Abstract
The rise of antimicrobial resistance necessitates the discovery and/or production of novel antibiotics. Isolated strains of Paenibacillus alvei were previously shown to exhibit antimicrobial activity against a number of pathogens, such as E. coli, Salmonella, and methicillin-resistant Staphylococcus aureus (MRSA). The responsible antimicrobial compounds were isolated from these Paenibacillus strains and a combination of low and high resolution mass spectrometry with multiple-stage tandem mass spectrometry was used for identification. A group of closely related cyclic lipopeptides was identified, differing primarily by fatty acid chain length and one of two possible amino acid substitutions. Variation in the fatty acid length resulted in mass differences of 14 Da and yielded groups of related MSn spectra. Despite the inherent complexity of MS/MS spectra of cyclic compounds, straightforward analysis of these spectra was accomplished by determining differences in complementary product ion series between compounds that differ in molecular weight by 14 Da. The primary peptide sequence assignment was confirmed through genome mining; the combination of these analytical tools represents a workflow that can be used for the identification of complex antibiotics. The compounds also share amino acid sequence similarity to a previously identified broad-spectrum antibiotic isolated from Paenibacillus. The presence of such a wide distribution of related compounds produced by the same organism represents a novel class of broad-spectrum antibiotic compounds.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the rate of antibiotic resistance increases, there is a growing need for the discovery of novel classes of antibiotics [1, 2]. Of particular interest is biocontrol of crop production for food safety applications, where pathogens, including fungal and bacterial growth, can be minimized or prevented by using a bacterial strain or compounds produced by bacteria that are native to the environment and exhibit antibiotic properties [3, 4]. Previously, two naturally-occurring gram-positive bacteria identified as Paenibacillus alvei were isolated from plants native to the Virginia Eastern Shore tomato growing region [5, 6]. Both strains showed broad-spectrum antimicrobial activity against gram-negative and gram-positive foodborne pathogens. However, the products responsible for this activity are unknown.
Identification of bacterially-produced antibiotics can be challenging because of a range of molecular diversity. Paenibacillus strains produce several classes of antibiotic compounds. For example, P. polymyxa produces polymyxins, which are cyclic, contain the amino acid diaminobutyric acid, and have an acylated side chain; these are typically used as a last resort against gram-negative bacteria [7–9]. Another class of compounds is lantibiotics; these antibiotics are ribosomally synthesized and can have a number of post-translational modifications [10–12]. In another investigation, P. alvei co-produced two different peptide antibiotics that were antagonistic against gram-positive or -negative pathogens [13]. Fusaricidins are cyclic lipopeptides that inhibit fungal growth [14]. Recently, a broad-spectrum antibiotic was discovered from Paenibacillus that is also a cyclic lipopeptide [15]. Furthermore, non-ribosomal peptides can contain diverse amino acids [16], such as ornithine, which is present in antifungal compounds [17] and cyclic antibiotics [18]. To further complicate classification of these antibiotic compounds, more than one class of compounds can be co-produced in a single strain of Paenibacillus, and the presence of multiple closely related structures can be common within a given class of compounds. Such complexity makes isolation and identification of the bioactive compound or compounds challenging.
Tandem mass spectrometry offers the capability to detect and sequence a wide range of compounds as well as differentiate between closely related forms of the same compound within a given bacterial strain. However, interpretation of the fragmentation spectra of cyclic peptides is nontrivial because of the initial ring opening event occurring at a distribution of sites within a given ion population, resulting in different fragment ion series within the same spectrum. A number of studies have aimed to reduce or handle the complexity of tandem mass spectra of cyclic peptides. Recently, this has been accomplished through database searching [19, 20], mass spectral interpretation tools [21, 22], or the development of automated data analysis approaches [23]. Searching against genome databases is promising but cannot be implemented when genome data is unavailable. The development of data analysis approaches to analyze complicated MS data is beneficial and will hopefully lead to the discovery and identification of novel compounds. Other approaches have included using specific MS techniques to aid in the identification of primary sequence assignments either by using a combination of different types of fragmentation data [24] or by sequential fragmentation to reduce the spectral complexity produced from multiple ring openings [25].
Here, we present the detection and identification of broad-spectrum antimicrobial compounds produced by an environmentally isolated Paenibacillus alvei strain from plants native in the U.S. to the Virginia Eastern Shore tomato growing region [5, 6]. Compounds were first fractionated by off-line ultra-performance liquid chromatography (UPLC), and the bioactivity of each fraction was assessed against gram-negative and -positive bacteria. Multiple analytical tools were used to identify these purified antibiotic compounds. A combination of matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) and high resolution MS was used to evaluate the complexity of molecular species in the bioactive fractions. A distribution of closely related cyclic lipopeptide compounds was identified with tandem MS, differing primarily by fatty acid chain length and one of two possible amino acid substitutions; the primary peptide sequence was confirmed through gene mining. With the developed workflow, a complex mixture of multiple related molecular species was identified from a single strain of Paenibacillus, which represents a novel class of antibiotic compounds.
Experimental
Bacterial Cell Culture and Sample Preparation
Paenibacillus alvei strains A6-6i and TS-15 were previously isolated from plant and soil native in the U.S. to the Virginia Eastern Shore tomato growing region [6]. The data presented here is for the strain TS-15, although the data collected from both strains are similar. Strain TS-15 was propagated on tryptic soy agar (TSA) at 35 ± 2°C for 18 ± 2 h. Stock cultures were grown overnight at 35 ± 2°C on TSA and then were resuspended in brain heart infusion (BHI) broth with 25% glycerol and stored at –80°C.
The sample preparation of the bacteria for fraction collection was a modified method based on Guo et al [15]. Bacterial cells were removed from Petri dishes using cell scrapers and deposited directly into Eppendorf Protein LoBind tubes, and acetonitrile was added in a volume of 100 μL per dish of isolated cells. Samples were shaken for 30 min and centrifuged at 7710 g for 15 min; the supernatant was removed and evaporated. These were reconstituted in water and filtered with a 0.22 μm Nylon filter. For genome sequencing, genomic DNA was isolated from an overnight culture of strain TS-15 using a Qiagen DNeasy blood and tissue kit (Qiagen Inc., Valencia, CA, USA).
Fraction Collection
Fraction collection by UPLC was achieved using a Shimadzu Nexera with a Kinetex C18 column (1.7 μ, 100 Å, 150 × 2.10 mm; Phenomenex, Torrance, CA, USA). The separation was performed with a column temperature of 60°C and a flow rate of 400 μL/min using water with 0.1% formic acid (v/v) and acetonitrile with 0.1% formic acid (v/v) with the following gradient: 5 min hold at 95% water, 50 min linear gradient from 95% to 5% water, 5 min equilibration at 95% water. The injection volume was 40 μL and fractions were collected in 1-min intervals across the 60 min LC gradient. Injections and fractionation were repeated and respective fractions were combined and concentrated to 30 μL for biological activity experiments and MS analysis.
Bioactivity Assay
Ten microliter volumes of each concentrated 1-min fraction collected from Paenibacillus alvei strain TS-15 were spotted directly on culture plates containing a lawn of 106 cells of Escherichia coli O157:H7 strain EDL933 or methicillin-resistant Staphylococcus aureus strain #12, respectively [26]. After incubation at 35 ± 2°C for 24 h, antimicrobial activity was observed as a clear zone of inhibition (ZI). Isolated fractions that exhibited a ZI were further characterized by mass spectrometry.
MALDI-TOF/MS Analysis
Isolated fractions with an observed ZI were diluted 1:30 in water and 1 μL was placed onto a MALDI target with 1 μL of prepared α-cyano-4-hydroxycinnamic acid matrix (20 mg/mL in 70% acetonitrile with 0.1% formic acid). The MALDI instrument used to analyze the samples was a 4800 MALDI TOF/TOF Analyzer (Applied Biosystems/MDS Sciex, Framingham, MA, USA). The laser power was optimized for each analysis to use the minimum power required for sufficient ionization. Tandem mass spectrometry (MS/MS) was also performed on ions of interest using post-source decay (PSD).
High-Resolution LC-MS Analysis
Concentrated fractions with an observed ZI were analyzed using the same UPLC conditions as fraction collection coupled to a high-resolution mass spectrometer (Q-Exactive, Thermo Scientific, San Jose, CA, USA). The Q-Exactive settings were: 140,000 resolution, 1e6 AGC target, maximum ion accumulation time of 60 ms, and a mass range of 300–4000 Da was monitored; the settings for the heated electrospray ionization probe (HESI-II) were: 4 kV spray voltage, 50 psi sheath gas, 15 (arbitrary units) auxiliary gas, 380°C capillary temperature, and 300°C heater temperature. The Elemental Composition and Isotope Simulation tools in Xcalibur were used for molecular formulae generation and calculations of theoretical masses.
MSn Analysis
Multiple-stage tandem mass spectrometry experiments (MSn) were performed using an Orbitrap Elite (Thermo Scientific, San Jose, CA, USA). For infusion experiments, fractions were diluted 1:30 in 70% methanol with 0.1% formic acid. Samples were infused via the Triversa Nanomate (Advion, Ithaca, NY, USA) chip-based nanospray source with 1.5 kV voltage and 0.3 psi gas pressure. A mass range of 225 to 2000 was monitored in full MS mode with 120,000 resolving power with a target ion number of 5e5 and a maximum ion accumulation time of 100 ms. MSn experiments presented in this work were collision-induced dissociation (CID) performed in the linear ion trap and detected at 120,000 resolving power in the Orbitrap with a target ion number of 1e5 and a maximum ion accumulation time of 150 ms. Precursor fragment ions were selected with an isolation width of 2–3 Da. For additional separation, MSn experiments of individual fractions were repeated by nano-LC-MSn on an Orbitrap Elite equipped with a PicoView nanospray source (New Objective, Woburn, MA, USA) and coupled to a Waters nanoAcquity UPLC system (Waters, Milford, MA, USA). Fractions were loaded onto an Acquity Symmetry C18 100 Å, 5 μm trapping column (180 μm × 20 mm) and separated with a C18 BEH 130 Å, 1.7 μm column (100 μm × 100 mm) (Waters) at 35°C. Samples were trapped for 3 min at 5 μL/min and eluted at 300 nL/min over a 60 min linear gradient from 5% to 95% acetonitrile with 0.1% formic acid.
Whole Genome Sequencing
Whole genome sequencing was performed to confirm MS assignments using the Pacific Biosciences Real-Time (PacBio RS) sequencing platform (Pacific Biosciences, Menlo Park, CA, USA). A single 10-kb library was sequenced using C2 chemistry on eight single-molecule real-time (SMRT) cells with a 90-min collection protocol on the PacBio RS. The 10-kb continuous-long-read (CLR) data were de novo assembled using the PacBio hierarchical genome assembly process (HGAP)/Quiver software package, followed by Minimus 2, and were polished with Quiver [27]. The assembled contigs were annotated with the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html) [5].
Gene Cluster Analysis
The presence of nonribosomal peptide synthetases (NRPS) in the P. alvei strain TS-15 genome were analyzed by NRPSpredictor2, a webserver for predicting NRPS A-domains and their corresponding substrate residues [28, 29]. In addition, epimerization (E) domains were identified to determine the presence of d-amino acids within the synthesized compound (PKS/NRPS analysis webserver at http://nrps.igs.umaryland.edu/nrps/) [30, 31].
Results and Discussion
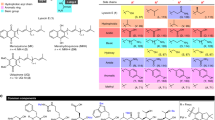
The P. alvei bacterial strains A6-6i and TS-15 were previously isolated from the environment and showed antibiotic properties against a number of pathogens such as Salmonella, E. coli, and methicillin-resistant Staphylococcus aureus (MRSA) [6]; however, the responsible antibiotic compounds were unknown. Assessment of the class and mechanism of antibiotic(s) produced by these Paenibacillus strains required identification of the bioactive compound(s). In this work, we used a combination of fraction collection, tandem mass spectrometry, and genome mining to characterize a series of related compounds that comprise a novel class of cyclic antibiotic peptides with fatty acids attached to the N-terminus of the amino acid ornithine. These peptides primarily differed by the length of the fatty acid and amino acid substitutions at two possible sites. The combination of these analytical tools enabled the identification of compounds illustrated in Scheme 1.
General structure of compounds discovered and identified in the current study. “FA” corresponds to a fatty acid attached to the N-terminus of ornithine (or Lys); molecular formulae for the different fatty acid chains are listed. The asterisks indicate molecular differences between the compounds discovered in this work and paenibacterin (Figure S1)
Identification of the Primary Peptide Sequence
To isolate bioactive compounds, bacterial cells were pooled, extracted, and separated by fraction collection. Seven of the 1-min fractions collected during a 60-min gradient exhibited bioactivity against both gram-negative E. coli and gram-positive MRSA (Figure 1a). Subsequent MALDI-TOF MS analysis revealed a number of compounds that were present in each of the seven bioactive fractions, as shown in Figure 1b. The MALDI spectra provided a view of the full complement of compounds from each bioactive fraction within a single spectrum where clusters of these compounds differed by 14 Da, indicated by red arrows. The compound with a molecular weight of 1623 will be referred to as the primary compound (ion indicated with an asterisk in Figure 1b), although multiple variants of similar abundances are present (Figure 1b).
Strain TS-15 1-minute bioactive fractions against gram-positive and -negative bacteria and resulting MALDI-TOF MS spectra from the bioactive fractions. (a) A 10 μL volume from the 1-min fractions of P. alvei strain TS-15 was spotted on a lawn of 106 cells of Escherichia coli O157:H7 strain EDL933 (left) and methicillin-resistant Staphylococcus aureus strain #12 (right). After incubation at 35 ± 2°C for 24 h, the antimicrobial activity exhibited by the 1-min fractions was observed as a clear zone of inhibition. (b) MALDI-TOF MS results of fractions with observed bioactivity. Red arrows indicate a mass difference of 14 Da, which correspond to a molecular difference of CH2; an asterisk indicates MW 1623, which is designated as the primary sequence; blue arrows show two examples of a mass difference of 2 and 16 Da, which correspond to the other molecular variants
The MALDI-TOF MS/MS analyses of these molecular species revealed similar fragmentation patterns, which confirmed that the compounds that differ by 14 Da were related. As illustrated in Figure 2a, comparison of MS/MS spectra of the primary compound (MW 1623) and the compound of MW 1637, which differ in molecular weight by 14 Da, revealed a series of product ions that shared the same mass and a second product ion series that differed by 14 Da, suggesting that the mass discrepancy between compounds was localized to one region of the molecule. This enabled identification of complementary product ion pairs, with one direction corresponding to the product ion series retaining the region of the molecule that contained the 14 Da mass-shift and the other direction corresponding to product ions that were identical between the two peptides (Figure 2b). Manual de novo sequencing resulted in a partial amino acid sequence, yielding a putative sequence assignment.
MALDI-TOF MS/MS characterization of antibiotic compounds. ( a ) MALDI-TOF MS/MS comparison of two compounds that differ by 14 Da in molecular weight. Mass-to-charge ratios in red with an asterisk indicate an observed mass difference of 14 Da in the comparison between the two spectra. (b) Partial sequence information from MALDI-TOF MS/MS spectrum of MW 1623 elucidated by manual de novo sequencing
Analysis of MALDI-TOF MS/MS spectra only revealed partial sequence tags. To improve sequence coverage, individual fractions were infused and analyzed with the Orbitrap Elite, allowing for the collection of MSn data. Again, pairs of MSn spectra were analyzed and complementary ion pairs were identified based on the presence of 14 Da mass differences. The combined MSn data allowed a complete amino acid sequence to be determined, which is illustrated in Scheme 1. This amino acid sequence was similar to a previously identified cyclic compound isolated from a different Paenibacillus strain, which also showed broad-spectrum activity against MRSA and E. coli [15, 32]; the structure of that compound, designated as paenibacterin, is shown in Supplementary Figure S1. The location of the molecular differences between this compound and the compounds described herein are highlighted by asterisks in Scheme 1.
While linearizing the molecule through alkaline hydrolysis simplified de novo sequencing of paenibacterin [15], Guo et al. benefited from a strain of Paenibacillus that produced a single dominant cyclic antibiotic. However, the TS-15 strain presented here produces a wide distribution of related cyclic compounds. Efforts to linearize these compounds further complicated analysis by the additional presence of ions with 18 and 22 Da mass differences, attributable to the linearization of the molecule and presence of sodium adducts, respectively. This necessitated the development of a workflow for the identification of cyclic peptides without linearization.
By determining which series of product ions did or did not contain the molecular component that results in the 14 Da difference, de novo sequencing by MSn analysis was more straightforward. This was particularly critical because the compounds were cyclic and resulting MSn spectra can be difficult to interpret because of multiple ring opening events occurring at a distribution of sites. An example is shown in the MS3 spectrum in Figure 3 where the ring opens at different amino acid positions, yielding a number of different sequence series within the same spectrum; thus, de novo sequencing of the primary sequence of cyclic peptides can be challenging. However, by using the described approach, the assignment of product ions and the identification of the primary sequence and sequence variants were accomplished without linearizing the molecule. The cumulative ion assignments for the primary amino acid sequence can be found in in Figure 4b.
Representative example of different product ion series in an MSn spectrum (MS3 6422+). The cyclic peptide structure can open at different amino acid positions, which can complicate interpretation. The corresponding sequences are color-coded to the amino acids in the inset schematic of the cyclic peptide structure
Product ion assignments as a result of combined interpretation of multiple MSn analyses. Representative examples of MS2 spectra are shown in (a) and (b) for two of the most abundant compounds, containing a Phe (MW 1607) or Tyr (MW 1623) at position 6, respectively. These product ion assignments led to the chemical structure shown in Scheme 1
Identification of Sequence Variants
Subsequent analysis with UPLC coupled to high resolution MS provided accurate mass data, which confirmed that there were actually three predominant compound series, with each series containing groups of compounds that differ by 14.02 Da. The most pronounced differences between the three observed series were either a decrease of 15.99 Da or an increase of 1.98 Da in mass compared with the primary compound series; examples are designated with blue arrows in Figure 1b. A comprehensive list of these compounds and their accurate mass molecular weights can be found in Table 1. These are designated as F, Y, and Y, –CH2+O in the table and throughout the figures; F and Y correspond to phenylalanine or tyrosine at position 6 in Scheme 1 and –CH2+O corresponds to a molecular difference in the fatty acid chain.
Two compound series in the bioactive fractions differed from each other by 15.99 Da and exhibited MS2 spectra with all the compounds in one series yielding a product ion at m/z 6572+ whereas the other series generates a product at m/z 6492+ (Figure 5a). Figure 5b illustrates the MSn spectra of three precursor ions that differed by 14 Da, all of which generated an MS2 product ion at m/z 6572+. Because these were conserved product ions within a precursor series that included compounds that differed in molecular weight by 14 Da, we were able to conclude that the region of the compound that yields product ions 6572+ or 6492+ does not contain the fatty acid. The MS3 spectra of 6572+ were consistent with one another, confirming that the region of the peptide that generated this sequence was conserved between these compounds (Figure 5b). The MS3 spectra from the ion series that generated an MS2 product ion at 6492+ were similar to the 6572+ MS3 spectra, except for a series of product ions that differed by 16 Da (red masses with asterisks in Figure 5a). This enabled the distinction between tyrosine and phenylalanine at position 6 (Scheme 1), designated as Y and F in the tables and figures, respectively; these amino acids differ in molecular weight by 16 Da. Assignments for the MS/MS spectrum for a peptide containing Phe are illustrated in Figure 4a, where a direct comparison can be observed between the peptides containing Phe and Tyr at position 6.
MS2 and resulting MS3 spectra. (a) Three series of compounds within the class of antibiotics differ by an amino acid or a difference in their attached fatty acid. Compounds that contain a Tyr at position 6 have m/z 6572+ as a consistent product ion in their resulting MS/MS spectra, whereas Phe at position 6 results in m/z 6492+. Red m/z values with an asterisk indicate a mass difference of 16 Da between the MS3 spectra, which is the mass difference between Phe and Tyr. The mass difference between MW 1623 and MW 1625 corresponds to one less CH2 group and an additional oxygen in the attached fatty acid (–CH2+O). (b) Three representative MS3 spectra demonstrate sequence similarity between compounds of molecular weights that differ by 14 Da and their corresponding complementary ion pair MS3 spectra. MS2 spectra are outlined in blue and MS3 spectra are outlined in orange, with product ions selected for MS3 highlighted in orange
Identification of the Attached Fatty Acid
The MS2 spectra of the series of compounds that contain a Tyr at position 6 were dominated by product ion 6572+ and its complementary ion pair (Figure 5a). While 6572+ was conserved within the Tyr compound series, its complementary ion contained the same 14 Da mass shift as its precursor (i.e., m/z 311, 325, and 339 in Figure 5b). The same trend was also present for the Phe ion series. A list of the multiple complementary ions for 6572+ and 6492+ are listed in Table 2A. When these complementary ions were dissociated (examples shown in Figure 5b), a loss of ornithine was observed. Subtracting the cyclized peptide sequence mass from the mass of the entire compound yields the mass attributed to an attached fatty acid; molecular formula generation of these masses yield the molecular formulae of the different fatty acid variants (Table 2B and Scheme 1). These fatty acids are similar to what was observed in Guo et al., although the lengths of the carbon chains differ and both the TS-15 and A6-6i strains presented in the current work exhibit a greater variability in chain length and composition [15].
As mentioned previously, three major series of compounds were determined: two series containing a tyrosine at position 6 and one containing phenylalanine. The two compound series containing Tyr differed by a molecular weight of 1.979 Da. Similar to the MS spectral analysis methodology shown in Figure 2, these compound series also had similar MS2 fragmentation patterns, where some product ion masses are conserved and others differ by 1.979 Da (Supplementary Figure S2). This mass difference corresponded to one less CH2 and an additional oxygen (labeled as –CH2+O) in the attached fatty acid compared to the tyrosine molecular series.
Multiple Compounds with the Same Molecular Weight
It was also observed that more than one of the complementary ion pairs were occasionally present within the same MS2 spectrum in the infusion experiment data. This corresponded to two compounds of the same precursor mass being fragmented within the same isolation window. There were multiple examples in the UPLC and NanoLC/MS data that showed several eluting peaks for entities with the same mass (example shown in Figure 6). The MS2 spectra of the ions in each of these chromatographic peaks showed small differences in the fragmentation pattern and, thus, the primary sequences of these respective peptides. Nearly identical amino acid sequences were confirmed for compounds with an identical molecular weight (1579): a compound with Lys at position 7 and a compound with ornithine at position 7 with an additional CH2 in the attached fatty acid (Lys and ornithine differ by a CH2 in their side chains). Specifically, the product ion at m/z 6492+ corresponds to Lys at position 7 and m/z 6422+ corresponds to ornithine at position 7. The two chromatographic peaks with Lys at position 7 may indicate a structural difference in the attached fatty acid resulting in the observed difference in retention time (Figure 6).
Different MS/MS spectra (highlighted in blue) for three compounds of the same molecular weight indicate Lys or Orn at Position 7 in Scheme 1 and potential diversity in the structure of the attached fatty acid attributable to distinct chromatographic peaks shown in the extracted ion chromatogram (EIC)
A similar substitution was also found at position 1. Some of the subsequent MS3 analyses of the fatty acid-containing fragment ions (m/z 3251+ and 3391+ in Figure 5b) indicated that Lys can also be present at position 1 rather than ornithine (m/z 1291+ in MS3 spectra in Figure 5b). A Lys at position 1 and a decrease of CH2 in the attached fatty acid resulted in identical molecular weights for each compound.
Genome Mining to Confirm Compound Identification
The sequence shown in Scheme 1 was confirmed by genome mining for nonribosomal peptide synthesis. Many pharmacologically important peptides in bacteria are synthesized by nonribosomal peptide synthetases (NRPS). NRPS machinery is composed of modular multi-domain enzymes that act as an assembly line to incorporate each amino acid monomer by one module [31]. A typical module in an NRPS contains an adenylation (A) domain, which possesses a conserved binding pocket for the recruitment of amino acid monomers that are to be incorporated into the final peptide product [31]. A single contig of 6784766 bp (G + C content, 46.69%) representing the complete chromosome for P. alvei strain TS-15 was generated. A total of six NRPSs were identified from the annotated genome, with one corresponding to the compounds characterized in this work. The responsible gene cluster, defined here as Pa-NRPS1, was present on a 49-kb DNA region and contained 13 modules (Table 3) corresponding to the 13 amino acids in the microbially-synthesized compounds.
The amino acid sequence generated by NRPSs is primarily determined by the specificity of the A domains and was predicted using NRPSpredictor2 [28, 29]. The predicted peptide agreed with the dominant amino acid sequence determined by MS/MS and MSn (Table 3 and Scheme 1). It is worth noting that genome mining did not predict the presence of abundant sequence variants. As Lys and ornithine differ by CH2, their binding affinities are likely to be similar, which may be contributing to the observed molecular diversity [33]. Likewise, Tyr and Phe differ by a hydroxyl group. Furthermore, NRPSpredictor 2 has lower single amino acid substrate prediction scores for Phe and Lys, which may indicate why these were not additionally predicted in the primary sequence [29]. NRPSpredictor2 also predicted that the epimerization (E) domain was in modules 1, 4, 7, and 8, which indicated that these resulting amino acid substrates may be in the d-form. These are specified in Scheme 1. As expected, NRPS analysis does not offer information about the presence or length of the alkyl chain.
The NRPS analysis confirmed both the presence and order of the amino acids of the peptide assignments made through the combination of MALDI-MS, high-resolution mass spectrometry, and MSn analysis. NRPS can be used as a screening technique to identify potential nonribosomal peptides, which may act as antibiotics; however, it does not yield information regarding any molecular variants that may be produced. Moving forward, NRPS analysis and mass spectrometry can be used to combine rapid prediction of candidate peptides with the molecular specificity of mass spectrometry to enable identification of cyclic antibiotics and their sequence and fatty acid variants despite the presence of molecular diversity and complicated spectra.
Implications for Bioactivity
It is striking that the peptides found in this study are amphiphilic with distinct hydrophilic and hydrophobic regions: hydrophobicity on one side, the other side being predominantly polar and charged amino acids, and a hydrophobic fatty acid chain (Scheme 1). Major differences between paenibacterin [15] and the compounds discovered in this work are the length of the attached fatty acid, the different combinations of lysine and ornithine at positions 1 and 7, and the amino acids at position 6, 11, and 12. This is particularly interesting because the amino acids at position 6 and 12 have different properties (e.g., hydrophobic, hydrophilic, or positively charged). Furthermore, the presence of d-amino acids influences the structure and properties of the peptide and will also make the compound more resistant to enzymatic degradation and thus inherently more stable [16].
Aspects of paenibacterin’s mode of action have been previously studied [32]. The results suggest that the compound has a high affinity to the negatively-charged outer membrane of gram-negative bacteria. This is likely due to the presence of positively charged amino acids in the molecule, which is similar to the mode of action of polymyxin [9]. Three positively charged amino acids were found in the molecules discovered here compared with four in paenibacterin (Scheme 1 and Supplementary Figure S1), which may result in varying degrees of effectiveness. However, the Lys to Pro substitution at position 12 also increases the hydrophobicity of that portion of the molecule, which may result in a better affinity to the hydrophobic core of cellular membranes, a characteristic that may aid in its disruption. Similarly, the presence of Tyr at position 6 contributes to a more polar region of the molecule. The mode of action may also be due to the amphiphilic nature of the compound, acting as a surfactant to disrupt cell membranes. It is notable that polymyxin also has distinct hydrophilic and hydrophobic domains [34, 35].
It was also determined that paenibacterin resulted in the permeabilization of both gram-positive and -negative cell membranes, which was probably disrupted by the attached fatty acid [32]. The chain length will likely affect observed antimicrobial activity, although it is uncertain if it will be less or more effective with a longer/shorter chain. For polymyxin, it is hypothesized that the fatty acyl chain disrupts the cellular membrane [34]. Studies on the fatty acid chain of polymyxin indicate that antimicrobial activity correlates with the length and bulkiness of this moiety [9]. However, reports are varied and subsequent experiments to design fatty acid analogues for the compounds in this study will yield insight into how this affects antimicrobial activity. It is also interesting that a single strain of bacteria can produce such a large number of molecular variants. This may enable the strain to exert a more concerted antimicrobial effect and may have resulted from extensive selection pressure in the community from which it was isolated.
Conclusion
A combination of different analytical techniques enabled the detection and identification of a class of cyclic antibiotic compounds. By analyzing similar MS/MS spectra of compounds that differ by 14 Da, sequence tags can be determined despite the difficulties inherent to cyclic MS/MS interpretation. The sequences elucidated through MS/MS and MSn data are supported by gene mining data, confirming the sequences of the bacterially produced chemical products. This analysis strategy could be readily applied for the identification of novel antibiotics and antibiotic mixtures. Molecular diversity is apparent in this newly identified compound class; the identified compounds appear to be cyclic with an attached fatty acid of varying lengths (indicated by the molecular weights that differ by 14 Da) and contain different amino acids compared with a previously published similar compound [15]. The molecular weights of the theoretical formulas from the deduced sequence and fatty acid variants were calculated (Scheme 1 and Table 1) and correspond to observed values (Table 2). Further characterization of these compounds will yield a better understanding of the mechanism responsible for antimicrobial activity. Whatever the mechanism, these compounds potentially represent important future weapons in the control of multi-drug-resistant bacterial pathogens, and understanding their chemical and biological properties is essential to this end.
References
Newman, D.J., Cragg, G.M.: Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335 (2012)
Clardy, J., Fischbach, M.A., Walsh, C.T.: New antibiotics from bacterial natural products. Nat. Biotechnol. 24, 1541–1550 (2006)
Gálvez, A., Abriouel, H., Benomar, N., Lucas, R.: Microbial antagonists to food-borne pathogens and biocontrol. Curr. Opin. Biotechnol. 21, 142–148 (2010)
Ongena, M., Jacques, P.: Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125 (2008)
Luo, Y., Wang, C., Allard, S., Strain, E., Allard, M.W., Brown, E.W., Zheng, J.: Draft genome sequences of Paenibacillus alvei A6-6i and TS-15. Genome Announc. 1(5), e00673–13 (2013)
Allard, S., Enurah, A., Strain, E., Millner, P., Rideout, S.L., Brown, E.W., Zheng, J.: In situ evaluation of Paenibacillus alvei in reducing carriage of Salmonella enterica serovar Newport on whole tomato plants. Appl. Environ. Microbiol. 80, 3842–3849 (2014)
Storm, D.R., Rosenthal, K.S., Swanson, P.E.: Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46, 723–763 (1977)
Vaara, M.: Polymyxins and their novel derivatives. Curr. Opin. Microbiol. 13, 574–581 (2010)
Velkov, T., Thompson, P.E., Nation, R.L., Li, J.: Structure–activity relationships of polymyxin antibiotics. J. Med. Chem. 53, 1898–1916 (2009)
Chatterjee, C., Paul, M., Xie, L.L., van der Donk, W.A.: Biosynthesis and mode of action of lantibiotics. Chem. Rev. 105, 633–683 (2005)
He, Z.G., Kisla, D., Zhang, L.W., Yuan, C.H., Green-Church, K.B., Yousef, A.E.: Isolation and identification of a Paenibacillus polymyxa strain that co-produces a novel lantibiotic and polymyxin. Appl. Environ. Microbiol. 73, 168–178 (2007)
He, Z.G., Yuan, C.H., Zhang, L.W., Yousef, A.E.: N-terminal acetylation in paenibacillin, a novel lantibiotic. FEBS Lett. 582, 2787–2792 (2008)
Anandaraj, B., Vellaichamy, A., Kachman, M., Selvamanikandan, A., Pegu, S., Murugan, V.: Co-production of two new peptide antibiotics by a bacterial isolate Paenibacillus alvei NP75. Biochem. Biophys. Res. Commun. 379, 179–185 (2009)
Li, J., Jensen, S.E.: Nonribosomal biosynthesis of fusaricidins by Paenibacillus polymyxa PKB1 involves direct activation of a d-amino acid. Chem. Biol. 15, 118–127 (2008)
Guo, Y., Huang, E., Yuan, C., Zhang, L., Yousef, A.E.: Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl. Environ. Microbiol. 78, 3156–3165 (2012)
Caboche, S., Leclère, V., Pupin, M., Kucherov, G., Jacques, P.: Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. J. Bacteriol. 192, 5143–5150 (2010)
Pathak, K., Keharia, H., Gupta, K., Thakur, S., Balaram, P.: Lipopeptides from the Banyan endophyte, Bacillus subtilis K1: mass spectrometric characterization of a library of fengycins. J. Am. Soc. Mass Spectrom. 23, 1716–1728 (2012)
Bionda, N., Pitteloud, J.P., Cudic, P.: Cyclic lipodepsipeptides: a new class of antibacterial agents in the battle against resistant bacteria. Future Med. Chem. 5, 1311–1330 (2013)
Mohimani, H., Liu, W.-T., Mylne, J.S., Poth, A.G., Colgrave, M.L., Tran, D., Selsted, M.E., Dorrestein, P.C., Pevzner, P.A.: Cycloquest: identification of cyclopeptides via database search of their mass spectra against genome databases. J. Proteome Res. 10, 4505–4512 (2011)
Mohimani, H., Liu, W.-T., Kersten, R.D., Moore, B.S., Dorrestein, P.C., Pevzner, P.A.: NRPquest: coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J. Nat. Prod. 77, 1902–1909 (2014)
Liu, W.-T., Ng, J., Meluzzi, D., Bandeira, N., Gutierrez, M., Simmons, T.L., Schultz, A.W., Linington, R.G., Moore, B.S., Gerwick, W.H., Pevzner, P.A., Dorrestein, P.C.: Interpretation of tandem mass spectra obtained from cyclic nonribosomal peptides. Anal. Chem. 81, 4200–4209 (2009)
Niedermeyer, T.H., Strohalm, M.: mMass as a software tool for the annotation of cyclic peptide tandem mass spectra. PLoS One 7, e44913 (2012)
Kavan, D., Kuzma, M., Lemr, K., Schug, K., Havlicek, V.: CYCLONE—a utility for de novo sequencing of microbial cyclic peptides. J. Am. Soc. Mass Spectrom. 24, 1177–1184 (2013)
Guan, F., Uboh, C., Soma, L., Rudy, J.: Sequence elucidation of an unknown cyclic peptide of high doping potential by ETD and CID tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 22, 718–730 (2011)
Ngoka, L.C.M., Gross, M.L.: Multistep tandem mass spectrometry for sequencing cyclic peptides in an ion-trap mass spectrometer. J. Am. Soc. Mass Spectrom. 10, 732–746 (1999)
Wanger, A.: Antibiotic susceptibility testing. In: Goldman, E., Green, L.H. (eds.) Practical Handbook of Microbiology, 2nd ed, pp. 149–158. CRC Press, Boca Raton (2008)
Chin, C.-S., Alexander, D.H., Marks, P., Klammer, A.A., Drake, J., Heiner, C., Clum, A., Copeland, A., Huddleston, J., Eichler, E.E., Turner, S.W., Korlach, J.: Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569 (2013)
Rausch, C., Weber, T., Kohlbacher, O., Wohlleben, W., Huson, D.H.: Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33, 5799–5808 (2005)
Röttig, M., Medema, M.H., Blin, K., Weber, T., Rausch, C., Kohlbacher, O.: NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367 (2011)
Bachmann, B.O., Ravel, J.: Methods for in silico prediction of microbial polyketide and nonribosomal peptide biosynthetic pathways from DNA sequence data. In: David A.H. (ed.) Methods in Enzymology, vol. 458, pp. 181–217. Academic Press, Waltham (2009)
Fischbach, M.A., Walsh, C.T.: Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106, 3468–3496 (2006)
Huang, E., Yousef, A.E.: The lipopeptide antibiotic paenibacterin binds to the bacterial outer membrane and exerts bactericidal activity through cytoplasmic membrane damage. Appl. Environ. Microbiol. 80, 2700–2704 (2014)
Challis, G.L., Ravel, J., Townsend, C.A.: Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224 (2000)
Pristovšek, P., Kidrič, J.: Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J. Med. Chem. 42, 4604–4613 (1999)
Meredith, J.J., Dufour, A., Bruch, M.D.: Comparison of the structure and dynamics of the antibiotic peptide polymyxin B and the inactive nonapeptide in aqueous trifluoroethanol by NMR spectroscopy. J. Phys. Chem. B 113, 544–551 (2008)
Acknowledgments
The authors thank Samantha Farris for assisting with the fraction collection and Christine Parker for the nanoLC/Orbitrap experiments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 147 kb)
Rights and permissions
About this article
Cite this article
Knolhoff, A.M., Zheng, J., McFarland, M.A. et al. Identification and Structural Characterization of Naturally-Occurring Broad-Spectrum Cyclic Antibiotics Isolated from Paenibacillus . J. Am. Soc. Mass Spectrom. 26, 1768–1779 (2015). https://doi.org/10.1007/s13361-015-1190-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1190-2