Abstract
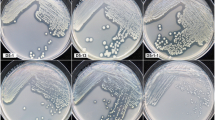
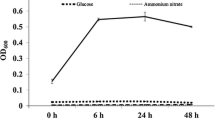
Three bacterial strains capable of degrading atrazine were isolated from Manfredi soils (Argentine) using enrichment culture techniques. These soils were used to grow corn and were treated with atrazine for weed control during 3 years. The strains were nonmotile Gram-positive bacilli which formed cleared zones on atrazine solid medium, and the 16S rDNA sequences indicated that they were Arthrobacter sp. strains. The atrazine-degrading activity of the isolates was characterized by the ability to grow with atrazine as the sole nitrogen source, the concomitant herbicide disappearance, and the chloride release. The atrazine-degrader strain Pseudomonas sp. ADP was used for comparative purposes. According to the results, all of the isolates used atrazine as sole source of nitrogen, and sucrose and sodium citrate as the carbon sources for growth. HPLC analyses confirmed herbicide clearance. PCR analysis revealed the presence of the atrazine catabolic genes trzN, atzB, and atzC. The results of this work lead to a better understanding of microbial degradation activity in order to consider the potential application of the isolated strains in bioremediation of atrazine-polluted agricultural soils in Argentina.




Similar content being viewed by others
References
Aislabie J, Hunter D, Ryburn J, Fraser R, Northcott GL, Di HJ (2004) Atrazine mineralization in New Zealand soils are affected by time since atrazina exposure. Aust J Soil Res 42:783–792
Aislabie J, Bej AK, Ryburn J, Lloyd N, Wilkins A (2005) Characterization of Arthrobacter nicotinovorans HIM, an atrazine-degrading bacterium, from agricultural soil of New Zeland. FEMS Microbiol Ecol 52:279–286
Alvey S, Crowly E (1996) Survival and activity of an atrazine-mineralizing bacterial consortium in rhizosphere soil. Environ Sci Technol 30:1596–1603
Anderson TA, Guthrie EA, Walton BT (1993) Bioremediation. Environ Sci Technol 27(13):2630–2636
Arbeli Z, Fuentes C (2010) Prevalence of the gene trzN and biogeographic patterns among atrazine-degrading bacteria isolated from 13 Colombian agricultural soils. FEMS Microbiol Ecol 73:611–623
Barriuso E, Houot S (1996) Rapid mineralization of the s-triazine ring of atrazine in soils in relation to soil management. Soil Biol Biochem 28:1341–1348
Bedmar F, Costa JL, Suero E, Gimenez D (2004) Transport of atrazine and metribuzin in three soils of the humid pampas of Argentina. Weed Technol 18:1–8
Behki R, Topp E, Dick W, Germon P (1993) Metabolism of the herbicide atrazine by Rhodococcus strains. Appl Environ Microbiol 59:1955–1959
Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cai B, Han Y, Liu B, Ren Y, Jiang S (2003) Isolation and characterization of an atrazine-degrading bacterium from industrial wastewater in China. Lett Appl Microbiol 36:272–276
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:141–145
De Schrijber A, De Mot R (1999) Degradation of pesticides by actinomycetes. Crit Rev Microbiol 25(2):85119
De Souza ML, Wackett LP, Boundy-Mills KL, Mandelbaum RT, Sadowsky MJ (1995) Cloning, characterization, and expression of a gene region from Pseudomonas sp. strain ADP involved in the dechlorination of atrazine. Appl Environ Microbiol 61:3373–3378
De Souza M, Seffernick J, Martinez B, Sadowsky M, Wackett LP (1998) The atrazine catabolism genes atzABC are widespread and highly conserved. J Bacteriol 180:1951–1954
Delmonte AA, Bedmar F, Mantecon JD, Echeverria H, Barassi CA (1997) Residual phytotoxicity and chemical persistence of atrazine in soils of the southeast of Buenos Aires Province, Argentina. J Environ Biol 18:201–207
Devers M, Soulas G, Martin-Laurent F (2004) Real-time reverse transcription PCR analysis of expression of atrazine catabolism genes in two bacterial strains isolated from soil. J Microbiol Methods 56:3–15
Fernández LA, Perotti EB, Sagardoy MA, Gómez MA (2008) Denitrification activity of Bradyrhizobium sp. isolated from Argentine soybean cultivated soils. W J Microbiol Biotech 24:2577–2585
Focht DD (1994) Microbiological procedures for biodegradation research. In: SSSA (eds). Methods of soil analysis. Part 2: Microbiological and Biochemical Properties. A. Klute, Madison, pp 407–425
Forlani G, Mangiagalli A, Nielsen E, Suardi CM (1999) Degradation of the phosphate herbicide glyphosate in soil: evidence for a possible involvement of unculturable microorganisms. Soil Biol Biochem 31:991–997
Fuscaldo F, Bedmar F, Monterubbianesi G (1999) Persistence of atrazine, metribuzin and simazine herbicides in two soils. Pesq Agropec Bras 34:2037–2044
Getenga ZM, Dörfler U, Iwobi A, Schmid M, Schroll R (2009) Atrazine and terbuthylazine mineralization by an Arthrobacter sp. isolated from a sugarcane-cultivated soil in Kenya. Chemosphere 77:534–539
Hang S, Barriuso E, Houot S (2003) Behavior of 14C-Atrazine in Argentinean Topsoils under Different Cropping Managements. J Environ Qual 32:2216–2222
Hang S, Barriuso E, Houot S (2005) Atrazine behaviour in the different pedological horizons of two Argentinean non-till soil profiles. Weed Res 45:130–139
Houot S, Topp E, Yassir A, Soulas G (2000) Dependence of accelerated degradation of atrazine on soil pH in French and Canadian soils. Soil Biol 32:615–625
Li Q, Zhu X, Cai B (2008) Isolation and characterization of atrazine-degrading Arthrobacter sp. AD26 and use of this strain in bioremediation of contaminated soil. J Environ Sci 20:1226–1230
Mandelbaum RT, Wackett LP, Allan DL (1993) Mineralization of the s-triazine ring of atrazine by stable bacterial mixed culture. Appl Environ Microbiol 59:1695–1701
Mandelbaum RT, Allan DL, Wackett LP (1995) Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide Atrazine. Appl Environ Microbiol 61:1451–1457
Marecik R, Kroliczak P, Czaczyk K, Bialas W, Olejnik A, Cyplik P (2008) Atrazine degradation by aerobic microorganisms isolated from the rhizosphere of sweet flag (Acorus calamus L.). Biodegradation 19:293–301
Martínez BJ, Tomkins LP, Wackett LP, Wing R, Sadowsky MJ (2001) Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J Bacteriol 183:5684–5697
Martin-Laurent F, Cornet L, Ranjard L, López-Gutiérrez JC, Philippot L, Schawartz C, Chaussod R, Catroux G, Soulas G (2004) Estimation of atrazine-degrading genetic potential and activity in three French agricultural soils. FEMS Microbiol Ecol 48:425–435
Picard C, Di Cello F, Vestura M, Fani R, Guckert A (2000) Frequency and biodiversity of 2,4 diacetylphloroglucinol producing bacteria isolated from the maize rhizosphere at different stages of plant growth. Appl Environ Microbiol 66:948–955
Piutti S, Semon E, Landry D, Hartmann A, Dousset S, Lichtfouse E, Topp E, Soulas G, Martin-Laurent F (2003) Isolation and characterization of Nocardioides sp. SP12, an atrazine-degrading bacterial strain possessing the gene trzN from bulk – and maize rhizosphere soil. FEMS Microbiol Lett 221:111–117
Popov VH, Cornish PS, Sultana K, Morris EC (2005) Atrazine degradation in soils: the role of microbial communities, atrazine application history, and soil carbon. Aust J Soil Res 43:861–871
Ralebits TK, Senior E, Verseveld HW (2002) Microbial aspects of atrazine degradation in natural environments. Biodegradation 13:11–19
Roberts TR (1998) 1,3,5-triazines. In: Roberts TR (ed) Metabolic pathway of agrochemicals. Part 1: Herbicides and plant growth regulators. The Royal Society of Chemistry, Cambridge, pp 623–635
Rousseaux S, Hartmann A, Soulas G (2001) Isolation and characterization of new Gram negative and Gram positive atrazine degrading bacteria from different French soils. FEMS Microbiol Ecol 36:211–222
Rousseaux S, Soulas G, Hartmann A (2002) Plasmid localisation of atrazine-degrading genes in newly described Chelatobacter and Arthrobacter strains. FEMS Microbiol Ecol 41:69–75
Sajjaphan K, Shapir N, Wackett LP, Palmer M, Blackmon B, Tomkins J, Sadowsky MJ (2004) Arthrobacter aurescens TC1 atrazine catabolism genes trzN, atzB, and atzC are linked on a 160-kb region and are functional in Escherichia coli. Appl Environ Microbiol 70:4402–4407
Sajjaphan K, Pimpak H, Sadowsky MJ, Boonkerd N (2010) Arthrobacter sp. strain KU001 Isolated from a Thai soil degrades atrazine in the presence of inorganic nitrogen sources. J Microbiol Biotech 20(3):602–608
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Satsuma K (2006) Characterisation of new strains of atrazine-degrading Nocardioides sp. isolated from Japanese riverbed sediment using naturally derived river ecosystem. Pest Manag Sci 62(4)
Singh P, Suri CR, Singh Cameotra S (2004) Isolation of a member of Acinetobacter species involved in atrazine degradation. Biochem Bioph Res Comm 317:697–702
Smith D, Alvey S, Crowley DE (2005) Cooperative catabolic pathways within an atrazine-degrading enrichment culture isolated from soil. FEMS Microbiol Ecol 53:265–273
Strong LC, Rosendahl C, Johnson G, Sadowsky MJ, Wackett LP (2002) Arthrobacter aurescens TC1 metabolizes diverse s-triazine ring compounds. Appl Environ Microbiol 68(12):5973–5980
Topp E, Nour SM, Cuppels D, Houot S, Lewis M (2000a) Characterization of an atrazine-degrading Pseudominabacter isolated from Canadian and French agricultural soils. App Environ Microbiol 66:3134–3141
Topp E, Mulbry WM, Zhu H, Nour SM, Cuppels D (2000b) Characterization of s-triazine herbicide metabolism by a Nocardioides sp. isolated from agricultural soils. App Environ Microbiol 66:3134–3141
Vibber LL, Pressler MJ, Colores GM (2007) Isolation and characterization of novel atrazine-degrading microorganisms from an agricultural soil. Appl Microbiol Biotechnol 75:921–928
Wenk M, Baumgartner T, Dobovsek J, Fuchs T, Kucsera J, Zopfi SG (1998) Rapid atrazine mineralisation in soil slurry and moist soil by inoculation of an atrazine-degrading Pseudomonas sp. strain. Appl Microbiol Biotechnol 49:624–630
Yassir A, Lagacherie B, Houot S, Soulas G (1999) Microbial aspects of atrazine biodegradation in relation to history of soil treatment. Pest Sci 55:799–809
Zhu X, Li Q, Cai B (2009) Isolation and identification of Arthrobacter sp. AD26 and joint degradation of atrazine by Arthrobacter sp. AD26 and Pseudomonas sp.ADP. J Agro-Environ Sci. doi: CNKI:SUN:NHBH.0.2009-03-042
Acknowledgments
The authors are thankful to Universidad Nacional del Sur for financial support of the research work, through PGI No. 24/A112 and to CONICET for providing a fellowship to Leticia A. Fernández. We would like to thank Peter Daniel from Facultad de Ciencias Agrarias, Universidad Nacional de Mar del Plata (Balcarce, Argentina) for permitting access to HPLC analysis and for helping to perform them. We also want to thank Fabrice Martin-Laurent from INRA-CMSE, Dijon Cedex (France) for providing the primers to detect atrazine degradation genes and Ana María Zamponi, CONICET (Argentina), for her helpful technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fernández, L.A., Valverde, C. & Gómez, M.A. Isolation and characterization of atrazine-degrading Arthrobacter sp. strains from Argentine agricultural soils. Ann Microbiol 63, 207–214 (2013). https://doi.org/10.1007/s13213-012-0463-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-012-0463-2