Abstract
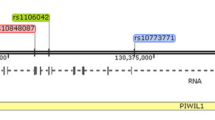
The kinesin-like factor 1 B (KIF1B) gene plays an important role in the process of apoptosis and the transformation and progression of malignant cells. Genetic variations in KIF1B may contribute to risk of epithelial ovarian cancer (EOC). In this study of 1,324 EOC patients and 1,386 cancer-free female controls, we investigated associations between two potentially functional single nucleotide polymorphisms in KIF1B and EOC risk by the conditional logistic regression analysis. General linear regression model was used to evaluate the correlation between the number of variant alleles and KIF1B mRNA expression levels. We found that the rs17401966 variant AG/GG genotypes were significantly associated with a decreased risk of EOC (adjusted odds ratio (OR) = 0.81, 95 % confidence interval (CI) = 0.68–0.97), compared with the AA genotype, but no associations were observed for rs1002076. Women who carried both rs17401966 AG/GG and rs1002076 AG/AA genotypes of KIF1B had a 0.82-fold decreased risk (adjusted 95 % CI = 0.69–0.97), compared with others. Additionally, there was no evidence of possible interactions between about-mentioned co-variants. Further genotype-phenotype correlation analysis indicated that the number of rs17401966 variant G allele was significantly associated with KIF1B mRNA expression levels (P for GLM = 0.003 and 0.001 in all and Chinese subjects, respectively), with GG carriers having the lowest level of KIF1B mRNA expression. Taken together, the rs17401966 polymorphism likely regulates KIF1B mRNA expression and thus may be associated with EOC risk in Eastern Chinese women. Larger, independent studies are warranted to validate our findings.

Similar content being viewed by others
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Yang N, Yan Y, Zheng R, Zhang S, Chen W. An analysis of incidence and mortality for ovarian cancer in China, 2009. China Cancer. 2013;22:617–21.
Hoskins WJ. Prospective on ovarian cancer: why prevent? J Cell Biochem Suppl. 1995;23:189–99.
Sato S, Yokoyama Y, Sakamoto T, Futagami M, Saito Y. Usefulness of mass screening for ovarian carcinoma using transvaginal ultrasonography. Cancer. 2000;89:582–8.
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300.
Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85.
Song H, Ramus SJ, Tyrer J, Bolton KL, Gentry-Maharaj A, Wozniak E, et al. A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat Genet. 2009;41:996–1000.
Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9.
Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–4.
Ratner E, Lu L, Boeke M, Barnett R, Nallur S, Chin LJ, et al. A kras-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70:6509–15.
Yarden RI, Friedman E, Metsuyanim S, Olender T, Ben-Asher E, Papa MZ. Single-nucleotide polymorphisms in the p53 pathway genes modify cancer risk in BRCA1 and BRCA2 carriers of Jewish-Ashkenazi descent. Mol Carcinog. 2010;49:545–55.
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7.
Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–96.
MacAskill AF, Kittler JT. Control of mitochondrial transport and localization in neurons. Trends Cell Biol. 2010;20:102–12.
Chen YY, Takita J, Chen YZ, Yang HW, Hanada R, Yamamoto K, et al. Genomic structure and mutational analysis of the human KIF1Balpha gene located at 1p36.2 in neuroblastoma. Int J Oncol. 2003;23:737–44.
Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M, et al. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem. 2008;283:24426–34.
Yeh IT, Lenci RE, Qin Y, Buddavarapu K, Ligon AH, Leteurtre E, et al. A germline mutation of the KIF1B beta gene on 1p36 in a family with neural and nonneural tumors. Hum Genet. 2008;124:279–85.
Dong Z, Xu X, Du L, Yang Y, Cheng H, Zhang X, et al. Leptin-mediated regulation of mt1-mmp localization is KIF1B dependent and enhances gastric cancer cell invasion. Carcinogenesis. 2013;34:974–83.
Henrich KO, Schwab M, Westermann F. 1p36 tumor suppression—a matter of dosage? Cancer Res. 2012;72:6079–88.
Aulchenko YS, Hoppenbrouwers IA, Ramagopalan SV, Broer L, Jafari N, Hillert J, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40:1402–3.
Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–8.
Li S, Qian J, Yang Y, Zhao W, Dai J, Bei JX, et al. Gwas identifies novel susceptibility loci on 6p21.32 and 21q21.3 for hepatocellular carcinoma in chronic hepatitis B virus carriers. PLoS Genet. 2012;8:e1002791.
Sawai H, Nishida N, Mbarek H, Matsuda K, Mawatari Y, Yamaoka M, et al. No association for Chinese HBV-related hepatocellular carcinoma susceptibility SNP in other East Asian populations. BMC Med Genet. 2012;13:47.
Zhong R, Tian Y, Liu L, Qiu Q, Wang Y, Rui R, et al. HBV-related hepatocellular carcinoma susceptibility gene KIF1B is not associated with development of chronic hepatitis B. PLoS One. 2012;7:e28839.
Zhang Z. Association between KIF1B rs17401966 polymorphism and hepatocellular carcinoma risk: a meta-analysis involving 17,210 subjects. Tumour Biol. 2014. doi:10.1007/s13277-13014-12192-13276.
Wang ZC, Gao Q, Shi JY, Yang LX, Zhou J, Wang XY, et al. Genetic polymorphism of the kinesin-like protein KIF1B gene and the risk of hepatocellular carcinoma. PLoS One. 2013;8:e62571.
He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP, et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–44.
Holm K, Melum E, Franke A, Karlsen TH. Snpexp—a web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinf. 2010;11:600.
Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, et al. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–20.
Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–6.
Theriault BL, Pajovic S, Bernardini MQ, Shaw PA, Gallie BL. Kinesin family member 14: an independent prognostic marker and potential therapeutic target for ovarian cancer. Int J Cancer. 2012;130:1844–54.
MacQuarrie KL, Fong AP, Morse RH, Tapscott SJ. Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet. 2011;27:141–8.
Shi TY, Zhu ML, He J, Wang MY, Li QX, Zhou XY, et al. Polymorphisms of the interleukin 6 gene contribute to cervical cancer susceptibility in eastern Chinese women. Hum Genet. 2013;132:301–12.
He BZ, Holloway AK, Maerkl SJ, Kreitman M. Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules. PLoS Genet. 2011;7:e1002053.
Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H, et al. The kinesin KIF1Bbeta acts downstream from egln3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884–93.
Yang HW, Chen YZ, Takita J, Soeda E, Piao HY, Hayashi Y. Genomic structure and mutational analysis of the human KIF1B gene which is homozygously deleted in neuroblastoma at chromosome 1p36.2. Oncogene. 2001;20:5075–83.
Galvan A, Ioannidis JP, Dragani TA. Beyond genome-wide association studies: genetic heterogeneity and individual predisposition to cancer. Trends Genet. 2010;26:132–41.
Acknowledgments
This work was supported by the funds from “China’s Thousand Talents Program” at Fudan University and by the funds from the Shanghai Committee of Science and Technology, China (Grant No.12DZ2260100, 12DZ2295100), as well as by the funds from China Recruitment Program of Global Experts at Fudan University, the Shanghai Committee of Science and Technology, China (Grant No. 12DZ2260100), Ministry of Science and Technology (2011BAI09B00), Ministry of Health (201002007), and the National Science Fund for Young Scholars (Grant No. 81402142). We would like to thank Wenjuan Tian and Wen Gao from Fudan University Shanghai Cancer Center and Xiaomei Zhang from Jiangsu Cancer Hospital for their support on clinical database.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Shi, TY., Jiang, Z., Jiang, R. et al. Polymorphisms in the kinesin-like factor 1 B gene and risk of epithelial ovarian cancer in Eastern Chinese women. Tumor Biol. 36, 6919–6927 (2015). https://doi.org/10.1007/s13277-015-3394-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3394-2