Abstract
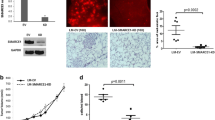
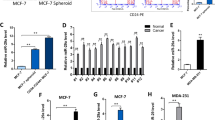
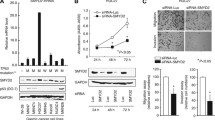
SMARCA5 partners with RSF-1 to compose the RSF complex, which belongs to the ISWI family of chromatin remodelers. Recent studies referred that SMARCA5 was overexpressed in some malignant tumors. However, expression pattern and biological roles of SMARCA5 in breast cancer have not been examined. In the present study, we found that SMARCA5 was overexpressed in breast cancer specimens by immunohistochemistry. Significant association was observed between SMARCA5 overexpression and TNM stage (p = 0.0199), tumor size (p = 0.0066), high proliferation index (p = 0.0366), and poor overall survival (p = 0.0141). SMARCA5 overexpression also correlated with Rsf-1 expression levels (p = 0.0120). Furthermore, colony formation assay and Matrigel invasion assay showed that knockdown of SMARCA5 expression in MDA-MB-231 and MDA-MB-435s cell lines with high endogenous expression decreased cell proliferation and cell invasion. Flow cytometry showed knockdown of SMARCA5-arrested cell cycle. Further analysis of cell cycle and invasion-related molecules showed that SMARCA5 downregulated cyclin A, MMP2 expression and upregulated p21 expression. In conclusion, our study demonstrated that SMARCA5 was overexpressed in human breast cancers and correlated with poor prognosis. SMARCA5 contributes to breast cancer cell proliferation and invasion.




Similar content being viewed by others
References
Chavarri-Guerra Y et al. Breast cancer in Mexico: a growing challenge to health and the health system. Lancet Oncol. 2012;13(8):e335–43.
Jemal A et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
Siegel R et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–41.
Ambrogi F et al. Molecular subtyping of breast cancer from traditional tumor marker profiles using parallel clustering methods. Clin Cancer Res. 2006;12(3 Pt 1):781–90.
Charpentier M, Martin S. Interplay of stem cell characteristics, EMT, and microtentacles in circulating breast tumor cells. Cancers (Basel). 2013;5(4):1545–65.
Cuppone F et al. Magnitude of risks and benefits of the addition of bevacizumab to chemotherapy for advanced breast cancer patients: meta-regression analysis of randomized trials. J Exp Clin Cancer Res. 2011;30:54.
Ahn S et al. The prognostic significance of tumor-associated stroma in invasive breast carcinoma. Tumour Biol. 2012;33(5):1573–80.
Kabbage M et al. Expression of the molecular chaperone alpha B-crystallin in infiltrating ductal breast carcinomas and the significance thereof: an immunohistochemical and proteomics-based strategy. Tumour Biol. 2012;33(6):2279–88.
Elfagieh M et al. Serum tumour markers as a diagnostic and prognostic tool in Libyan breast cancer. Tumour Biol. 2012;33(6):2371–7.
Kurbel S. In search of triple-negative DCIS: tumor-type dependent model of breast cancer progression from DCIS to the invasive cancer. Tumour Biol. 2013;34(1):1–7.
Kokavec J et al. Chromatin remodeling and SWI/SNF2 factors in human disease. Front Biosci. 2008;13:6126–34.
LeRoy G et al. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282(5395):1900–4.
Poot RA et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19(13):3377–87.
Strohner R et al. NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20(17):4892–900.
Aihara T et al. Cloning and mapping of SMARCA5 encoding hSNF2H, a novel human homologue of Drosophila ISWI. Cytogenet Cell Genet. 1998;81(3–4):191–3.
Buetow KH et al. Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1989;86(22):8852–6.
Lazzaro MA, Picketts DJ. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem. 2001;77(4):1145–56.
Chong S et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39(5):614–22.
Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc Natl Acad Sci U S A. 2003;100(24):14097–102.
Gigek CO et al. SMARCA5 methylation and expression in gastric cancer. Cancer Invest. 2011;29(2):162–6.
Stopka T et al. Chromatin remodeling gene SMARCA5 is dysregulated in primitive hematopoietic cells of acute leukemia. Leukemia. 2000;14(7):1247–52.
Reis ST et al. The role of micro RNAs let7c, 100 and 218 expression and their target RAS, C-MYC, BUB1, RB, SMARCA5, LAMB3 and Ki-67 in prostate cancer. Clinics (Sao Paulo). 2013;68(5):652–7.
Sheu JJ et al. The roles of human sucrose nonfermenting protein 2 homologue in the tumor-promoting functions of Rsf-1. Cancer Res. 2008;68(11):4050–7.
Sheu JJ et al. Rsf-1, a chromatin remodeling protein, induces DNA damage and promotes genomic instability. J Biol Chem. 2010;285(49):38260–9.
Loyola A et al. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol Cell Biol. 2003;23(19):6759–68.
Gerdes J et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133(4):1710–5.
Brown DC, Gatter KC. Monoclonal antibody Ki-67: its use in histopathology. Histopathology. 1990;17(6):489–503.
Cher ML et al. Cellular proliferation in prostatic adenocarcinoma as assessed by bromodeoxyuridine uptake and Ki-67 and PCNA expression. Prostate. 1995;26(2):87–93.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jin, Q., Mao, X., Li, B. et al. Overexpression of SMARCA5 correlates with cell proliferation and migration in breast cancer. Tumor Biol. 36, 1895–1902 (2015). https://doi.org/10.1007/s13277-014-2791-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2791-2