Abstract
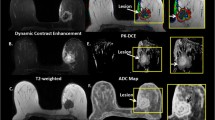
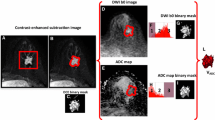
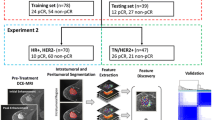
To explore radiomic features of pharmacokinetic dynamic contrast-enhanced (Pk-DCE) MRI on the extensive Tofts model to diagnose breast cancer and predict molecular phenotype. Breast lesions enrolled must undergo Pk-DCE MRI before treatment or puncture, and be identified as primary lesions by pathology. Ktrans, Kep, Ve and Vp were generated on the extensive Tofts model. Radiomic features (histogram, geometry and texture features) were extracted from parametric maps and selected by LASSO. The subjects were divided into training and validation cohort with a ratio of 4:1 to construct model in diagnosis of breast cancer. Feature analysis was made to predict the molecular phenotype. Area under curve (AUC), sensitivity, specificity and accuracy were used to evaluate radiomic features. DeLong’s test was performed to compare AUC values. 228 breast lesions met the criteria were used to discrimination and 126 malignant lesions were used to study molecular phenotypes. The number of training cohort and validation cohort were 182 and 46, respectively. The AUC of Ktrans, Kep, Ve, and Vp was 0.95, 0.93, 0.89, and 0.96, and their accuracy was 85%, 89%, 89%, 94% respectively in diagnosis of breast lesions, while their AUC was 0.71 to 0.77, 0.61 to 0.68, and 0.67 to 0.74 to predict ER/PR, Her-2, and Ki-67. There was no significant difference among parameters (P > 0.05). Radiomic features based on Pk-DCE MRI have an advantage to diagnose breast cancer and less ability to predict molecular phenotypes, which are beneficial to guide clinical treatment of breast lesions in some extent.


Similar content being viewed by others
Abbreviations
- Pk-DCE MRI:
-
Pharmacokinetic dynamic contrast-enhanced magnetic resonance imaging
- HR:
-
Hormone receptor
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- Her-2:
-
Human epidermal growth factor receptor 2
- ROC curve:
-
Receiver operating characteristic curve
- OR:
-
Odds ratio
- AUC:
-
Area under the curve
- ROI:
-
Region of interest
- VIF:
-
Variance inflation factor
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics. CA 68(6):394–424. https://doi.org/10.3322/caac.21442
Harbeck N, Gnant M (2017) Breast cancer. Lancet 389(1):1134–1150
DeSantis CE, Ma J, Goding Sauer A, Newman LA, Jemal A (2017) Breast cancer statistics, 2017, racial disparity in mortality by state. CA Cancer J Clin 67(6):439–448. https://doi.org/10.3322/caac.21412
Fusco R, Petrillo A, Petrillo M, Sansone M (2013) Use of tracer kinetic models for selection of semi-quantitative features for DCE-MRI data classification. Appl Magn Reson 44(11):1311–1324. https://doi.org/10.1007/s00723-013-0481-7
Sourbron S (2014) A tracer-kinetic field theory for medical imaging. IEEE Trans Med Imaging 33(4):935–946. https://doi.org/10.1109/TMI.2014.2300450
Jena A, Taneja S, Singh A, Negi P, Mehta SB, Sarin R (2017) Role of pharmacokinetic parameters derived with high temporal resolution DCE MRI using simultaneous PET/MRI system in breast cancer: a feasibility study. Eur J Radiol 86:261
Shin JK, Kim JY (2017) Dynamic contrast-enhanced and diffusion-weighted MRI of estrogen receptor-positive invasive breast cancers: associations between quantitative MR parameters and Ki-67 proliferation status. J Magn Reson Imaging 45(1):94–102
Sutton EJ, Dashevsky BZ, Oh JH, Veeraraghavan H, Apte AP, Thakur SB, Morris EA, Deasy JO (2016) Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging 44(1):122–129. https://doi.org/10.1002/jmri.25119
Khalifa F, Soliman A, El-Baz A, Abou El-Ghar M, El-Diasty T, Gimel'farb G, Ouseph R, Dwyer AC (2014) Models and methods for analyzing DCE-MRI: a review. Med Phys 41(12):124301. https://doi.org/10.1118/1.4898202
Thng CH, Koh TS, Collins DJ, Koh DM (2010) Perfusion magnetic resonance imaging of the liver. World J Gastroenterol 16(13):1598–1609
Park H, Lim Y, Ko ES, Cho HH, Lee JE, Han BK, Ko EY, Choi JS, Park KW (2018) Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res 24(19):4705–4714. https://doi.org/10.1158/1078-0432.CCR-17-3783
Wu J, Tha KK, Xing L, Li R (2018) Radiomics and radiogenomics for precision radiotherapy. J Radiat Res 59(suppl_1):i25–i31 Doi: 10.1093/jrr/rrx102.
Aerts HJ (2016) The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol 2(12):1636–1642. https://doi.org/10.1001/jamaoncol.2016.2631
Lambin P, Rth L, Deist TM, Peerlings J, Eec DJ, Van TJ, Sanduleanu S, Rthm L, Ajg E, Jochems A (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14(12):749
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278(2):563–577
Huang SY, Franc BL, Harnish RJ, Liu G, Mitra D, Copeland TP, Arasu VA, Kornak J, Jones EF, Behr SC, Hylton NM, Price ER, Esserman L, Seo Y (2018) Exploration of PET and MRI radiomic features for decoding breast cancer phenotypes and prognosis. NPJ Breast Cancer 4:24. https://doi.org/10.1038/s41523-018-0078-2
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Cavalho S, Bussink J, Monshouwer R, Haibekains B (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Agner SC, Soman S, Libfeld E, Mcdonald M, Thomas K, Englander S, Rosen MA, Chin D, Nosher J, Madabhushi A (2011) Textural kinetics: a novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J Digit Imaging 24(3):446–463
Fan M, Li H, Wang S, Zheng B, Zhang J, Li L (2017) Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLoS ONE 12(2):e0171683. https://doi.org/10.1371/journal.pone.0171683
Nie K, Chen J-H, Yu HJ, Chu Y, Nalcioglu O, Su M-Y (2008) Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad Radiol 15(12):1513–1525
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ, Panel m (2013) Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol 24(9):2206–2223. https://doi.org/10.1093/annonc/mdt303
Ahmed AR (2016) HER2 expression is a strong independent predictor of nodal metastasis in breast cancer. J Egypt Natl Canc Inst 28(4):219–227. https://doi.org/10.1016/j.jnci.2016.09.002
Filice S, Crisi G (2016) Dynamic contrast-enhanced perfusion MRI of high grade brain gliomas obtained with arterial or venous waveform input function. J Neuroimaging 26(1):124–129
Ning J, Schubert T, Johnson KM, Roldán-Alzate A, Chen H, Yuan C, Reeder SB (2017) Vascular input function correction of inflow enhancement for improved pharmacokinetic modeling of liver DCE-MRI. Magn Reson Med 79(6):3093–3102
Yang JF, Zhao ZH, Zhang Y, Zhao L, Yang LM, Zhang MM, Wang BY, Wang T, Lu BC (2016) Dual-input two-compartment pharmacokinetic model of dynamic contrast-enhanced magnetic resonance imaging in hepatocellular carcinoma. World J Gastroenterol 22(13):3652–3662. https://doi.org/10.3748/wjg.v22.i13.3652
Donaldson SB, West CM, Davidson SE, Carrington BM, Hutchison G, Jones AP, Sourbron SP, Buckley DL (2010) A comparison of tracer kinetic models for T1-weighted dynamic contrast-enhanced MRI: application in carcinoma of the cervix. Magn Reson Med 63(3):691–700. https://doi.org/10.1002/mrm.22217
El Khouli RH, Macura KJ, Kamel IR, Jacobs MA, Bluemke DA (2011) 3-T dynamic contrast-enhanced MRI of the breast: pharmacokinetic parameters versus conventional kinetic curve analysis. AJR Am J Roentgenol 197(6):1498–1505. https://doi.org/10.2214/AJR.10.4665
Li L, Wang K, Sun X, Wang K, Sun Y, Zhang G, Shen B (2015) Parameters of dynamic contrast-enhanced MRI as imaging markers for angiogenesis and proliferation in human breast cancer. Med Sci Monit 21:376–382. https://doi.org/10.12659/MSM.892534
Vallieres M, Boustead A, Laberge S, Levesque IR, Naqa IE (2015) A machine learning approach for creating texture-preserved MRI tumor models from clinical sequences. Med Phys 42(6):3323–3324
Damerjian V, Tankyevych O, Guellich A, Damy T, Petit E (2016) Ultrasound image texture characterization with Gabor wavelets for cardiac hypertrophy differentiation. In: IEEE International Symposium on Biomedical Imaging.
Parekh VS, Jacobs MA (2017) Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. NPJ Breast Cancer 3:43. https://doi.org/10.1038/s41523-017-0045-3
Mammoto A, Mammoto T, Kanapathipillai M, Wing YC, Jiang E, Jiang A, Lofgren K, Gee EP, Ingber DE (2013) Control of lung vascular permeability and endotoxin-induced pulmonary oedema by changes in extracellular matrix mechanics. Nat Commun 4(2):1759
Acknowledgements
This study was supported by the grant from Natural Science Foundation of Jiangsu Province (Grant No. BK20170626 to G. F.) and Jiangsu Provincial Natural Science Foundation (Grant No. 19KJB520025 to Xiaoyu Zhou). All authors thank Mr. Zhoushe Zhao, a scientist at GE Healthcare.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Ethical approval
This study was approved by the Ethics Committee of local hospital.
Informed consent
Informed consent was obtained from all study participants from October 2016 to June 2018.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhou, X., Gao, F., Duan, S. et al. Radiomic features of Pk-DCE MRI parameters based on the extensive Tofts model in application of breast cancer. Phys Eng Sci Med 43, 517–524 (2020). https://doi.org/10.1007/s13246-020-00852-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-020-00852-9