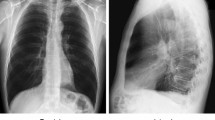
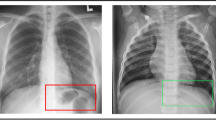
Abstract
Chest X-rays are used for mass screening for the early detection of lung cancer. However, lung nodules are often overlooked because of bones overlapping the lung fields. Bone suppression techniques based on artificial intelligence have been developed to solve this problem. However, bone suppression accuracy needs improvement. In this study, we propose a convolutional neural filter (CNF) for bone suppression based on a convolutional neural network which is frequently used in the medical field and has excellent performance in image processing. CNF outputs a value for the bone component of the target pixel by inputting pixel values in the neighborhood of the target pixel. By processing all positions in the input image, a bone-extracted image is generated. Finally, bone-suppressed image is obtained by subtracting the bone-extracted image from the original chest X-ray image. Bone suppression was most accurate when using CNF with six convolutional layers, yielding bone suppression of 89.2%. In addition, abnormalities, if present, were effectively imaged by suppressing only bone components and maintaining soft-tissue. These results suggest that the chances of missing abnormalities may be reduced by using the proposed method. The proposed method is useful for bone suppression in chest X-ray images.











Similar content being viewed by others
References
American Cancer Society (2018) Cancer facts and figures
Kobayashi T, Tsubura S, Katsuhara S, Kasai S, Sasano Y (2015) Development of bone suppression processing as an application of computer aided detection of nodules in chest radiographs. KONICA MINOLTA Technol Rep 12:71–76 (in Japanese)
Kelcz F, Zink FE, Peppler WW, Kruger DG, Ergun DL, Mistretta CA (1994) Conventional chest radiography vs. dual-energy computed radiography in the detection and characterization of pulmonary nodules. AJR Am J Roentgenol 162:271–278
Vock P, Szucs-Farkas Z (2009) Dual energy subtraction: principles and clinical applications. Eur J Radiol 72:231–237
Ishigaki T, Sakuma S, Horikawa Y, Ikeda M, Yamaguchi H (1986) One-shot dual-energy subtraction imaging. Radiology 161:271–273
Ishigaki T, Sakuma S, Ikeda M (1988) One-shot dual-energy subtraction chest imaging with computed radiography: clinical evaluation of film images. Radiology 168:67–72
Stewart BK, Huang HK (1990) Single-exposure dual-energy computed radiography. Med Phys 17:866–875
Ergun DL, Mistretta CA, Brown DE, Bystrianyk RT, Sze WK, Kelcz F, Nadich DP (1990) Single-exposure dual-energy computed radiography: improved detection and processing. Radiology 174:243–249
Whitman GJ, Niklason LT, Pandit M, Oliver LC, Atkins EH, Kinnard O, Alexander AH, Weiss MK, Sunku K, Schulze ES, Greene RE (2002) Dual-energy digital subtraction chest radiography: technical considerations. Curr Probl Diagn Radiol 31:48–62
Suzuki K, Abe H, Li F, Doi K (2004) Suppression of the contrast of ribs in chest radiographs by means of massive training artificial neural network. In: Proc SPIE, medical imaging 2004: image processing. 5370:1109–1119.
Suzuki K, Abe H, MacMahon H, Doi K (2006) Image-processing technique for suppressing ribs in chest radiographs by means of massive training artificial neural network (MTANN). IEEE Trans Med Imaging 25:406–416.
Chen S, Suzuki K (2014) Separation of bones from chest radiographs by means of anatomically specific multiple massive-training ANNs combined with total variation minimization smoothing. IEEE Trans Med Imaging 33:246–257
Zarshenas A, Liu J, Forti P, Suzuki K (2019) Separation of bones from soft tissue in chest radiographs: anatomy-specific orientation-frequency-specific deep neural network convolution. Med Phys 46:2232–2242
Chen S, Zhong S, Yao L, Shang Y, Suzuki K (2016) Enhancement of chest radiographs obtained in the intensive care unit through bone suppression and consistent processing. Phys Med Biol 61:2283–2301
Yang W, Chen Y, Liu Y, Zhong L, Qin G, Lu Z, Feng Q, Chen W (2017) Cascade of multi-scale convolutional neural networks for bone suppression of chest radiographs in gradient domain. Med Image Anal 35:421–433
Chen Y, Gou X, Feng X, Liu Y, Qin G, Feng Q, Yang W, Chen W (2019) Bone suppression of chest radiographs with cascaded convolutional networks in wavelet domain. IEEE Access 7:8346–8357
Zhou B, Lin X, Eck B, Hou J, Wilson D (2018) Generation of virtual dual energy images from standard single-shot radiographs using multi-scale and conditional adversarial network. arXiv:1810.09354v1
Oh DY, Yun ID (2018) Learning bone suppression from dual energy chest X-rays using adversarial networks. arXiv:1811.02628v1
Lo SB, Lou SA, Lin J, Freedman MT, Chien MV, Mun SK (1995) Artificial convolutional neural network techniques and applications for lung nodule detection. IEEE Trans Med Imaging 14:711–718
Cha KH, Hadjiiski L, Samala R, Chan H, Caoili EM, Cohan RH (2016) Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Med Phys 43:1882–1896
Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S (2016) Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging 35:1207–1216
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35:1240–1251
Dou Q, Chen H, Yu L, Zhao L, Qin J, Wang D, Mok VCT, Shi L, Heng P (2016) Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 35:1182–1195
Samala RK, Chen H, Hadjiiski L, Helvie MA, Wei J, Cha K (2016) Mass detection in digital breast tomosynthesis: deep convolutional neural network with transfer learning from mammography. Med Phys 43:6654–6666
Miki Y, Muramatsu C, Hayashi T, Zhou X, Hara T, Katsumata A, Fujita H (2017) Classification of teeth in cone-beam CT using deep convolutional neural network. Comput Biol Med 80:24–29
Ghafoorian M, Karssemeijier N, Heskes T, Bergkamp M, Wissink J, Obels J, Keizer K, de leeuw F, van Ginneken B, Marchiori E, Platel B (2017) Deep multi-scale location-aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. Neuroimage Clin 14:391–399
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Han X (2107) MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys 44:1408–1419.
Teramoto A, Fujita H, Yamamuro O, Tamaki T (2016) Automated detection of pulmonary nodules in PET/CT images: ensemble false-positive reduction using a convolutional neural network technique. Med Phys 43:2821–2827
Teramoto A, Yamada A, Kiriyama Y, Tsukamoto T, Yan K, Zhang L, Imaizumi K, Saito K, Fujita H (2019) Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inf Med Unlocked 16:100205
Matsubara N, Teramoto A, Saito K, Fujita H (2019) Generation of pseudo chest X-ray images from computed tomographic images by nonlinear transformation and bone enhancement. Med Image Inf Sci 36:141–146
Clark K, Vendt B, Smith K, Freymann J, Kirby JS, Koppel P, Moore SM, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F (2013) The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057
Armato III SG, McLennan G, Bidaut L, McNitt‐Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA (2011) The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38:915–931
Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. IEEE CVPR. arXiv:1705.02315
Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu K, Matsui M, Fujita H, Kodera Y, Doi K (2000) Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists' detection of pulmonary nodules. AJR Am J Roentgenol 174:71–74
Wang Z, Bovik AC, Sheikh HR, Simoncelli EP (2004) Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 13:1–14
Suzuki K, Horiba I, Sugie N (2002) Efficient approximation of neural filters for removing quantum noise from images. IEEE Trans Signal Process 50:1787–1799
Suzuki K, Horiba I, Sugie N, Nanki M (2002) Neural filter with selection of input features and its application to image quality improvement of medical image sequences. IEICE Trans Inf Syst 85:1710–1718.
Kobayashi T, Xu XW, MacMahon H, Metz CE, Doi K (1996) Effect of a computer-aided diagnosis scheme on radiologists' performance in detection of lung nodules on radiographs. Radiology 199:843–848.
Abe H, MacMahon H, Engelmann R, Li Q, Shiraishi J, Katsuragawa S, Aoyama M, Ishida T, Ashizawa K, Metz CE, Doi K (2003) Computer-aided diagnosis in chest radiography: results of large-scale observer tests at the 1996–2001 RSNA scientific assemblies. Radiographics 23:255–265.
Kakeda S, Moriya J, Sato H, Aoki T, Watanabe H, Nakata H, Oda N, Katsuragawa S, Yamamoto K, Doi K (2004) Improved detection of lung nodules on chest radiographs using a commercial computer-aided diagnosis system. AJR Am J Roentgenol 182:505–510.
Xu XW, Doi K, Kobayashi T, MacMahon H, Giger ML (1997) Development of an improved CAD scheme for automated detection of lung nodules in digital chest images. Med Phys 24:1395–1403
Matsumoto T, Yoshimura H, Doi K, Giger ML, Kano A, MacMahon H, Abe K, Montner SM (1992) Image feature analysis of false-positive diagnoses produced by automated detection of lung nodules. Invest Radiol 27:587–597
Acknowledgements
This research was partially supported by a Grant-in-Aid for Scientific Research on Innovative Areas (Multidisciplinary Computational Anatomy, No. 26108005) and a Grant-in-Aid for Scientific Research (No. 17K09070), MEXT, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
This article does not contain patient data.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Matsubara, N., Teramoto, A., Saito, K. et al. Bone suppression for chest X-ray image using a convolutional neural filter. Phys Eng Sci Med 43, 97–108 (2020). https://doi.org/10.1007/s13246-019-00822-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-019-00822-w