Abstract
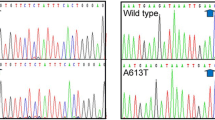
Human DNA polymerase β (polβ) is a small monomeric protein that is essential for short-patch base excision repair. It plays an important role in regulating the sensitivity of tumor cells to chemotherapy. We have previously identified a G to C point mutation at nucleotide 648 (G648C) of polβ in esophageal cancer (EC). In this study, we evaluated the mutation of polβ in a larger cohort of EC patients by RT-PCR and sequencing analysis. The function of the mutation was evaluated by MTT, in vivo tumor growth, and flow cytometry assays. The G648C mutation occurred in 15 (3.45 %) of 435 EC patients. In addition, patients with this mutation had significantly longer survival time than those without, following postoperative chemotherapy. Cell lines with G648C mutation in polβ gene were more sensitive to treatment with 5-fluorouracil and cisplatin than those with wild-type polβ. These results suggest that polβ gene with G648C mutation in surgically resected esophagus may be clinically useful for predicting responsiveness to chemotherapy in patients with EC. The polβ gene alteration may serve as a prognostic biomarker for EC.





Similar content being viewed by others
References
Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50.
Nowak R, Woszczynski M, Siedlecki JA. Changes in the DNA polymerase beta gene expression during development of lung, brain, and testis suggest an involvement of the enzyme in DNA recombination. Exp Cell Res. 1990;191:51–6.
Krahn JM, Beard WA, Wilson SH. Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004;12(10):1823–32.
Prasad R, Batra VK, Yang XP, Krahn JM, Pedersen LC, Beard WA, et al. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair (Amst). 2005;4(12):1347–57.
Friedberg EC. DNA damage and repair. Nature. 2003;421:436–40.
Kidane D, Jonason AS, Gorton TS, Mihaylov I, Pan J, Keeney S, et al. DNA polymerase beta is critical for mouse meiotic synapsis. EMBO J. 2010;29:410–23.
Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase b (Pol4)-dependent pathway. J Biol Chem. 1999;274:23599–609.
Dalal S, Chikova A, Jaeger J, Sweasy JB. The Leu22Pro tumor-associated variant of DNA polymerase beta is dRP lyase deficient. Nucleic Acids Res. 2008;36:411–22.
Canitrot Y, Hoffmann JS, Calsou P, Hayakawa H, Salles B, Cazaux C. Nucleotide excision repair DNA synthesis by excess DNA polymerase beta: a potential source of genetic instability in cancer cells. FASEB J. 2000;14:1765–74.
Srivastava DK, Husain I, Arteaga CL, Wilson SH. DNA polymerase beta expression differences in selected human tumors and cell lines. Carcinogenesis. 1999;20:1049–54.
Bhattacharyya N, Chen HC, Comhair S, Erzurum SC, Banerjee S. Variant forms of DNA polymerase beta in primary lung carcinomas. DNA Cell Biol. 1999;18:549–54.
Dobashi Y, Shuin T, Tsuruga H, Uemura H, Torigoe S, Kubota Y. DNA polymerase beta gene mutation in human prostate cancer. Cancer Res. 1994;54:2827–39.
Miyamoto H, Miyagi Y, Ishikawa T, Ichikawa Y, Hosaka M, Kubota Y. DNA polymerase beta gene mutation in human breast cancer. Int J Cancer. 1999;83:708–19.
Wang L, Patel U, Ghosh L, Banerjee S. DNA polymerase beta mutations in human colorectal cancer. Cancer Res. 1992;52:4824–7.
Zhao GQ, Wang T, Zhao Q, Yang HY, Tan XH, Dong ZM. Mutation of DNA polymerase beta in esophageal carcinoma of different regions. World J Gastroenterol. 2005;11:4618–22.
Feng L, Ma YY, Zhao GQ, Li M, Sun SJ, Dong ZM, et al. Establishment and characterization of DNA pol beta knockout human esophageal carcinoma cell line EC9706. Life Sci J. 2010;7:13–8.
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29.
Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80:827–41.
Li M, Zang W, Wang Y, Ma Y, Xuan X, Zhao J, et al. DNA polymerase beta mutations and survival of patients with esophageal squamous cell carcinoma in Linzhou City, China. Tumour Biol. 2014;35:553–9.
Li M, Zang W, Wang Y, Li Y, Ma Y, Wang N, et al. DNA polymerase beta promoter mutations and transcriptional activity in esophageal squamous cell carcinoma. Tumour Biol. 2013;34:3259–63.
Hiwasa T, Tokita H, Ike Y. Differential chemosensitivity in oncogene-transformed cells. J Exp Ther Oncol. 1996;1:162–70.
Vogt U, Falkiewicz B, Bielawski K, Bosse U, Schlotter CM. Relationship of c-myc and erbB oncogene family gene aberrations and other selected factors to ex vivo chemosensitivity of ovarian cancer in the modified ATP-chemosensitivity assay. Acta Biochim Pol. 2000;47:157–64.
Falkiewicz B, Schlotter CM, Bosse U, Bielawski K, Vogt U. c-myc oncogene gene dosage, serum CEA and CA-15.3 antigen levels, and cellular DNA values in relation to ex vivo chemosensitivity of primary human breast cancer. Acta Biochim Pol. 2000;47:149–56.
Wang Y, Chen X, Hu X, Zhang R, Du Y, Zang W, et al. Enhancement of silencing DNA polymerase β on the radiotherapeutic sensitivity of human esophageal carcinoma cell lines. Tumour Biol. 2014;35(10):10067–74.
Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA repair (Amst). 2005;4:1442–9.
McCullough AK, Dodson ML, Lloyd RS. Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem. 1999;68:255–85.
Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702.
Starcevic D, Dalal S, Sweasy JB. Is there a link between DNA polymerase beta and cancer? Cell Cycle. 2004;3:998–1001.
Poltoratsky V, Prasad R, Horton JK, Wilson SH. Down-regulation of DNA polymerase beta accompanies somatic hypermutation in human BL2 cell lines. DNA Repair (Amst). 2007;6:244–53.
Singhal RK, Prasad R, Wilson SH. DNA polymerase beta conducts the gap-filling step in uracil-initiated base excision repair in a bovine testis nuclear extract. J Biol Chem. 1995;270:949–57.
Sweasy JB, Lang T, DiMaio D. Is base excision repair a tumor suppressor mechanism? Cell Cycle. 2006;5:250–9.
Acknowledgments
This study was supported by National Natural Science Foundation of China (No. 81272188).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, Y., Sun, Q., Guo, W. et al. G648C variant of DNA polymerase β sensitizes esophageal cancer to chemotherapy. Tumor Biol. 37, 1941–1947 (2016). https://doi.org/10.1007/s13277-015-3978-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3978-x