRα LBD. Protein Sci. 16, 2491–2501 (2007)
Chalmers, M.J., Busby, S.A., Pascal, B.D., Southern, M.R., Griffin, P.R.: A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J. Biomol. Tech. 18, 194–204 (2007)
Dai, S.Y., Chalmers, M.J., Bruning, J., Bramlett, K.S., Osborne, H.E., Montrose-Rafizadeh, C., Barr, R.J., Wang, Y., Wang, M., Burris, T.P., Dodge, J.A., Griffin, P.R.: Prediction of the tissue-specificity of selective estrogen receptor modulators by using a single biochemical method. Proc. Natl. Acad. Sci. U.S.A. 105, 7171–7176 (2008)
Dai, S.Y., Burris, T.P., Dodge, J.A., Montrose-Rafizadeh, C., Wang, Y., Pascal, B.D., Chalmers, M.J., Griffin, P.R.: Unique Ligand Binding Patterns between Estrogen Receptor α and β Revealed by Hydrogen-Deuterium Exchange. Biochemistry 48, 9668–9676 (2009)
Acknowledgments
The authors are grateful for support from Bruce Pascal for software analyzing the HDX data. They thank John Bruning for providing the overlays of the co-crystal structures of rosiglitazone and MRL24. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of General Medical Sciences [grant R01-GM084041 to P.R.G.].
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 1294 kb)
Rights and permissions
About this article
Cite this article
Landgraf, R.R., Chalmers, M.J. & Griffin, P.R. Automated Hydrogen/Deuterium Exchange Electron Transfer Dissociation High Resolution Mass Spectrometry Measured at Single-Amide Resolution. J. Am. Soc. Mass Spectrom. 23, 301–309 (2012). https://doi.org/10.1007/s13361-011-0298-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-011-0298-2