Abstract
In the past decades, many neuroimaging studies have aimed to improve the scientific understanding of human neurodegenerative diseases using MRI and PET. This article is designed to provide an overview of the major classes of brain imaging and how/why they are used in this line of research. It is intended as a primer for individuals who are relatively unfamiliar with the methods of neuroimaging research to gain a better understanding of the vocabulary and overall methodologies. It is not intended to describe or review any research findings for any disease or biology, but rather to broadly describe the imaging methodologies that are used in conducting this neurodegeneration research. We will also review challenges and strategies for analyzing neuroimaging data across multiple sites and studies, i.e., harmonization and standardization of imaging data for multi-site and meta-analyses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As magnetic resonance imaging (MRI) and positron emission tomography (PET) have become more widely available, the field has seen a proliferation of imaging studies designed to improve our scientific understanding of neurodegenerative diseases. Readers of findings from these studies who are not themselves in the brain imaging field may have a harder time understanding them because of the wide range of imaging and analyses types employed. In this review, we describe these most popular types of imaging used in neurodegeneration research and what types of analyses they are primarily used for. We do not attempt to review or summarize any research findings, but only to describe the major methodologies that drive them. We will also review and discuss one of the largest challenges in this field: heterogeneity across scanners, sites, and studies—both of the images themselves and how research groups analyze them.
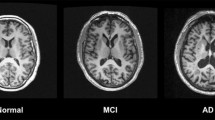
We begin by discussing each image type individually: describing its reason for inclusion in neuroimaging research and how it is analyzed. These are arranged by modality: first with MRI, then PET, then computed tomography (CT). We also provide a summary in Table 1 and some examples in Fig. 1. Lastly, we describe the challenge of data heterogeneity and the major strategies employed to combat it.
MRI
Broadly speaking, brain MRI scans are used in neurodegeneration studies to provide high-resolution imaging of brain structure and physiology. A participant is placed in a very strong magnetic field that forces the magnetic moments of protons in water and fat to become aligned, which allows these protons to absorb and emit pulses of radiofrequency (RF) waves. These pulses of energy are emitted by the scanner in various “pulse sequences” that are absorbed and emitted by body protons, which are then detected and localized by the scanner to image the density and locations of water and fat. By varying these pulse sequences, a wide variety of imaging contrasts may be obtained (which we describe below), making MRI highly versatile.
MRI is generally more expensive than CT, but less expensive than PET. All three modalities are generally considered minimal risk to participants when performed within approved protocols, but they may require mild sedation for participants with severe claustrophobia, and some participants may be excluded by physical size/weight limitations. Unlike PET and CT, MRI has additional contraindications for participants with metallic implants and foreign bodies such as pacemakers, aneurism clips, neurostimulators, shrapnel/bullets, insulin pumps, metallic intraocular foreign bodies, and some classes of tattoos or permanent make-up. However, unlike both PET and CT, MRI does not require the use of ionizing radiation that has been associated with an increased risk of cancer. In the following sections, we describe each of the major classes of MRI pulse sequences used in brain neuroimaging research and the types of analyses in which they are typically used.
T1-Weighted (Structural) MRI
T1-weighted (T1-w) scans are the most standard of MRI contrasts/sequences. These show tissue anatomy and density, where roughly white matter (WM) is white, gray matter (GM) is gray, and cerebrospinal fluid (CSF) is dark (i.e., CSF < GM < WM). In neuroimaging research, these scans have two major uses: (1) localization of brain regions and (2) estimation of tissue density. We show example images of these processes in Fig. 2.
Examples of processing of T1-w MRI. The top row depicts images in “native space” and the bottom row depicts images in “template space”. All images in the top row, after the original input image, were automatically produced by the segmentation and nonlinear registration (warping) using the pre-defined template/atlas information in the bottom row
Localization of Brain Regions
Arguably, the most fundamental research use of T1-w images is to automatically calculate a mapping between every voxel (3D volumetric pixel) location and its analogous location in a standard template brain. This process is called registration or normalization, and its mappings can be either linear (e.g., rigid or affine) or nonlinear (sometimes called warping), depending on the application and accuracy needed. These mappings allow automatic localization of any number of named regions of interest (ROIs) from an atlas (each drawn/defined on the template brain) by propagating them through the mapping onto the participant scan, enabling comparison of analogous brain regions across participants. Such mappings can be generated by a wide variety of software, most of which were designed specifically for brain images [1,2,3,4].
The most ubiquitous brain template is MNI152 aka ICBM152, which was generated from averaging 152 scans at the Montreal Neuroimaging Institute (MNI) as part of an International Consortium for Brain Mapping (ICBM) initiative [5,6,7]. Many other templates have been created (e.g., for specific ages/populations [8,9,10] or image types [11,12,13]) but the most ubiquitous are MNI152 or MNI305 (its predecessor). These “MNI coordinate spaces” are fundamental to each of the most popular software packages for MRI analyses, including FreeSurfer [14], SPM (Statistical Parametric Mapping [15]), FSL (FMRIB Software Library [16]), and AFNI (Analysis of Functional NeuroImages [17]).
Estimation of Tissue Density (Segmentation)
Localizing brain regions via T1-w images enables many analyses of all kinds of MRI and PET, but analyses of T1-w images themselves are principally focused on estimating tissue density in each voxel (i.e., its estimated probability of being gray matter, white matter, CSF, etc.). This process is typically called tissue segmentation and commonly performed by many of the same software toolkits, e.g., SPM, FreeSurfer, and FSL. These softwares typically use T1-w image intensity (brightness) in conjunction with tissue probability maps (TPMs: segmentation maps that are defined in template space and used as statistical priors that guide the segmentation). Mostly, these segmentations are used to measure gray matter volume: the total of all gray matter in a region or across the whole brain, in mm3 or cm3. The loss of a person’s brain tissue over time, due to healthy aging and/or disease, is called atrophy. Measuring atrophy is the second core use of T1-w images; this can also include measurements of white matter volumes/atrophy, or atrophy of both GM and WM via expansion of CSF volume [18]. Comparisons of volume measurements across individuals are inherently confounded by head size (which itself is heavily correlated with sex). To avoid these biases, analyses typically normalize by or regress out a measurement of total intracranial volume (TIV or ICV) [19, 20], which is also typically measured from T1-w images. Another alternative is to measure cortical thickness, which removes the surface area component of volume measurements to directly capture the width of the cortical ribbon in millimeters [21,22,23], avoiding volumes’ confound with TIV/head size. However, the resolution of most MRI (approximately 1 mm) limits the accuracy and precision of cortical thickness measurements relative to volume [24, 25], and it prevents measuring thickness in regions where the cortex is too small to resolve, e.g., hippocampus, amygdala, and cerebellum.
Comparative Analyses
By combining estimates of tissue density with localization, researchers can compare measurements across participants. Analyses typically occur either in “native space” or “template space”. In native space analyses, software automatically propagates atlas regions from the template space to the “native” space of the individual participant scan. From there, one can sum the volume in each brain region to produce numeric values (i.e., spreadsheets) and compare them across participants. In template space analyses, software transforms the “native space” MRI into the standard “template space” where every voxel location across all participants/scans should be anatomically analogous. After this transformation, comparisons are performed across participants in each voxel (e.g., voxel-based morphometry (VBM)) [26]. Typically, these transformations between native and template space are invertible, so for each scan, they can be computed once and then used in both directions for both types of analyses. Some software pipelines instead analyze transformations themselves (either participant-to-template or within-participant across time); these analyses are known as tensor-based morphometry (TBM), and they are considered the most powerful way to measure within-participant atrophy over time [27, 28]. Some analyses define their template space as a cortical surface, an “unfolded” model of the gray matter ribbon; these surface-based analyses co-register and compare scans in each vertex of the surface model space, rather than in each voxel of an image-based template space [29,30,31].
Analyses of Other Sequences and Modalities
In the previous paragraphs, we described how T1-w images are used both to measure a quantity of interest (tissue density) and to localize and compare these quantities in analogous brain regions. These same processes can be used to measure other quantities from other brain images (other MRI sequences, or PET, CT, etc.), by aligning (linear or affine registration) each image to the T1-w image and then using the existing T1w-to-template voxel mapping to perform the comparisons. In the research setting, analyses of most other image types described below are typically performed first by registering the image to the T1-w image and using this information to perform native-space or template-space analyses. Some image types have spatial distortions that can challenge achieving good alignment with T1, such as echo-planar imaging sequences (e.g., dMRI/fMRI); analyses of these modalities either use specialized distortion corrections to better match T1-w images or bypass T1-w and use modality-specific templates directly [32].
T2-Weighted and FLAIR MRI
While T1-weighted MRI is typically used to measure atrophy and localize tissue, T2-weighted MRI measures different tissue properties that are mostly associated with inflammation and edema. In T2-w, contrast is reversed from T1-w: in order of brightness, CSF > WM > GM. Inflammation and edema typically appear bright (brighter than or similar to CSF), along with white matter hyperintensities (WMH) that are associated with small vessel disease and demyelination in aging [33,34,35] or multiple sclerosis [36, 37]. To separate the signals of CSF from the pathologies of interest (both appear bright), FLAIR (fluid-attenuated inversion recovery) sequences were introduced. In this variant of T2-w imaging, CSF signal is suppressed (appears dark). Consequently, the signal of interest (WMH, inflammation, etc.) is mostly brighter than the rest of the image, aiding both visual and quantitative analyses. FLAIR’s CSF suppression is particularly helpful at the borders between tissue and ventricles, where WMH are very common and cannot be separated from CSF in standard T2-w images. FLAIR images have thus become the most popular sequence for clinical neuroradiology for general pathology detection and cerebrovascular disease [38].
Compared with T2-w sequences, FLAIR scan times may be longer and/or noise levels may be higher. In research imaging studies of aging and dementia, FLAIR or T2-w sequences are typically used for (a) measuring WMH and (b) monitoring for exclusionary criteria or adverse events, such as infarcts or immune/inflammatory responses to treatment [39, 40]. White matter hyperintensities are typically measured from FLAIR scans (sometimes in conjunction with T1-w scans) using automated or semi-automated algorithms that compute regional or whole-brain volumes using tools such as Lesion Segmentation Tool and FSL BIANCA [36, 37, 41]. Another use of T2-w (but not FLAIR) images is to better estimate TIV than with T1-w alone because T2-w images have signal in extracranial CSF, which is suppressed in FLAIR and T1-w sequences but should be included in TIV measurements [42, 43].
Iron-Sensitive Sequences: T2*, SWI, and QSM
MRI scans contain susceptibility artifacts in areas where the magnetic field is disrupted by magnetic materials; these occur in areas of interface between bone, blood, and air, and around metallic implants such as those common in dentistry. In the brain, hemorrhages and microhemorrhages also produce these local artifacts due to iron depots from the blood. To measure the prevalence of these hemorrhages and microhemorrhages, MRI sequences were developed that highlight these regions of magnetic susceptibility. These sequences are called T2*-w (“T2 star weighted”) or gradient-recalled echo (GRE) imaging. Later variants were developed that enhance resolution and sensitivity; these are known as susceptibility-weighted imaging (SWI) [44]. Still-newer sequences allow quantitative susceptibility mapping (QSM), which have the additional benefit of providing quantitative measures of susceptibility rather than only relative intensities within the image (as in T1-w, T2-w, SWI, and T2*-w/GRE) [45]. All of these sequences are principally used in neurodegeneration research to measure microhemorrhages, primarily through manual marking/counting by image analysts and radiologists [38, 46,47,48].
Diffusion MRI
Diffusion MRI or diffusion-weighted MRI (dMRI or DWI) measures the strength and direction of water molecules’ movement during the scan [49]. Clinically, dMRI is primarily used to image pathologies that reduce diffusion (e.g., ischemic strokes). In the research setting, it is used primarily to image the structure and integrity of white matter. Water molecules travel predominantly along axons, and their motion becomes less one-directional (more omnidirectional or isotropic) as axons degrade. These diffusion changes can reflect multiple etiologies at the molecular level (below the resolution of MRI), and thus changes measured in dMRI are referred to nonspecifically as changes in tissue integrity or microstructure [50].
In dMRI, multiple 3D images are acquired sequentially that each measure motion in one specific direction and at a particular strength or b-value. These sets of diffusion volumes are analyzed jointly to compute derived scalar quantities for each 3D voxel location. In its most basic form, typically six directions are acquired at a single b-value, and these are averaged to produce a trace image, which is a mixture of diffusion-weighted signal with underlying T2-w signal. This is the form most used clinically to image strokes, and in clinical contexts, the term DWI is sometimes used specifically to refer to these images.
In the research setting, more volumes (e.g., 30+ directions) are acquired to increase angular (directional) resolution, and this allows tensor-based analyses; such sequences are referred to as diffusion-tensor imaging (DTI). These sequences have become so standard in research neuroimaging that sometimes all diffusion sequences are called DTI, but really DTI should refer only to this specific class of diffusion sequences and their tensor-based analyses. In DTI analyses, a tensor model is estimated at each voxel location, and these are used to compute several derived scalars at each location. The most common are fractional anisotropy (FA) and mean diffusivity (MD). FA is designed to measure the degree (fraction from 0 to 1) of anisotropy (predominance in one direction). Functionally, FA images provide a map of major white matter tracts, which degrade (reduces FA) with age and pathology. In FA images, the order of brightness/intensity is CSF < GM « WM. Sometimes, directional information is added to FA images to produce a synthetic color FA image that coarsely shows whether each FA voxel flows primarily anterior–posterior, superior-inferior, or right-left. White matter hyperintensities (WMH), typically imaged in T2-w and FLAIR imaging, also reduce WM FA [51]. MD estimates the per-voxel average rate of diffusion across all directions, rather than its directionality. CSF is bright in MD, and WMH and other degradation cause an increase in MD signal toward resembling CSF [50]. Together, FA and MD are typically used to investigate nonspecific mostly vascular WM pathology associated with aging and neurodegenerative diseases [52,53,54,55]. Another class of dMRI analyses is tractography, which attempts to trace or follow diffusion along axons to produce maps of their locations (tractograms) [50]. Tractograms can be used to estimate the structural connectivity (i.e., connection through WM) between pairs of cortical regions. The minimal or optimal configuration of directions/strengths/spatial resolution needed for accurate and reproducible tractography is a matter of considerable debate [56,57,58,59].
Several more-advanced forms of diffusion imaging exist in the research setting; these increase the number of directions and strengths (b-values) to enable more complex analyses. These include multi-shell (multiple “shells” of b-values) dMRI, diffusion spectrum imaging (DSI), diffusion kurtosis imaging (DKI), high angular resolution diffusion imaging (HARDI), and others. Such more-advanced sequences enable more-advanced (than tensors) diffusion models like neurite orientation dispersion and density imaging (NODDI) [60]. These models can enhance voxel-, region-, and tractography-based analyses by allowing better separation of diffusion-related changes from general tissue atrophy and better separation of signals from multiple WM tracts in regions where they overlap.
In the research setting, diffusion images are quantified using automated software pipelines. Some of the most popular include DTI-TK [61], DSI Studio [62], and multiple tools within the DIPY [63] and FSL packages [16, 64].
BOLD: Functional and Resting State MRI
MRI sequences in the previous sections all image brain structure. By contrast, functional MRI (fMRI) images function, by using a mechanism called the blood-oxygen-level-dependent (BOLD) signal. Oxygenated and deoxygenated blood have different magnetic susceptibility, and this can be used to track the hemodynamic response function (HRF), where oxygenated blood is delivered to replenish recently fired neurons [65]. BOLD fMRI sequences rapidly image the brain while a stimulus is presented repeatedly, and statistical analyses (temporal correlations of regional activations) can use this signal to infer which regions of the brain were involved (“activated”) during that stimulus by measuring the HRF (i.e., the BOLD signal after a delay of several seconds).
In resting state (RS-) or task-free (TF-) fMRI, these same scans are performed without any stimulus; the participant is instructed to lie awake in the scanner without any particular thought. These analyses are used to identify and measure the strengths of the brain’s functional networks, large spatially disconnected regions of co-activation which typically cycle and change throughout the total imaging time. The strength of the temporal correlations between pairs of brain regions is called their functional connectivity [66]. Together, functional connections and structural connections (measured from dMRI) are called the brain’s connectome and their study is called connectomics. The most well-known functional network is the default-mode network (DMN) and its variants, thought to be active when the brain is at wakeful rest [67, 68]. Measurable changes in DMN integrity have been associated with Alzheimer’s disease, schizophrenia, autism-spectrum disorders, and others [68]. How long of a time series (scanning time) is required to produce clinically meaningful RS-fMRI data is itself an area of debate, but typical times in the research setting may be between approximately 5 and 20 min [69]. In recent years, fMRI and RS-fMRI analyses have come under some controversy due to concerns that their statistical significance may be inflated or findings may be unreproducible [70,71,72].
Other MRI Sequences
MRI is extremely versatile and there are a variety of more experimental, less common sequences that also play a role in neurodegeneration research. Arterial spin labeling (ASL) is one such sequence, where blood flowing into the brain through the neck is “tagged” with an RF pulse, which changes its MRI signal. After a variable delay of several seconds, subtraction between tagged and un-tagged images can be used to produce a map of cerebral blood flow (CBF) or perfusion [73, 74]. Others include magnetic resonance angiography (MRA) and 4-D flow MRI, used to image blood vessels and blood flow [75, 76], and magnetic resonance elastography (MRE), which measures tissue stiffness, akin to palpitation [77]. MRI with intravenous gadolinium-based contrast agents (GBCA) has also been proposed for measuring the integrity of the blood-brain barrier, which may be implicated in several neurodegenerative diseases, but this is rarely included in large imaging studies due to participant discomfort, concerns of adverse reactions, and potential long-term gadolinium retention in the brain [78].
Nuclear Medicine (PET and SPECT)
Positron emission tomography (PET) scans use radioactive tracers or ligands that are injected intravenously and co-localize in the body with a specific target of interest. These tracers then release positrons that collide with local electrons to produce gamma rays that are imaged by the scanner. These are used to image the density and location of specific targets, e.g., amyloid-beta, tau, and metabolic demand, which is not (currently) possible with MRI.
Compared with MRI, PET is significantly more expensive, has much lower resolution (approximately 5–8 mm vs. approximately 1 mm with clinical MRI), and it carries different participant risks in the form of ionizing radiation. However, these scans are currently the only way to image amyloid, tau, metabolism, and dopamine transporters in living brains, which are of critical interest to neurodegeneration research and clinical trials [79,80,81].
PET measurements can be quantitative if they are acquired using full-dynamic scans (participant is in the scanner for approximately 1–2 h, depending on the tracer used) in conjunction with arterial sampling. These scans allow better accuracy and precision (particularly important for longitudinal measurements) by avoiding the influences of perfusion and blood flow [82,83,84], but the costs of these scans are prohibitive in terms of participant burden and risk (via arterial sampling), and the practical costs of using the PET scanners for so long. More commonly, research studies acquire static or late-uptake scans, where after tracer injection, participants wait (in a waiting room, not in the scanner) for an uptake time period (e.g., 30–90 min depending on the tracer) before being scanned for approximately 20 min [85]. These late-uptake scans are quantified using standardized uptake value ratio (SUVR), which measures PET signal as a ratio over the signal in a reference region that is thought to be free of the target pathology. SUVR measurements are only semiquantitative [83, 84], in contrast to quantitative distribution volume ratio (DVR) measurements from full-dynamic PET scans. There is much debate over the best SUVR reference region for each tracer and type of analyses, but most common choices are cerebellar, pontine, or white matter [86,87,88,89,90]. Middle-ground approaches between quantitative and late-uptake scans include full-dynamic scans without arterial sampling [91], or coffee-break protocols where participants are in the scanner for the first 5–10 min after injection, but then leave and return in time to acquire the typical late-uptake periods. For some tracers, this “coffee break” period is sufficient to allow scanning another participant, while producing DVR measurements closer in quality to full-dynamic than late-uptake scans [84, 92].
FDG PET
Fluorodeoxyglucose (FDG) is a form of glucose modified to emit positrons. When used in brain PET imaging, FDG measures the glucose metabolism associated with neuronal and synaptic activity. In the brain, reduced FDG signal (hypometabolism) is associated with neurodegeneration from all causes. FDG is highly effective at capturing spatial patterns that discriminate between diseases and clinical subtypes of diseases at the individual level [93,94,95,96], making it especially useful in clinical differential diagnosis. In the research setting, FDG can also be used to measure non-specific neurodegeneration (atrophy) when MRI is unavailable [79]. It is also relatively inexpensive (compared with other PET tracers) and widely available due to its widespread use in cancer imaging. However, as more research imaging studies begin using amyloid and tau PET imaging, their use of FDG is declining because study participants can only tolerate (for both practical and radiation-safety reasons) a finite number of PET scans.
Amyloid PET
With the introduction of the Pittsburgh Compound B (PiB) tracer in 2004, researchers first became able to image brain amyloid pathology in living people [97], and amyloid PET has become a cornerstone of the imaging and diagnosis (in the research setting) of Alzheimer’s disease (AD) and related dementias [79, 80]. PiB uses the 11-C isotope, which has a half-life of approximately 20 min. Consequently, facilities using PiB must manufacture it on-site, which requires a costly cyclotron and significant nuclear expertise. Since then, many other amyloid PET tracers have been developed using F-18, which has a half-life of 109.7 min and is thus more practical to manufacture centrally and distribute to smaller facilities. These tracers include Florbetapir (aka FBP, AV-45, Amyvid), Florbetaben (aka FBB), Flutemetamol (aka FMT), and Flutafuranol (aka AZD4694, NAV4694) [98]. Each of these F-18-based amyloid tracers has comparable utility to PiB but their measurements are confounded by relatively more off-target binding (nuisance signal) in WM; the exception is Flutafuranol, which is relatively recent and has approximately identical WM binding with PiB [99]. Because the distribution of amyloid in the brain is relatively homogeneous, amyloid PET is often analyzed to produce only a single numeric measurement of total “global” amyloid [100, 101], which can be thresholded to produce a binary measure of amyloid positivity [79, 100, 102]. Individuals typically become amyloid-positive 10–15 years before the clinical onset of AD symptoms [103, 104]. In AD, amyloid PET and other amyloid biomarkers (such as from cerebral spinal fluid obtained via lumbar puncture) typically become abnormal prior to tau biomarkers (PET or CSF), and both typically occur prior to related neurodegeneration (FDG PET, MRI, or CSF) [79, 103,104,105].
Tau PET
Approximately a decade after the introduction of PiB, tracers were developed to image tau proteins associated with Alzheimer’s disease. First-generation tracers include Flortaucipir (aka FTP, AV-1451, Tauvid, T-807) [106], THK5317, THK5351, and PBB3. Second-generation tracers include MK-6240, RO-948, PI-2620, GTP1, and PM-PBB3 [107]. All of these tracers (in both generations) are F-18-based, and thus, they can be transported more practically than PiB. FTP and MK-6240 have emerged as arguably the most common tau tracers in AD research. Each of these tracers has varying binding affinity (i.e., signal strength) to AD-associated tau and varying levels of problematic off-target binding in other regions, such as the basal ganglia, choroid plexus, and even skull/bone surrounding the brain, which can “bleed into” regions of interest and make quantification/interpretation a challenge [107, 108]. Despite these challenges, tau PET has become widespread and crucial to AD research [79, 109,110,111,112,113]. Spatial patterns of tau PET are much more heterogenous across individuals than amyloid PET [114,115,116]. Within the time continuum of Alzheimer’s disease, highly elevated levels of tau PET signal typically occur only after substantial elevation of amyloid PET, and compared with amyloid they are more strongly associated (both in time and spatially within the brain) with neurodegeneration (atrophy) and with clinical symptoms [105, 117].
The use of current tau PET tracers for non-AD tauopathies, such as primary supranuclear palsy (PSP), corticobasal degeneration (CBD), chronic traumatic encephalopathy (CTE), and some subtypes of Frontotemporal lobar degeneration (FTLD), is much more controversial. Studies with early tracers (mainly Flortaucipir) have shown relatively weak signal in these diseases that is detectable on the group-level, but individual measurements are in ranges much lower than in AD patients [112, 118, 119]. The location of this weak signal is typically in white matter regions that are adjacent to the cortex where signal would be expected [119], and autoradiography studies have repeatedly found minimal binding of these tracers to non-AD tau ex-vivo [120, 121]; consequently, the specific pathology underlying this signal in non-AD tauopathies is uncertain and its related findings must be interpreted with caution [107, 120]. Some tracers (PI-2620, PM-PBB3) have reported relatively more signal in these diseases [122, 123], but no tau PET tracers have been widely accepted for non-AD tau [107].
Other Tracers
Although none has become as common as amyloid, tau, and FDG, several other classes of PET tracers are being explored to study the molecular pathology of dementing illnesses. These include tracers for synaptic density (e.g., UCB-J) [124] and neuroinflammation (e.g., ER176, PBR28, SMBT-1) [125126, 127].
Alpha-synuclein is the key protein of interest for Parkinson’s disease, Dementia with Lewy bodies (DLB), and other parkinsonian disorders. There are currently no released PET tracers for imaging alpha-synuclein in vivo. Some early results from in-development tracers (e.g., ACI-3847) have been presented at research meetings, but none is yet available. However, DaTScan (aka Ioflupane) is a single photon emission computed tomography (SPECT) tracer that images dopamine transporters (DaT); the appearance of these images in the striatum can be used to differentiate “true” parkinsonian disorders from essential tremor or drug-induced parkinsonism [128, 129]. These scans are also sometimes used for differential diagnosis in the research setting, particularly in imaging studies of parkinsonian disorders, but they do not allow directly studying the underlying alpha-synuclein pathology. Similarly, tracers for TDP-43 (TAR DNA-binding protein 43) would be of high interest for neurodegeneration research, but none have yet become available.
Quantifying PET Scans
Quantifying PET scans in neurodegeneration research is typically based on registration with T1-w MRI (which is almost always also performed) to allow the same types of template- and atlas-driven region- and voxel-based approaches as above, with largely the same sets of software packages, e.g., SPM, FreeSurfer, and FSL [31, 100]. Once PET scans are registered to T1-w MRI, nonlinear registrations between MRI and standard templates allow propagation of PET scans into template space, or atlas regions onto the native PET images. There are also specialized pipelines for PET-only measurements, and these can achieve similar performance with MRI-based approaches [130, 131], but MRI-based approaches are considered the gold standard because they have higher resolution to more accurately segment tissue and localize regions, and they can better adjust for partial volume (described below).
A large concern in PET quantification is partial volume: PET signal is reduced in regions of tissue loss (atrophy), and it is impossible from PET alone to determine whether a target (e.g., glucose, amyloid, or tau) is sparsely present within dense underlying tissue vs. densely present within sparse underlying tissue (i.e., regions of atrophy). There are many algorithms for partial volume correction (PVC) that use corresponding T1-w MRI to estimate hypothetical PET signal in each voxel or region as if there were no atrophy. PVC methods inherently boost measurement noise because they amplify signal in areas with relatively low signal. Determining which (if any at all) PVC method should be applied for a given study is a very active area of research and discussion [132133,134,135]. Applying PVC to PET analyses comparing two groups typically increases the statistical power of their differences; proponents of PVC argue that this supports it having corrected the biasing effects of atrophy from the PET signal. However, opponents of PVC argue that it effectively multiplies PET signal with MRI, so increased group differences come from increased information content from combining the MRI (atrophy) and the PET (molecular) signals together. Studies examining correlations between in vivo amyloid scans and quantitative pathology at autopsy have found that these correlations were reduced by application of all major classes of PVC [134], which works against the pro-PVC hypothesis that its increased group-differences are the result of more accurately quantifying the underlying molecular pathology.
CT
Computed tomography (CT) scans have a limited role in neurodegeneration research because CT has poor contrast for differentiating brain tissues when compared with MRI. However, most PET scanners are actually PET/CT scanners that acquire a low-dose CT used for attenuation correction (compensation for signal loss due to structurally dense areas such as bone) during reconstruction of the PET images. These CT scans are typically not analyzed after on-scanner PET reconstruction is completed. PET/MRI scanners have been slowly growing in popularity; these replace the low-dose CT with MRI- and artificial intelligence-based substitutes to eliminate the CT’s dose of radiation to the participant. However, achieving CT-like bone contrast with MRI is not straightforward [136], and in neurodegeneration research studies PET/CT scanners still far outnumber PET/MRI scanners.
Challenges to Multi-site and Meta-analyses: Data Standardization and Harmonization
One huge challenge in neuroimaging is the inability to directly compare images and imaging-derived measurements across differences in imaging protocols (pulse sequences, PET tracers, scan times), imaging hardware (scanner vendor/model, head coil, on-board reconstruction software/version), and analyses software/measurements. Each imaging site or research group typically performs these tasks according to their own preferences and hardware availability, which makes the images and measurements unable to be compared directly without some transformation for harmonization [100, 137] to remove non-biologic sources of variability. In an ideal study, every participant would be imaged on the same scanner with the same protocols and data would be analyzed identically, but very large studies are typically multi-site and even within a single site, practical concerns often necessitate the use of multiple scanners. With longitudinal studies, maintaining participants on the same scanner over time can become impossible over long periods, as scanners naturally age, break down, and are replaced with newer technology. In multi-site trials, each typically has a different mix of scanners of varying ages and manufacturer/models, and images from these are rarely directly comparable.
To reduce these challenges, researchers first reduce as many technical sources of variation as they can, i.e., using the same hardware and methods as much as possible. Pulse sequences and imaging protocols have been designed that attempt to produce relatively more-comparable results across a wide range of manufacturers and models for many imaging sequences [138, 139]; these protocols have been shared for use by other research studies and are designed to reduce compatibility issues, but they do not eliminate them. Scanners can also be validated (repeatedly) for use in a research study, such as by scanning standardized hardware phantoms and ensuring that resulting measurements are within expected ranges [140]. Within a study, analyses software/pipelines can also be harmonized; images can always be re-analyzed retrospectively with consistent software, but this is costly in both human and computational resources. Even from identical images, differences in software/analyses (even very small ones like minor version or operating system changes) can produce very different findings [141,142,143], which is a large challenge for scientific reproducibility and meta-analyses.
When standardizing all technical aspects of imaging and analyses is not feasible, statistical approaches can reduce (but not eliminate) the effects of these confounding factors. For example, a coalition of amyloid PET researchers has made specific efforts to correct for the effects of different tracers and analyses by using linear regressions to transform every individual combination to a standard “centiloid” 0–100 scale [87, 144]. Nonlinear mappings for amyloid PET have also been proposed to increase agreement across tracers and methods [145]. Other techniques involve post-processing images to better match each other, e.g., blurring or adding random noise to images of higher quality to match a common lower standard, which can be effective but inherently reduces the quality of the data that is deliberately degraded [146, 147]. Researchers are also increasingly applying techniques designed for generic numeric data (i.e., not specific for images) to reduce non-biologic factors, such as ComBat [148]. These methods can reduce, but do not eliminate, the effects of non-biologic sources of variance [149].
Conclusion
In this review, we have summarized the major types of brain images acquired in neurodegeneration research studies, their major purposes, and how they are analyzed. We also discussed some of the larger technical challenges in this area and how researchers are working to address them. We hope that this work will help readers who are relatively less familiar with neuroimaging technology to gain a better understanding of the wide breadth of imaging-based neurodegeneration research studies.
References
Klein A, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage, 2009;46:786–802.
Avants BB, Epstein CL, Grossman M, Gee JC. “Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain,” Med. Image Anal 2008;12:26–41.
Ashburner J. “A fast diffeomorphic image registration algorithm,” Neuroimage, 2007;38:95–113.
Andersson J, Smith S, Jenkinson M. “FNIRT-FMRIB’s non-linear image registration tool,” in Annual Meeting of the Organization for Human Brain Mapping (OHBM), 2008.
Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. “A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM).,” Neuroimage, 1995;2:89–101.
Mazziotta J, et al. “A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM),” Philos. Trans. R. Soc. B Biol. Sci. 2001;356:1293–1322.
Evans AC, Janke AL, Collins DL, Baillet S. “Brain templates and atlases,” Neuroimage, 2012;62:911–922.
Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL. “Unbiased average age-appropriate atlases for pediatric studies.,” Neuroimage, 2011;54:313–327.
Schwarz CG, et al. “The Mayo Clinic Adult Lifespan Template: Better Quantification Across the Lifespan,” Alzheimer’s Dement. 2017;13:P792.
Tang Y, et al. “The construction of a Chinese MRI brain atlas: A morphometric comparison study between Chinese and Caucasian cohorts,” Neuroimage, 2010;51:33–41.
Varentsova A, Zhang S, Arfanakis K. “Development of a High Angular Resolution Diffusion Imaging Human Brain Template,” Neuroimage, 2014.
Horn A, Blankenburg F, “Toward a standardized structural-functional group connectome in MNI space,” Neuroimage, 2016;124:310–322.
Zhou L, et al. “MR-less surface-based amyloid assessment based on 11C PiB PET,” PLoS One, 2014;9:e84777.
Fischl B, et al. “Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain,” Neuron, 2002;33:341–355.
Ashburner J, Friston KJ. “Unified segmentation,” Neuroimage, 2005;26:839–851.
Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. “FSL,” Neuroimage, 2012;62:782–90.
Cox RW. “AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages.,” Comput. Biomed. Res., 1996;29:162–173.
Freeborough PA, Fox NC. “The boundary shift integral: An accurate and robust measure of cerebral volume changes from registered repeat MRI,” IEEE Trans. Med. Imaging, 1997;16:623–629.
Barnes J, et al. “Head size, age and gender adjustment in MRI studies: A necessary nuisance?,” Neuroimage, 2010;53:1244–1255.
Buckner RL, et al. “A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: Reliability and validation against manual measurement of total intracranial volume,” Neuroimage, 2004;23:724–738.
Fischl B, Dale AM. “Measuring the thickness of the human cerebral cortex from magnetic resonance images,” Proc. Natl. Acad. Sci. U. S. A. 2000;97:11050–11055.
Das SR, Avants BB, Grossman M, Gee JC. “Registration based cortical thickness measurement,” Neuroimage, 2009;45:867–879.
Lerch JP, Evans AC. “Cortical thickness analysis examined through power analysis and a population simulation,” Neuroimage, 2005;24:163–173.
Cardinale F, et al. “Validation of FreeSurfer-Estimated Brain Cortical Thickness: Comparison with Histologic Measurements,” Neuroinformatics, 2014;12:535–542.
Schwarz CG, et al. “A large-scale comparison of cortical thickness and volume methods for measuring Alzheimer’s disease severity,” NeuroImage Clin., 2016;11:802–812.
Ashburner J, Friston KJ. “Voxel-Based Morphometry—The Methods,” Neuroimage, 2000;11:805–821.
Hua X, et al. “Accurate measurement of brain changes in longitudinal MRI scans using tensor-based morphometry,” Neuroimage, 2011;57:5–14.
Cash DM, et al. “Assessing atrophy measurement techniques in dementia: Results from the MIRIAD atrophy challenge,” Neuroimage, 2015;123:149–164.
Dale AM, Fischl B, Sereno MI. “Cortical Surface-Based Analysis,” Neuroimage, 1999;9:179–194.
Greve DN. “An Absolute Beginner’s Guide to Surface- and Voxel-based Morphometric Analysis,” Proc. ISMRM, 2011;19.
Greve DN, et al. “Cortical surface-based analysis reduces bias and variance in kinetic modeling of brain PET data,” Neuroimage, 2014;92:225–236.
Calhoun VD, et al. “The impact of T1 versus EPI spatial normalization templates for fMRI data analyses,” Hum. Brain Mapp., 2017;38:5331–5342.
Carmichael O, et al. “Longitudinal Changes in White Matter Disease and Cognition in the First Year of the Alzheimer Disease Neuroimaging Initiative,” Arch. Neurol., 2010;67:1370–1378.
Brickman AM, et al. “Regional White Matter Hyperintensity Volume, Not Hippocampal Atrophy, Predicts Incident Alzheimer Disease in the Community.,” Arch. Neurol., 2012;69:1621–1627.
de Groot JC, de Leeuw FE, Oudkerk M, Hofman A, Jolles J, Breteler MMB. “Cerebral white matter lesions and subjective cognitive dysfunction: the Rotterdam Scan Study.,” Neurology, 2001;56:1539–1545.
van Leemput K, Maes F, Vandermeulen D, Colchester A, Suetens P. “Automated segmentation of multiple sclerosis lesions by model outlier detection,” Med. Imaging, IEEE Trans., 2001;20:677–688.
Schmidt P, et al. “An automated tool for detection of FLAIR-hyperintense white-matter lesions in Multiple Sclerosis,” Neuroimage, 2012;59:3774–3783.
Jack CRJ, et al. “Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative,” Alzheimer’s Dement., 2010;6:212–220.
Sperling R, et al. “Amyloid-related imaging abnormalities in patients with Alzheimer’s disease treated with bapineuzumab: A retrospective analysis,” Lancet Neurol., 2012;11:241–249.
Sperling RA, et al. “Amyloid-related imaging abnormalities in amyloid-modifying therapeutic trials: Recommendations from the Alzheimer’s Association Research Roundtable Workgroup,” Alzheimer’s Dement., 2011;7:367–385.
Griffanti L, et al. “BIANCA (brain intensity AbNormality classification algorithm): A new tool for automated segmentation of white matter hyperintensities,” Neuroimage, 2016.
Hansen TI, Brezova V, Eikenes L, Håberg A, Vangberg XTR. “How does the accuracy of intracranial volume measurements affect normalized brain volumes? sample size estimates based on 966 subjects from the HUNT MRI cohort,” Am. J. Neuroradiol., 2015;36:1450–1456.
Vuong P, Drucker D, Schwarz C, Fletcher E, Decarli C, Carmichael O. “Effects of T2-weighted MRI based cranial volume measurements on studies of the aging brain,” in Progress in Biomedical Optics and Imaging - Proceedings of SPIE, 2013;8669.
Haacke EM, Xu Y, Cheng YCN, Reichenbach JR. “Susceptibility weighted imaging (SWI),” Magn. Reson. Med., 2004;52:612–618.
Wang Y, Liu T. “Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker,” Magn. Reson. Med., 2015;73:82–101.
Cordonnier C, Van Der Flier WM. “Brain microbleeds and Alzheimer’s disease: Innocent observation or key player?,” Brain, 2011;134:335–344.
Yates PA, Sirisriro R, Villemagne VL, Farquharson S, Masters CL, Rowe CC. “Cerebral microhemorrhage and brain β-amyloid in aging and Alzheimer disease,” Neurology, 2011;77:48–54.
Goos JDC et al. “Patients with alzheimer disease with multiple microbleeds: Relation with cerebrospinal fluid biomarkers and cognition,” Stroke, 2009;40:3455–3460.
Le Bihan D, Breton E. “Imagerie de self-diffusion in vivo par résonance magnétique nucléaire,” C. R. Académie Sci., 1986;7:713–720.
O’Donnell LJ, Westin CF. “An introduction to diffusion tensor image analysis,” Neurosurg. Clin. N. Am., 2011;22:185–196.
Maillard P, et al. “White matter hyperintensities and their penumbra lie along a continuum of injury in the aging brain,” Stroke, 2014;45:1721–1726.
Sullivan EV, Pfefferbaum A. “Diffusion tensor imaging and aging,” Neurosci. Biobehav. Rev., 2006;30:749–761.
Westlye LT, et al. “Life-span changes of the human brain white matter: Diffusion tensor imaging (DTI) and volumetry,” Cereb. Cortex, 2010;20:2055–2068.
Maniega SM, et al. “White matter hyperintensities and normal-appearing white matter integrity in the aging brain,” Neurobiol. Aging, 2014.
Lebel C, Gee M, Camicioli R, Wieler M, Martin W, Beaulieu C. “Diffusion tensor imaging of white matter tract evolution over the lifespan,” Neuroimage, 2012;60:340–352.
Hoefnagels FWA, de Witt Hamer PC, Pouwels PJW, Barkhof F, Vandertop WP. “Impact of Gradient Number and Voxel Size on Diffusion Tensor Imaging Tractography for Resective Brain Surgery,” World Neurosurg., 2017;105:923–934.e2.
Correia MM, Carpenter TA, Williams GB. “Looking for the optimal DTI acquisition scheme given a maximum scan time: are more b-values a waste of time?,” Magn. Reson. Imaging, 2009;27:163–175.
Liebrand LC, van Wingen GA, Vos FM, Denys D, Caan MWA. “Spatial versus angular resolution for tractography-assisted planning of deep brain stimulation,” NeuroImage Clin., 2020;25:102116.
Lebel C, Benner T, Beaulieu C. “Six is enough? Comparison of diffusion parameters measured using six or more diffusion-encoding gradient directions with deterministic tractography,” Magn. Reson. Med., 2012;68:474–483.
Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. “NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain,” Neuroimage, 2012;61:1000–1016.
Zhang H, Yushkevich PA, Alexander DC, Gee JC. “Deformable registration of diffusion tensor MR images with explicit orientation optimization,” Med. Image Anal., 2006;10:764–785.
Yeh FC, Verstynen TD, Wang Y, Fernández-Miranda JC, Tseng WYI. “Deterministic diffusion fiber tracking improved by quantitative anisotropy,” PLoS One, 2013;8:1–16.
Garyfallidis E, et al. “Dipy, a library for the analysis of diffusion MRI data,” Front. Neuroinform., 2014;8:8.
Smith SM, et al. “Tract-Based Spatial Statistics: Voxelwise Analysis of Multi-Subject Diffusion Data,” Tech. Rep., 2005;1:1–26.
Ogawa S, Lee TM, Nayak AS, Glynn P. “Oxygenation‐sensitive contrast in magnetic resonance image of rodent brain at high magnetic fields,” Magn. Reson. Med., 1990;14:68–78.
van den Heuvel MP, Hulshoff Pol HE. “Exploring the brain network: A review on resting-state fMRI functional connectivity,” Eur. Neuropsychopharmacol., 2010;20:519–534.
Greicius MD, Krasnow B, Reiss AL, Menon V. “Functional connectivity in the resting brain: A network analysis of the default mode hypothesis,” Proc. Natl. Acad. Sci. U. S. A., 2003;100:253–258.
Buckner RL, Andrews-Hanna JR, Schacter DL. “The brain’s default network: Anatomy, function, and relevance to disease,” Ann. N. Y. Acad. Sci., 2008;1124:1–38.
Birn RM, et al. “The effect of scan length on the reliability of resting-state fMRI connectivity estimates,” Neuroimage, 2013;83:550–558.
Eklund A, Nichols TE, Knutsson H. “Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates,” Proc. Natl. Acad. Sci., 2016;113:7900–7905.
Bennett CM, Baird AA, Miller MB, Wolford GL. “Neural correlates of interspecies perspective taking in the post-mortem Atlantic Salmon: An argument for multiple comparisons correction Craig,” in Organization for Human Brain Mapping, 2009.
Lyon L. “Dead salmon and voodoo correlations: should we be sceptical about functional MRI?,” Brain, 2017;140:e53–e53.
Alsop DC, et al. “Recommended implementation of arterial spin-labeled Perfusion mri for clinical applications: A consensus of the ISMRM Perfusion Study group and the European consortium for ASL in dementia,” Magn. Reson. Med., 2015;73:102–116.
Alsop DC, Dai W, Grossman M, Detre JA. “Arterial spin labeling blood flow MRI: its role in the early characterization of Alzheimer’s disease.,” J. Alzheimer’s Dis., 2010;20:871–880.
Rivera-Rivera LA, et al. “4D flow MRI for intracranial hemodynamics assessment in Alzheimer’s disease,” J. Cereb. Blood Flow Metab., 2016;36:1718–1730.
Markl M, Frydrychowicz A, Kozerke S, Hope M, Wieben O. “4D flow MRI,” J. Magn. Reson. Imaging, 2012;36:1015–1036.
Murphy MC, Huston J, Ehman RL. “MR elastography of the brain and its application in neurological diseases,” Neuroimage, 2019;187:176–183.
Montagne A, Toga AW, Zlokovic BV. “Blood-brain barrier permeability and gadolinium benefits and potential pitfalls in research,” JAMA Neurol., 2016;73:13–14.
Jack CR, et al. “NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease,” Alzheimer’s Dement., 2018;14:535–562.
Villemagne VL, et al. “Aβ Imaging: feasible, pertinent, and vital to progress in Alzheimer’s disease,” Eur. J. Nucl. Med. Mol. Imaging, 2012;39:209–219.
Mathis CA, Lopresti BJ, Ikonomovic MD, Klunk WE. “Small-molecule PET Tracers for Imaging Proteinopathies,” Semin. Nucl. Med., 2017;47:553–575.
van Berckel BNM, et al. “Longitudinal amyloid imaging using 11C-PiB: methodologic considerations,” J. Nucl. Med., 2013;54:1570–1576.
Ottoy J, et al. “Validation of the semi-quantitative static SUVR method for [18 F]-AV45 PET by pharmacokinetic modeling with an arterial input function,” J. Nucl. Med., 2017;58:1483–1489.
Timmers T, et al. “Test-retest repeatability of [18F] Flortaucipir PET in Alzheimer’s disease and cognitively normal individuals,” J. Cereb. Blood Flow Metab., 2019;40:2464–2474.
Jagust WJ, et al. “The Alzheimer’s Disease Neuroimaging Initiative 2 PET Core: 2015,” Alzheimer’s Dement., 2015;11:757–771.
Landau S, Fero A, Baker S, Jagust W. “Modeling longitudinal florbetapir change across the disease spectrum,” in Alzheimer’s Association International Conference (AAIC), 2014.
Klunk WE, et al. “The Centiloid Project: standardizing quantitative amyloid plaque estimation by PET,” Alzheimer’s Dement., 2015;11:1–15.
Schwarz CG, et al. “Optimizing PiB-PET SUVR change-over-time measurement by a large-scale analysis of longitudinal reliability, plausibility, separability, and correlation with MMSE.,” Neuroimage, 2017;144:113–127.
Chiao P, et al. “Impact of Reference/Target Region Selection on Amyloid PET Standard Uptake Value Ratios in the Phase 1b PRIME Study of Aducanumab,” J. Nucl. Med., 2018;1–23.
Brendel M, et al. “Improved longitudinal [18F]-AV45 amyloid PET by white matter reference and VOI-based partial volume effect correction,” Neuroimage, 2015;108:450–459.
Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, Alexoff DL. “Distribution volume ratios without blood sampling from graphical analysis of PET data,” J. Cereb. Blood Flow Metab., 1996;16:834–840.
Tuncel H, et al. “Effect of Shortening the Scan Duration on Quantitative Accuracy of [18F]Flortaucipir Studies,” Mol. Imaging Biol., 2021.
Kato T, Inui Y, Nakamura A, Ito K. “Brain fluorodeoxyglucose (FDG) PET in dementia,” Ageing Res. Rev., 2016;30:73–84.
Cerami C, et al. “Different FDG-PET metabolic patterns at single-subject level in the behavioral variant of fronto-temporal dementia,” Cortex, 2016;83:101–112.
Graff-Radford J, et al. “Dementia with Lewy bodies: Basis of cingulate island sign,” Neurology, 2014;83:801–809.
Jones D, et al. “Patterns of neurodegeneration in dementia reflect a global functional state space,” medRxiv, 2020.
Klunk WE, et al. “Imaging Brain Amyloid in Alzheimer’s Disease with Pittsburgh Compound-B,” Ann. Neurol., 2004;55:306–319.
Villemagne VL. “Amyloid imaging: Past, present and future perspectives,” Ageing Res. Rev., 2016;30:95–106.
Rowe CC, et al. “Head-to-head comparison of11C-PiB and18F-AZD4694 (NAV4694) for β-amyloid imaging in aging and dementia,” J. Nucl. Med., 2013;54:880–886.
Klunk WE. “Standardization of Amyloid PET: The Centiloid Project,” in Alzheimer’s Association International Conference (AAIC), 2014.
Jack CRJ, et al. “Brain β-amyloid load approaches a plateau,” Neurology, 2013;80:890–896.
Villeneuve S, et al. “Existing Pittsburgh Compound-B positron emission tomography thresholds are too high: Statistical and pathological evaluation,” Brain, 2015;138:2020–2033.
Villemagne VL, et al. “Amyloid β deposition, neurodegeneration, and cognitive decline in sporadic Alzheimer’s disease: a prospective cohort study,” Lancet Neurol., 2013;12:357–367.
Bateman RJ, et al. “Clinical and Biomarker Changes in Dominantly Inherited Alzheimer’s Disease,” N. Engl. J. Med., 2012;367:795–804.
Jack CRJ, et al. “Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers,” Lancet Neurol., 2013;12:207–216.
Xia CF, et al. “[(18)F] T807, a novel tau positron emission tomography imaging agent for Alzheimer’s disease,” Alzheimer’s Dement., 2013;9:666–676.
Leuzy A, et al. “Tau PET imaging in neurodegenerative tauopathies—still a challenge,” Mol. Psychiatry, 2019.
Baker SL, Harrison TM, Maass A, La Joie R, Jagust WJ. “Effect of Off-Target Binding on 18 F-Flortaucipir Variability in Healthy Controls Across the Life Span,” J. Nucl. Med., 2019;60:1444–1451.
Johnson KA, et al. “Tau positron emission tomographic imaging in aging and early Alzheimer disease,” Ann. Neurol., 2016;79:110–119.
Schwarz AJ, et al. “Regional profiles of the candidate tau PET ligand 18 F-AV-1451 recapitulate key features of Braak histopathological stages,” Brain, 2016;aww023.
Gordon BA, et al. “Tau PET in autosomal dominant Alzheimer’s disease: relationship with cognition, dementia and other biomarkers,” Brain, 2019.
Ossenkoppele R, et al. “Discriminative Accuracy of [18F] flortaucipir Positron Emission Tomography for Alzheimer Disease vs Other Neurodegenerative Disorders.,” JAMA, 2018;320:1151–1162.
Harrison TM, et al. “Longitudinal tau accumulation and atrophy in aging and alzheimer disease,” Ann. Neurol., 2019;85:229–240.
Lockhart SN, et al. “Amyloid and Tau PET Demonstrate Region-Specific Associations in Normal Older People,” Neuroimage, 2017.
Lowe VJ, et al. “Widespread brain tau and its association with ageing, Braak stage and Alzheimer’s dementia.,” Brain, 2018;141:271–287.
Vemuri P, et al. “Tau-PET uptake: Regional variation in average SUVR and impact of amyloid deposition,” Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit., 2017;6:21–30.
Ossenkoppele R, et al. “Tau PET patterns mirror clinical and neuroanatomical variability in Alzheimer’s disease,” Brain, 2016;139:1551–1567.
Smith R, et al. “In vivo retention of 18 F-AV-1451 in corticobasal syndrome,” Neurology, 2017;0:1–10.
Josephs KA, et al. “[18F] AV-1451 tau-PET uptake does correlate with quantitatively measured 4R-tau burden in autopsy-confirmed corticobasal degeneration,” Acta Neuropathol., 2016;132:931–933.
Lowe VJ, et al. “An autoradiographic evaluation of AV-1451 Tau PET in dementia,” Acta Neuropathol. Commun., 2016;4:1–19.
Marquié M, et al. “Validating novel tau positron emission tomography tracer [F-18]-AV-1451 (T807) on postmortem brain tissue,” Ann. Neurol. 2015;78:787–800.
Brendel M, et al. “Assessment of 18 F-PI-2620 as a Biomarker in Progressive Supranuclear Palsy,” JAMA Neurol., 2020:1–12.
Oh M, et al. “Clinical Evaluation of 18 F-PI-2620 as a Potent PET Radiotracer Imaging Tau Protein in Alzheimer Disease and Other,” Clin. Nucl. Med., 2020;00.
Koole M, et al. “Quantifying SV2A density and drug occupancy in the human brain using [ 11 C]UCB-J PET imaging and subcortical white matter as reference tissue,” Eur. J. Nucl. Med. Mol. Imaging, 2019;46:396–406.
Kreisl WC, Kim MJ, Coughlin JM, Henter ID, Owen DR, Innis RB. “PET imaging of neuroinflammation in neurological disorders,” Lancet Neurol., 2020;19:940–950.
Kim MJ, et al. “Neuroinflammation in frontotemporal lobar degeneration revealed by 11C-PBR28 PET,” Ann. Clin. Transl. Neurol., 2019;6:1327–1331.
Harada R, et al. “18 F-SMBT-1: A Selective and Reversible Positron-Emission Tomography Tracer for Monoamine Oxidase-B Imaging,” J. Nucl. Med., 2020;jnumed.120.244400.
Papathanasiou ND, Boutsiadis A, Dickson J, Bomanji JB. “Diagnostic accuracy of 123I-FP-CIT (DaTSCAN) in dementia with Lewy bodies: A meta-analysis of published studies,” Park. Relat. Disord., 2012;18:225–229.
Sadasivan S, Friedman JH. “Experience with DaTscan at a tertiary referral center,” Park. Relat. Disord., 2015;21:42–45.
Bourgeat P, et al. “Implementing the centiloid transformation for 11C-PiB and β-amyloid 18F-PET tracers using CapAIBL,” Neuroimage, 2018;183:387–393.
Bourgeat P, et al. “Comparison of MR-less PiB SUVR quantification methods,” Neurobiol. Aging, 2015;36:S159–S166.
Thomas BA, et al. “PETPVC: a toolbox for performing partial volume correction techniques in positron emission tomography,” Phys. Med. Biol., 2016;61:7975–7993.
Schwarz CG, et al. “A Comparison of Partial Volume Correction Techniques for Measuring Change in Serial Amyloid PET SUVR,” J. Alzheimer’s Dis. 2018;67:181–195.
Minhas DS, et al. “Impact of partial volume correction on the regional correspondence between in vivo [C-11]PiB PET and postmortem measures of Aβ load,” NeuroImage Clin., 2018;19:182–189.
Greve DN, et al. “Different partial volume correction methods lead to different conclusions: An 18F-FDG PET Study of aging,” Neuroimage, 2016;132:334–343.
Vandenberghe S, Marsden PK. “PET-MRI: A review of challenges and solutions in the development of integrated multimodality imaging,” Phys. Med. Biol., 2015;60:R115–R154.
Schnack HG, et al. “Mapping reliability in multicenter MRI: Voxel-based morphometry and cortical thickness,” Hum. Brain Mapp., 2010;31:1967–1982.
Jack CRJ, et al. “The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods,” J. Magn. Reson. Imaging JMRI, 2008;27:685–691.
Duchesne S, et al. “The Canadian Dementia Imaging Protocol: Harmonizing National Cohorts,” J. Magn. Reson. Imaging, 2019;49:456–465.
Gunter JL, et al. “Measurement of MRI scanner performance with the ADNI phantom,” Med. Phys., 2009;36:2193.
Gronenschild EHBM, et al. “The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements,” PLoS One, 2012;7.
Jones DK, et al. “What Happens When Nine Different Groups Analyze the Same DT-MRI Data Set Using Voxel-Based Methods ?,” Proc. Int. Soc. Magn. Reson. Med., 2007;15:2007.
Poline JB, Strother SC, Dehaene-Lambertz G, Egan GF, Lancaster JL. “Motivation and synthesis of the FIAC experiment: Reproducibility of fMRI results across expert analyses,” Hum. Brain Mapp., 2006;27:351–359.
Tudorascu DL, et al. “The use of Centiloids for applying [11C] PiB classification cutoffs across region-of-interest delineation methods,” Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit., 2018:1–8.
Properzi MJ, et al. “Nonlinear Distributional Mapping (NoDiM) for harmonization across amyloid-PET radiotracers,” Neuroimage, 2019;186:446–454.
Ligero M, et al. “Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis,” Eur. Radiol., 2020.
Joshi A, Koeppe RA, Fessler JA. “Reducing between scanner differences in multi-center PET studies,” Neuroimage, 2009;46:154–159.
Johnson WE, Li C, Rabinovic A. “Adjusting batch effects in microarray expression data using empirical Bayes methods,” Biostatistics, 2007;8:118–127.
Radua J, et al., “Increased power by harmonizing structural MRI site differences with the ComBat batch adjustment method in ENIGMA,” Neuroimage, 2020;218.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schwarz, C.G. Uses of Human MR and PET Imaging in Research of Neurodegenerative Brain Diseases. Neurotherapeutics 18, 661–672 (2021). https://doi.org/10.1007/s13311-021-01030-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13311-021-01030-9