Abstract
Paenibacillus polymyxa-M1 is a potent producer of bioactive compounds, such as lipopeptides, polyketides, and lantibiotics of biotechnological and medical interest. Genome sequencing revealed nine gene clusters for nonribosomal biosynthesis of such agents. Here we report on the investigation of the fusaricidins, a complex of cyclic lipopeptides containing 15-guanidino-3-hydroxypentadecanoic acid (GHPD) as fatty acid component by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). More than 20 variants of these compounds were detected and characterized in detail. Mass spectrometric sequence analysis was performed by MALDI-LIFT-TOF/TOF fragment analysis. The obtained product ion spectra show a specific processing in the fatty acid part. GHPD is cleaved between the α- and ß-position yielding two fragments a and b, one bearing the end-standing guanidine group and another one comprising the residual two C-atoms of GHPD with the attached peptide moiety. The complete sequence of all fusaricidins was derived from sets of bn- and yn-ions. The fusaricidin complex can be divided into four lipopeptide families, three of them showing variations of the amino acid in position 3, Val or Ile for the first and Tyr or Phe for families 2 and 3, respectively. A collection of novel fusaricidins was detected differing from those of families 1–3 by an additional residue of 71 Da (family 4). LIFT-TOF/TOF fragment spectra of these species imply that in their peptide moiety, an Ala-residue is attached by an ester bond to the free hydroxyl group of Thr4. More than 10 novel fusaricidins were characterized mass spectrometrically.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Paenibacillus is a rich source of bioactive compounds [1]. The most frequent products are lipopeptides [2–16], peptide/polyketide hybrids [17, 18], as well as lantibiotics [19, 20]. Such products qualify Paenibacillus strains for applications in agriculture, medicine, and biotechnology. In recent times the genomes of numerous Paenibacillus strains have been sequenced, revealing the wealth of attractive secondary metabolites produced by these organisms [21–29]. In this work, also gene clusters for the biosynthesis of polyketide products were annotated, which have not yet been identified [21, 22, 25, 27, 28]. This report is part of exploring the biosynthetic capacity of Paenibacillus polymyxa-M1, a strain found among 202 bacterial isolates from surface sterilized wheat plants collected from Beijing and Henan Province, China. M1 is an efficient plant growth promoting rhizobacterium (PGPR) interacting in the soil with plant roots, thus stimulating plant growth and productivity as a biofertilizer and protecting plants by biocontrol against phytopathogenic microorganisms using its powerful antimicrobial potential [30].
The complete genome of Paenibacillus-M1 was sequenced and nine giant gene clusters involved in nonribosomal synthesis of lipopeptides and polyketides were detected [21], which have to be explored now by genome mining in combination with efficient, advanced methodology for structure elucidation, such as mass spectrometry and NMR-spectroscopy. The prominent products of this strain are two lipopeptide families, the polymyxins and the fusaricidins, which both are of industrial and medical interest. Recently, we investigated its polymyxin products in detail [4]. They efficiently inhibit Gram-negative bacteria. M1 synthesizes two variants of polymyxin P which were revealed as efficient agents against phytopathogenic Erwinia strains in biocontrol [4, 31].
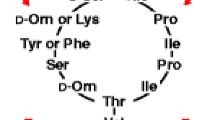
This research is focussed on the structural characterization of the fusaricidins, which appear as a complex of more than 20 variants. Fusaricidins are a class of cyclic lipohexapeptides with potent antifungal activities [5–11]. They contain 15-guanidino-3-hydroxypentadecanoic acid (GHPD) integrated in a ring of six amino acids. Cyclization of the peptide chain is accomplished by forming a lactone bond between the carboxyl group of D-Ala6 and the OH-group of Thr1. The biosynthesis of fusaricidin is catalyzed by a multifunctional enzyme FusA comprising six modules, which are equipped with the domains for activation, modification, and condensation of the amino acid substrates [32, 33]. Fusaricidins and the related LI-F-antibiotics were discovered and investigated by Kurusu et al. [5], Kajimura et al. [6, 7], and Kuroda et al. [8, 9]. Their structure has been elucidated mainly by two-dimensional-NMR methodology. They were detected by fast atom bombardment mass spectrometry (FAB MS). Sequence data of some well-known fusaricidin species were derived from CID-tandem mass spectrometry by Kuroda et al. [9] and Debois et al. [11], but the entire fusaricidin family shows a much higher complexity. Therefore, in this paper we initiated an extensive mass spectrometric study of these compounds by MALDI-TOF MS presenting numerous novel fusaricidin species. A breakthrough in the structural characterization of these lipohexapeptides was achieved by MALDI-LIFT-TOF/TOF MS affording a thorough, sensitive fragment analysis also of numerous minor fusaricidins.
Materials and Methods
Chemicals
The matrix α-cyano-hydroxycinnamic acid was obtained from Bruker (Bremen, Germany). Acetonitrile (HPLC grade) was from Merck (Darmstadt, Germany). Trifluoroacetic acid (TFA) was purchased from Sigma Aldrich (Steinheim, Germany).
Growth of Organism
Paenibacillus polymyxa strain M-1 isolated from surface sterilized wheat roots in China was kept frozen at –70 oC with 15% glycerol as laboratory stock and cultivated in glucose-starch-CaCO3 (GSC) liquid broth at 30 oC for 24 h, as described previously [4, 34]. In addition, strain M1 was grown on GSC-agar at 30 oC for at least 24 h.
MALDI-TOF Mass Spectrometric Analysis
Aliquots of culture supernatants of strain M-1 were directly used for the investigation of fusaricidins by MALDI-TOF mass spectrometry after dilution 1:10 (v/v) with 50% aqueous acetonitrile/0.1 trifluoroacetic acid. In addition, surface extracts of M-1 were prepared by extraction of cells picked from GSC-agar plates with 50% aqueous acetonitrile/0.1 trifluoroacetic acid.
MALDI-TOF mass spectra of the lipopeptide products of Paenibacillus polymyxa M-1 were recorded using a Bruker Autoflex Speed TOF/TOF mass spectrometer with smartbeam-laser technology using a 1 kilohertz Nd-YAG laser (excitation wavelength: 355 nm) for desorption and ionization. Samples of 2 μL culture supernatant or surface extract were mixed with the same volume of matrix solution (a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% aqueous acetonitrile containing 0.1% (v/v) trifluoroacetic acid), spotted on the target, air dried, and measured. Spectra were obtained by positive ion detection and reflector mode. Monoisotopic masses were detected.
Sequence analysis of the fusaricidins was performed by MALDI-LIFT-TOF/TOF mass spectrometry in CID mode [35]. The following instrumental parameters were used:
-
(a)
for recording of mass spectra in the range of 0.7–3.5 kDa: acceleration voltages: ion source 1: 18.97 kV; ion source 2: 16.72 kV. Reflector voltages: reflector 1: 21.05 kV; reflector 2: 9.54 kV. Matrix ions were suppressed up to 500 kDa.
-
(b)
for the generation of LIFT-TOF/TOF fragment spectra: acceleration voltages: ion source 1: 5.99 kV; ion source 2: 5.30 kV. Reflector voltages: reflector 1: 27.05 kV; reflector 2: 11.74 kV. LIFT 1: 18.98 kV; LIFT 2: 4.40 kV.
Results and Discussion
This paper is focussed on an extensive mass spectrometric characterization of the fusaricidins, a complex of lipohexapeptides with potent antifungal activities. In this study numerous novel fusaricidins were identified by MALDI-TOF mass spectrometry.
The Fus Gene Cluster and the Modular Organization of FusA Synthetase
Biosynthesis of fusaricidins is encoded by the fus gene cluster, which is located in the P. polymyxa-M1 genome [21] between base positions 80.646 and 111.506 in close vicinity to the origin of replication. Similar to P. polymyxa PKB1 [32], the cluster consists of eight genes and covers around 32 kb (Figure 1a).
Module organization of fusaricidin synthetase FusA. FusA comprises 7909 amino acids and is composed of six amino acid activating modules. The organization of domains is indicated. A, adenylation domain; C, condensation domain; PCP, peptidyl carrier protein (thiolation domain); E, epimerization domain; TE, thioesterase domain. The assignment of the amino acid components activated and processed within the modules was taken from the literature [5–11]
The predicted products of six genes (fusB-fusG) are presumably involved in synthesis of the fatty acid moiety of fusaricidin. The genes for immunity, regulation, and transport are missing in the cluster. The nonribosomal peptide synthetase FusA consists of 6 modules, each containing the three core units for condensation (C), adenylation (A), and thiolation (peptidyl carrier protein, PCP). Modules 2, 4, and 5 comprise an additional epimerization (E) domain, located downstream from the PCP. The E-domain directs conversion of amino acids in D-configuration. Interestingly, module 6 directing D-Ala does not contain an E-domain. It has been shown that the A6 domain activates D-Ala directly [32]. The cyclization reaction between D-Ala6 and L-Thr1 is most likely accomplished by the thioesterase (TE) domain located at the carboxy terminus of FusA (Figure 1b). This structure is identical to that found in other P. polymyxa strains producing fusaricidins [21–28, 32].
Demonstration of the Fusaricidin Complex by MALDI-TOF- MS
The fusaricidins were studied extensively by MALDI-TOF MS, which is the method of choice for rapid in situ detection of these compounds in culture supernatants and surface extracts of Paenibacilli without the need for further purification prior to mass spectrometric analysis. This technique affords simple preparation and efficient investigation of complex families of natural compounds, such as the fusaricidins, with high sensitivity and accuracy in a minimum of time.
To find out the optimal growth conditions for the production of secondary metabolites, strain M1 was grown in various liquid cultivation media. The best results were obtained (a) in the Landy medium [36] supplemented with yeast extract in addition with D, L-alanine, and/or phenylalanine, and (b) in the glucose-starch-CaCO3 (GSC) medium [34].
Figure 2a shows the MALDI-TOF mass spectrum of a culture supernatant of Paenibacillus polymyxa M-1 cultivated in liquid GSC medium in the range of m/z 860–1250 exhibiting the mass peaks of the fusaricidins (m/z 860–1050) and the polymyxins (m/z 1170–1230), which appeared in similar intensities. In Figure 2b and c, the mass spectrum of the fusaricidins obtained from a surface extract of P. polymyxa M-1 cells grown on GSC-agar is presented in expanded form. It was divided into two parts from m/z 860 to 970 (Figure 2b) and from m/z 970 to 1050 (Figure 2c) to make visible the mass peaks of all detected variants. The obtained mass data are summarized in Table 1. More than 20 fusaricidin species were observed, which appeared in protonated form and as their alkali adducts. The mass peaks of the protonated fusaricidins [M + H]+ dominate in the MALDI-TOF mass spectra shown in Figure 2. In addition, for all variants, minor mass peaks appeared at 18 mass units higher than those found for the protonated forms ([M + H + H2O]+), which can be attributed to the open chain fusaricidins. The main components of the fusaricidin complex, which have already been investigated by other authors, are indicated in Table 1. They are fusaricidins A–D, which have been characterized in detail by Kajimura et al. [6, 7] and those designated by the LI-F-nomenclature introduced by Kurusu et al. [5] and Kuroda et al. [8, 9]. All other fusaricidin variants in Table 1 were detected and characterized by us for the first time. This wide spectrum of structurally modified agents equips Paenibacillus with a potent antifungal arsenal enabling this rhizobacterium to attack competitors in the soil in a specific directed manner. This has been demonstrated by Debois et al. [11] by MALDI-FTICR MS imaging for the inactivation of the plant pathogen Fusarium oxysporum by Paenibacillus polymyxa strain Pp56. Here, mainly fusaricidin B and a mixture of LI-F05b/06b/08b were involved in the defense mechanism of Pp56.
MALDI-TOF mass spectra of a culture filtrate of Paenibacillus polymyxa M-1 cultivated in the GSC medium (a) and of a surface extract from cells of this organism grown on GSC-agar (b) and (c). The mass spectrum of the culture filtrate in the range of m/z 860–1250 (a) exhibits the fusaricidins (m/z 860–1050) and the polymyxins (m/z 1170–1230). The mass spectrum of the fusaricidins was divided into two parts, (m/z 860–970 (b) and m/z 970–1050 (c) for a better presentation of the mass peaks
Structural Characterization of Fusaricidins from LIFT-MALDI-TOF/TOF Spectra
Structural characterization of fusaricidins listed in Table 1 was performed by MALDI-TOF/TOF-LIFT mass spectrometry, a modern technique introduced in proteomics for efficient mass spectrometric peptide mapping of proteins [35]. In our studies, the CID-mode was applied. Advantages of this technique compared with PSD-MALDI-TOF fragment analysis are (a) a higher accuracy and resolution of the mass peaks, which is high enough that monoisotopic masses can be assigned to product ions; (b) much better signal-to-noise ratios of product ion peaks; (c) a higher number of fragment peaks; and d) a rapid collection of fragmentation data. While PSD-MALDI-TOF MS requires the fragment ion spectrum to be recorded in 10–20 steps, each step measured at a lower reflector voltage than the last one, with the LIFT-technique the fragment spectrum is obtained rapidly in only one scan. The acquisition of the PSD-data needs at least 5–10 min per spectrum. In contrast, the LIFT- fragment spectrum can be recorded in 10–20 s.
Fusaricidins are distinguished by characteristic features that are related to the guanidine group attached N-terminally to its GHPD fatty acid constituent, which qualify them for efficient CID- and PSD-based mass spectrometric structure analysis. Fusaricidins are protonated preferentially at the guanidine group. Therefore, precursor ions for mass spectrometric sequencing are predominantly available in protonated form in contrast to many other cyclic lipopeptides, such as the surfactins, fengycins, and iturins [37, 38] with a neutral fatty acid side chain, which mainly appear as the alkali adducts when investigated by MALDI-TOF MS. The intensities of the protonated precursor ions of fusaricidins are 10- to 100-fold higher than their corresponding alkali counterparts. Generally, protonated forms are better amenable to fragmentation in CID- and LIFT-TOF/TOF-experiments than the alkali adducts showing a wealth of product ions that can be utilized for structure analysis. This is illustrated by the LIFT-TOF/TOF spectrum of the fusaricidin A precursor ion [M + H]+ = 883.89 presented in Supplementary Figure S1. Supplementary Figure S1a shows the complete product ion spectrum. Here, the dominant precursor ions of high intensity were cut off for a better presentation of the fragment peaks. In Supplementary Figure S1b and S1c, this spectrum was expanded and divided up into two parts from m/z 40 to 240 (b) and from m/z 280 to 840 (c) for higher resolution and better labelling of the product ion mass peaks.
The LIFT-TOF/TOF spectra of all investigated fusaricidins show the following general characteristic features as illustrated in the fragmentation scheme for fusaricidin A, for example (Scheme 1).
Remarkably, collision-induced dissociation of the linearized fusaricidins formed by cleavage of the lactone bond between the OH-group of Thr1 and the carboxyl group of the C-terminal alanine6 shows an intensive processing of the GHPD side chain. The mass peak exhibiting the highest intensity in the LIFT-TOF/TOF spectra of all investigated fusaricidins was always found at m/z 256.2. It can be attributed to fragment ion a formed by collision induced cleavage of the GHPD chain between the α- and ß-position. The remaining fragment ionb containing the first two carbon atoms of the fatty acid chain with the attached peptide part was found for fusaricidins A and D at m/z 628.6 and 706.4, respectively. Presumably, this cleavage is facilitated by the positively charged guanidine group of GHPD. Such an effect was not observed for related lipodepsipeptides, such as the surfactins and fengycins produced by numerous B. subtilis and B. amyloliquefaciens strains also equipped with a ß-hydroxy fatty acid component, but showing a neutral side chain [37, 38].
In addition, elimination of H2O and NH3 from the fatty acid chain was observed. The fragment patterns of fusaricidins A (m/z 883.8) and D (m/z 961.8) in Figures 3 and 4 imply that water is eliminated involving the ß-hydroxyl group forming the α,ß-unsaturated GHPD. NH3 is split off the guanidine group. The modified species were found a 17, 18, and 35 mass units lower than the mass of the precursor ion. Elimination of NH3 was also observed for fragment ion a defined in the fragmentation scheme (m/z 239.3).
Sequence of fusaricidin A derived from the LIFT-TOF/TOF spectrum of the precursor ion [M + H]+ = 883.89 shown in Supplementary Figure 1S. Structure analysis was based (a) on three sets of bn-ions of fusaricidin A starting with unmodified fatty acid residue GHPD; with α,ß-unsaturated GHPD and with GHPD after elimination of both H2O and NH3, and (b) of bn′(–H2O) – ions obtained for fragment ion b specified in the fragmentation scheme. It was corroborated by the yn-ions. Amino acid components of fusaricidin A were detected by their immonium ions. Nearest neighbor relationships were determined by detection of dipeptide ions of amino acids adjacent in the peptide sequence
Sequence of fusaricidin D derived from the LIFT-TOF/TOF spectrum of the precursor ion [M + H]+ = 961.72 (not shown). Structure analysis was based on four sets of bn-ions and the corresponding yn-ions as outlined in the legend of Figure 3
Because after the decay of the precursor ions of fusaricidins the positive charge is preferentially located at the guanidine group of GHPD N-terminal fragment ions, mainly those of the bn-type are dominating, as is apparent from Supplementary Figure S1.
The product ions used for fusaricidin sequence determination in Figures 3, 4, and 5 were classic bn- and yn-type fragments. From the LIFT-TOF/TOF spectra of all fusaricidins, four sets of bn-ions were derived, as demonstrated for two prominent species, fusaricidins A and D, in Figures 3 and 4. Set 1 bn was obtained from fusaricidin species with unmodified GHPD precursor starting with the b1-ion at m/z 298.4. Set 2 bn(–H2O) originates from the α,ß- unsaturated GHPD molecule (b1(–H2O): m/z 280.4). A characteristic feature of the fragment patterns is the loss of NH3 from the guanidine group of the α,ß-unsaturated GHPD leading to set 3 bn (–H2O;-NH3) starting with (b1(–H2O;–NH3): m/z 263.4). Remarkably, some bn-ions of set 3 belong to the dominating fragment ions in the LIFT-TOF/TOF spectra of all fusaricidins. A set of bn-ions starting from a species showing the loss of NH3 from the GHPD component alone was not observed. These results imply that H2O and NH3 are eliminated from GHPD successively. In addition, a set of bn-ions designated as bn′-ions (set 4) was found for fragment ion b of all fusaricidins defined in the fragmentation scheme. Such data are shown in Figures 3b and 4b for fusaricidins A and D. Here, a prominent set of bn′-ions starting with the b1′-ion at m/z 43.0 was detected always after elimination of H2O from threonine1 of the fusaricidin sequence forming a dehydro-alanine (precursor ions m/z 610.6 and 688.4 for fusaricidins A and D).
Sequence of the fusaricidin variant derived from the LIFT-TOF/TOF spectrum of the precursor ion [M + H]+ = 1032.8 (not shown). Structure analysis was based on four sets of bn-ions and the corresponding yn-ions as outlined in the legend of Figure 3
From Figures 3 and 4 it is apparent that in the case of fusaricidins A and D almost complete bn-data were found for sets 2–4. In set 1 with an unmodified GHPD, some bn-ions were missing. For these fusaricidins, complete sets of yn-ions were also obtained for both the entire lipohexapeptide precursor ion and fragment ion b. Our results achieved by mass spectrometric sequencing are corroborated by amino acid analysis of fusaricidins by detection of the immonium ions of their amino acid components as well as by a study of nearest neighbor relationships detected by di-, tri-, and tetrapeptide fragment ions, which presumably are second generation fragments. Such data are of great value for the first steps in de novo sequence determination of unknown peptide compounds. From our results, the sequence of fusaricidins could be determined efficiently. They are in excellent agreement with previous structure analysis of fusaricidins performed by other authors [5–11]. However, in our MALDI-TOF-MS study of the fusaricidins, it was not possible to discriminate between Leu, Ile, and allo-Ile in positions 2 and 3 of the fusaricidin sequence because the corresponding w-ions were missing in the product ion spectra. We also could not distinguish optical isomers by this technique. For this reason, we did not specify the configuration of the amino acid components in the fragmentation patters determined mass spectrometrically in Figures 3, 4, and 5. However, in Figures 1 and 6 their chirality was assigned because here we refer to the knowledge compiled in the literature [5–11].
Classification of Fusaricidins into Four Families
A general feature of our results summarized in Table 2 is that in the peptide sequence of any fusaricidin, the Thr in position 1 and the C-terminal Ala6 are strictly conserved. All other amino acid components are variable. From previous work of other authors [5–9] it is well known that the valine residues in positions 2 and 3 can be replaced by Ile or allo-Ile. Position 5 can be occupied either by Asn or Gln. Until now Thr in position 4 was assumed to be invariable, but our data show that it can be replaced by a serine, which became obvious from mass spectrometric sequencing of the fusaricidin found at m/z 869.7. According to the literature data [5–11], the amino acid components in position 2, 4, 5, and 6 appear as the D-isomers. This is consistent with the module organization of fusaricidin synthetase (FusA) shown in Figure 1. Modules 2, 4, and 5 contain epimerization domains catalyzing the conversion of the L- into the D-isomer. An exception of this rule is module 6, which directly activates D-Ala.
GHPD, the fatty acid component of fusaricidins, is strictly conserved. This finding is in contrast to many other lipopeptides reported in the literature, where the fatty acid part is a major target of structural variation. Prominent examples are the surfactins, iturins, and fengycins produced by numerous B. subtilis and B. amyloliquefaciens strains [37, 38]. From our mass spectrometric sequence analysis, we classified the fusaricidins into three main families (series 1–3 in Table 2). Series 1 comprises the variants containing Val, Ile, or allo-Ile in positions 2 and 3. Evaluation of the LIFT-TOF/TOF spectra revealed that each of the fusaricidins found at m/z 897.8 and 911.8 represent mixtures of two isomers with different substitutions at positions 2, 3, and 5. Val3 can also be substituted either by Tyr (series 2) or Phe (series 3). Interestingly, a yet unknown family of fusaricidins was detected (series 4) including the species found at m/z 954.8, 968.8, 982.8, 996.8, 1002.8, 1016.8, and 1032.8, which are distinguished from the fusaricidins of series 1–3 by a mass difference of 71 Da.
As a representative example of these groups, we investigated the fusaricidin with a molecular mass [M + H]+ = 1032.8, which is 71 Da higher than that of fusaricidin D (m/z 961.8), in order to structurally characterize and locate the position of the unknown residue. The LIFT-TOF/TOF spectrum of this compound is shown in Figure 5. Complete sets 1–4 of bn-ions and of the yn-ions were obtained. In comparison with the fragment pattern for fusaricidin D (Figure 4), we found the same data for b1–b4 (from GHPD until Tyr3) as well as for y1 and y2 (from Ala6 to Gln5), however, b5 and y3 including residue Thr4 are by 71 mass units higher than the corresponding product ions of fusaricidin D. These results imply that the unknown residue of 71 Da is either attached to Thr4 or integrated into the lipopeptide chain between Thr4 and Gln5. Similar results have been obtained for the bn′-ions for fragment ion b of this fusaricidin in Figure 5b. To clarify the location of this residue, we performed nearest neighbor relationship studies (see Figure 5a). The mass data for the di-, tri-, and tetrapeptide fragment ions are listed in Figure 5a. We detected the mass peaks for the tripeptide ions Tyr-Thr-71 (m/z 336.32) and Thr-71-Gln (m/z 301.27) as well as the tetrapeptide ions Val-Tyr-Thr-71 (m/z 435.22), Tyr-Thr-71-Gln (m/z 464.39), and Thr-71-Gln-Ala (m/z 372.27), whereas the tripeptide fragment ions 71-Gln-Ala and Gln-Ala-71 (for both m/z 271.14 was calculated) were missing. These data indicate that the unknown residue of m/z 71 is residing on Thr4 of fusaricidin D, as shown in Figure 6.
The mass of 71 Da matches with an Ala-residue. Therefore, we hypothesize that in the biosynthesis of all fusaricidins, Ala can be attached to Thr4 in a side reaction. From the biosynthetic view, this conclusion seems to be reasonable for the following reasons: (1) in the assembly line of FusA there is no additional module between modules 4 and 5 that would accomplish the integration of Ala between Thr4 and Gln5 in the fusaricidin sequence, and (2) the C-terminal module of fusaricidin synthetase (see Figure 1) is distinguished by an affinity for Thr because in the termination process of fusaricidin biosynthesis ring closure occurs by lactonization of Ala6 with the hydroxyl-group of Thr1. Similarly, it may also show a significant affinity for Thr in position 4. In this way, Ala would be attached to this constituent by formation of an ester bond. Furthermore, this conclusion is corroborated by the fact that in the LIFT-TOF/TOF spectrum of the fusaricidin found at m/z 1032.8, the same sequence of product ions can be derived as observed for fusaricidin D in Figure 4. This would happen when by CID the ester bond between Thr4 and the attached Ala is cleaved.
The Ala-specific module 6 may be used iteratively. In nonribosomal peptide biosynthesis peptide synthetases (NRPSs) of type B use modules and/or domains more than once [39]. Manifold variations of such iterative mechanisms have been reported. Frequently, modules are used repetitively when the final product is composed of repeated smaller units, as, for example, in the case of gramicidin S, which consists of two identical pentapeptides with the consequence that all modules along the assembly line were used twice in the biosynthetic process [40]. Other prominent examples are enniatin [41] and related depsipeptides, which are cyclic trimers of dipeptidols as well as the siderophore enterobactin [42] produced by Escherichia coli, which is a cyclic trimer of dihydroxybenzoylserine units. On the other hand, iterative phenomena have also been observed for specific domains. Nice examples are the repeated use of an unique adenylation domain involved in the biosynthesis of the antibiotic congocidine in Streptomyces ambofaciens [43] and the iterative bimodular PCP-thioesterase (TE) domain of gramicidin S synthetase B, which catalyzes the ligation of the pentapeptide intermediates prior to product release by cyclization [44]. In our work, fusaricidin synthetase exhibits an interesting iterative potential of the C-terminal alanine-specific module, which is able to perform both cyclization and modification of fusaricidins.
To verify this hypothesis further, efforts are needed to isolate fusaricidin species of family 4 in pure form by separation techniques of high resolution for their structural characterization applying advanced MS- and NMR-methodology.
Conclusions
In this paper, we investigated the diversity, structure, and biosynthesis of the fusaricidins, prominent antifungal products of Paenibacillus polymyxa strain M1. The fusaricidin complex comprises more than 20 structural variants, which differ in their amino acid composition and sequence. They were efficiently detected by MALDI-TOF MS in culture filtrates and surface extracts of this organism. The structure of all variants was characterized extensively by MALDI-LIFT-TOF/TOF fragment ion analysis, a novel, powerful method extending the potential of MALDI-TOF MS, particularly for proteomics and structural characterization of natural compounds. From our mass spectrometric data, the fusaricidins could be divided into three families showing specific substitutions mainly in position 3 of the lipopeptide chain. Of particular relevance is the discovery of a fourth family of novel fusaricidins, which are distinguished from those of families 1–3 by an increase in the molecular mass of 71 Da. From our data we infer that the 71 Da-residue can be attributed to an alanine that is attached to the hydroxyl group of Thr4 via an ester bond. This location is compatible with the module organization of fusaricidin synthetase and indicates an iterative usage of the terminal, alanine-specific module of FusA catalyzing both the cyclization and modification of fusaricidins. The great variety in the physicochemical and antimicrobial properties of fusaricidins may contribute to a high flexibility of strain M1 in the defense of competitors in the rhizosphere.
References
Cochrane, S.A., Vederas, J.C.: Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med. Res. Rev. (2014). doi:10.1002/med 21321
Velkov, T., Thompson, P.E., Nation, R.L., Li, J.: Structure-activity relationships of polymyxin antibiotics. J. Med. Chem. 53, 1898–1916 (2010)
Shasheen, M., Li, J., Ross, A.C., Vederas, J.C., Jensen, S.E.: Paenibacillus polymyxa PKB1 produces variants of polymyxin B-type antibiotics. Chem. Biol. 18, 1640–1648 (2011)
Niu, B., Vater, J., Rueckert, C., Blom, J., Lehmann, M., Ru, J.-J., Chen, X.-H., Wang, Q., Borriss, R.: Polymyxin P is the active principle in suppressing phytopathogenic Erwinia spp. by the biocontrol rhizobacterium Paenibacillus polymyxa M-1. BMC Microbiol. 13, 137–150 (2013)
Kurusu, K., Ohba, K., Arai, T., Fukushima, K.: New peptide antibiotics LI-F03, F04, F05, F07, and F08, produced by Bacillus polymyxa. Isolation and characterization. J Antibiot (Tokyo) 40, 1506–1514 (1987)
Kajimura, Y., Kameda, M.: Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. Taxonomy, fermentation, isolation, structure elucidation, and biological activity. J Antibiot (Tokyo) 49, 129–135 (1996)
Kajimura, Y., Kameda, M.: Fusaricidin B, C and D, new depsipeptide antibiotics produced by Bacillus polyma KT-8. Isolation, structure elucidation, and biological activity. J Antibiot (Tokyo) 50, 220–228 (1997)
Kuroda, J., Fukai, T., Konishi, M., Ono, J., Kurusu, K., Nomura, T.: LI-F antibiotics, family of antifungal cyclic depsipeptides produced by Bacillus polymyxa L-1129. Heterocycles 53, 1533–1549 (2000)
Kuroda, J., Fukai, T., Nomura, T.: Collision-induced dissociation of ring-opened cyclic depsipeptides with a guanidino group by electrospray ionization/ion trap mass spectrometry. J. Mass Spectrom. 36, 30–37 (2001)
Choi, S.-K., Park, S.-Y., Kim, R., Lee, C.-H., Kim, J.F., Park, S.-H.: Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 365, 89–95 (2008)
Debois, D., Ongena, M., Cawoy, H., De Pauw, E.: MALDI-FTICR MS imaging as a powerful tool to identify Paenibacillus antibiotics involved in the inhibition of plant pathogens. J. Am. Soc. Mass Spectrom. 24, 1202–1213 (2013)
Wu, X.C., Shen, X.B., Ding, R., Qian, C.D., Fang, H.H., Li, O.: Isolation and partial characterization of antibiotics produced by Paenibacillus elgii B69. FEMS Microbiol. Lett. 310, 32–38 (2010)
Ding, R., Wu, X.C., Qian, C.D., Teng, Y., Li, O., Zhan, Z.J., Zhao, Y.H.: Isolation and identification of lipopeptide antibiotics from Paenibacillus elgii B69 with inhibitory activity against methicillin-resistant Staphylococcus aureus. J. Microbiol. 49, 942–949 (2011)
Guo, Y., Huang, E., Yuan, C., Zhang, L., Yousef, A.E.: Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl. Environ. Microbiol. 78, 3156–3165 (2012)
Huang, E., Guo, Y., Yousef, A.E.: Biosynthesis of the new broad-spectrum lipopeptide antibiotic paenibacterin in Paenibacillus thiaminolyticus OSY-SE. Res. Microbiol. 165, 243–251 (2014)
Sood, S., Steinmetz, H., Beims, H., Mohr, K., Stadler, M., Djukic, M., von der Ohe, W., Steinert, M., Daniel, R., Müller, R.: Paenilarvins: Iturin family lipopeptides from the honey bee pathogen Paenibacillus larvae. ChemBioChem 15, 1947–1955 (2014)
Garcia-Gonzalez, E., Müller, S., Hertlein, G., Heid, N., Süssmuth, R.D., Genersch, E.: Biological effects of paenilamicin, a secondary metabolite antibiotic produced by the honey bee pathogenic bacterium Paenibacillus larvae. Microbiol Open 3, 642–656 (2014)
Müller, S., Garcia-Gonzalez, E., Mainz, A., Hertlein, G., Heid, N.C., Mösker, E., van den Elst, H., Overkleeft, H.S., Genersch, E., Süssmuth, R.D.: Paenilamicin: structure and biosynthesis of a hybrid nonribosomal peptide/polyketide antibiotic from the bee pathogen Paenibacillus larvae. Angew. Chem. Int. Ed. 53, 1–6 (2014)
He, Z., Kisla, D., Zhang, L., Yuan, C., Green-Church, K.B., Yousef, A.E.: Isolation and identification of a Paenibacillus polymyxa strain that coproduces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73, 168–178 (2007)
Teng, Y., Zhao, W., Qian, C., Li, O., Zhu, L., Wu, X.: Gene cluster analysis for the biosynthesis of elgicins, novel lantibiotics produced by Paenibacillus elgii B69. BMC Microbiol. 12, 45 (2012)
Niu, B., Rueckert, C., Blom, J., Wang, Q., Borriss, R.: The genome of the plant growth-promoting rhizobacterium Paenibacillus polymyxa M-1 contains nine sites dedicated to nonribosomal synthesis of lipopeptides and polyketides. J. Bacteriol. 193, 5862–5863 (2011)
Li, S., Yang, D., Qiu, M., Shao, J., Guo, R., Shen, B., Yin, X., Zhang, R., Zhang, N., Shen, Q.: Complete genome sequence of Paenibacillus polymyxa SQR-21, a plant growth-promoting rhizobacterium with antifungal activity and rhizosphere colonization ability. Genome Announc. 2, 1–2 (2014)
Huang, E., Guo, Y., Yousef, A.E.: Draft genome sequence of Paenibacillus sp. OSY-SE, a bacterium producing the novel broad-spectrum lipopeptide antibiotic paenibacterin. J. Bacteriol. 194, 6306 (2012)
Ding, R., Li, Y., Qian, C., Wu, X.: Draft genome sequence of Paenibacillus elgii B69, a strain with broad antimicrobial activity. J. Bacteriol. 193, 4537 (2011)
Jeong, H., Park, S.Y., Chung, W.-H., Kim, S.H., Kim, N., Park, S.-H., Kim, J.F.: Draft genome sequence of the Paenibacillus polymyxa type strain (ATCC 842T), a plant growth-promoting bacterium. J. Bacteriol. 193, 5026–5027 (2011)
Jeong, H., Choi, S.-K., Park, S.-Y., Kim, S.H., Park, S.-H.: Draft genome sequence of Paenibacillus peoriae strain KCTC 3763. J. Bacteriol. 194, 1237–1238 (2012)
Kim, J.F., Jeong, H., Park, S.-Y., Kim, S.-B., Park, Y.K., Choi, S.-K., Ryu, C.-M., Hur, C.-G., Ghim, S.-Y., Oh, T.K., Kim, J.J., Park, C.S., Park, S.-H.: Genome sequence of the polymyxin-producing plant-probiotic rhizobacterium Paenibacillus polymyxa E681. J. Bacteriol. 192, 6103–6104 (2010)
Ma, M., Wang, C., Ding, Y., Li, L., Shen, D., Jiang, X., Guan, D., Cao, F., Chen, H., Feng, R., Wang, X., Ge, Y., Yao, L., Bing, X., Yang, X., Li, J., Du, B.: Complete genome sequence of Paenibacillus polymyxa SC2, a strain of plant growth-promoting rhizobacterium with broad-spectrum antimicrobial activity. J. Bacteriol. 193, 311–312 (2011)
Djukic, M., Brzuszkiewicz, E., Fünfhaus, A., Voss, J., Gollnow, K., Poppinga, L., Liesegang, H., Garcia-Gonzalez, E., Genersch, E., Daniel, D.: How to kill the honey bee larva: genomic potential and virulence mechanisms of Paenibacillus larvae. Plos One 9, e90914 (2014)
Yao, L.J., Wang, Q., Fu, X.C., Mei, R.H.: Isolation and identification of endophytic bacteria antagonistic to wheat sharp eye disease. Chin J Biol Control 24, 53–57 (2008)
Kimura, Y., Murai, E., Fujisawa, M., Tatsuki, T., Nobue, F.: Polymyxin P, new antibiotics of polymyxin group. J Antibiot (Tokyo) 22, 449–450 (1969)
Li, J., Jensen, S.E.: Nonribosomal biosynthesis of fusaricidins by Paenibacillus polymyxa PKB1 involves direct activation of a D-amino acid. Chem. Biol. 15, 118–127 (2008)
Choi, S.K., Park, S.Y., Kim, R., Kim, S.B., Lee, C.H., Kim, J.F., Park, S.H.: Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J. Bacteriol. 191, 3350–3359 (2009)
Eliasson Lantz, A., Jorgensen, P., Poulsen, E., Lindemann, C., Olsson, L.: Determination of cell mass and polymyxin using multi-wavelength fluorescence. J. Biotechnol. 121, 544–554 (2006)
Suckau, D., Resemann, A., Schuerenberg, M., Hufnagel, P., Franzen, J., Holle, A.: A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 376, 952–965 (2003)
Landy, M., Warren, G.H., Rosemann, S.B., Colio, L.G.: Bacillomycin, an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 67, 539–541 (1948)
Vater, J., Gao, X., Hitzeroth, G., Wilde, C., Franke, P.: “Whole cell”-matrix-assisted laser desorption ionization-time of flight-mass spectrometry, an emerging technique for efficient screening of biocombinatorial libraries of natural compounds-present state of research. Comb. Chem. High Throughput Screen. 6, 557–567 (2003)
Koumoutsi, A., Chen, X.-H., Henne, A., Liesegang, H., Hitzeroth, G., Franke, P., Vater, J., Borriss, R.: Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 186, 1084–1096 (2004)
Mootz, H.D., Schwarzer, D., Marahiel, M.A.: Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3, 490–504 (2002)
Kohli, R.M., Trauger, J.W., Schwarzer, D., Marahiel, M.A., Walsh, C.T.: Generality of peptide cyclization catalyzed by isolated thioesterase domains of nonribosomal peptide synthetases. Biochemistry 40, 7099–7108 (2001)
Haese, A., Schubert, M., Hermann, M., Zocher, R.: Molecular characterization of the enniatin synthetase gene encoding a multi-functional enzyme catalyzing N-methyl-depsipeptide formation in Fusarium scirpi. Mol. Microbiol. 7, 905–914 (1993)
Gehring, A.M., Mori, I., Walsh, C.T.: Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37, 2648–2659 (1998)
Juguet, M., Lautru, S., Francou, F.-X., Nezbedova, S., Leblond, P., Gondry, M., Pernodet, J.-L.: An iterative nonribosomal peptide synthetase assembles the pyrrole-amide antibiotic congocidine in Streptomyces ambofaciens. Chem. Biol. 16, 421–431 (2009)
Hoyer, K., Mahlert, C., Marahiel, M.A.: The iterative gramicidin S thioesterase catalyses peptide ligation and cyclization. Chem. Biol. 14, 13–22 (2007)
Acknowledgments
The authors thank Dr. P. Lasch from the Robert Koch-Institut Berlin for making available for us the Bruker Autoflex Speed instrument to perform the MALDI-TOF MS measurements.
K.D. and R.B. gratefully acknowledge the support given by the European’s Seventh Framework Programme (FP/2007-2013) under Grant Agreement no 312117.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
(DOCX 403 kb)
Rights and permissions
About this article
Cite this article
Vater, J., Niu, B., Dietel, K. et al. Characterization of Novel Fusaricidins Produced by Paenibacillus polymyxa-M1 Using MALDI-TOF Mass Spectrometry. J. Am. Soc. Mass Spectrom. 26, 1548–1558 (2015). https://doi.org/10.1007/s13361-015-1130-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1130-1