Abstract
Anti-coronavirusdisease-2019 (COVID-19; anti-severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2)) antibodies against nucleoprotein (NP) were purified from pig sera. Following the separation of the antibody fraction using a protein-A column, the final yield of the purified antibodies against SARS-CoV-2 NPs was estimated to be 0.26 ± 0.05 % (absolute amount of 143.4 ± 25.2 ng, n=5) from 1 mL of pig sera. The binding activities of the isolated antibodies were confirmed using immunoassay and immunostaining. Based on the specific binding activity to NPs, a quantitative assay was performed using a surface plasmon resonance (SPR) biosensor. From the doseresponse curve, the binding constant (Kd) was calculated to be 185 pM and the limit of detection was estimated to be 1.02 pM. The SPR biosensor with the isolated antibodies against SARS-CoV-2 NPs was applied for the detection of SARS-CoV-2, MERS-CoV, and CoV strain 229E in culture fluid.
Similar content being viewed by others
Introduction
Coronavirus (CoV) has been classified into four genera: alpha, beta, gamma, and delta, with humans being infected by the former two coronaviruses1. CoVs such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), SARS-CoV, and Middle East respiratory syndrome–related coronavirus (MERS-CoV), which are all beta viruses, cause acute respiratory diseases such as coronavirus disease-2019 (COVID-19), SARS, and MERS, respectively. These beta-CoVs also infect pigs (alpha-CoV and beta-CoV), dogs (alpha-CoV), cats (alpha-CoV), bats (beta-CoV), horses (beta-CoV), and cows (beta-CoV)2. Specifically, the porcine epidemic diarrhea virus (PEDV; alpha-CoV), the transmissible gastroenteritis virus (TGEV; alpha-CoV), and the porcine hemagglutinating encephalomyelitis virus (PHEV; beta-CoV) are known to affect pigs.
The CoV genome encodes for four major structural proteins—spike protein, envelope protein, membrane protein, and nucleoprotein (NP). Among these, NP is known to be highly immunogenic and is abundantly expressed during infections3–6. It contains N- and C-terminal binding domains with a linkage region. As a result, it is frequently used in vaccine development and serological assays7. NP enters the host cell with viral RNA and participates in viral replication, particle assembly, and release8.
The SARS-CoV-2 NP is 419 amino acid long and has a molecular weight of 50 kDa9. The amino acid homology between SARS-CoV-2 and the pig-infecting beta-CoV was compared for PEDV, TGEV, and PHEV using the Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov). As summarized in Table 1, we analyzed PHEV (beta-CoV), with 334 amino acid residues to be 40% identical (amino acids) and 55% similar (chemical properties) to SARS-CoV-2 NP. The other two pig-infecting alpha-CoVs, TGEV and PEDV, were 28% (48%), and 27% (40%) identical (similar), respectively. This sequence analyses demonstrate the highly homologous amino acid sequences across NP in different porcine CoVs and that of SARS-CoV-2 (Table 2, 3)10, 11.
Recently, the purification of antibodies against a specific strain of bacteria according to the type of lipopolysaccharides (LPS) on the cell membrane was reported using the bacteria as an affinity material, and the purified antibodies were demonstrated to be used for the detection of the specific bacteria strain12. Here, we demonstrated the use of SARS-CoV-2 NP as an affinity material for the isolation of anti-SARS-CoV-2 antibodies from pig sera, to aid in the diagnosis. First, SARS-CoV-2 NP, highly homologous with the virus affecting pigs was immobilized on magnetic beads. The antibodies against SARS- CoV-2 NP were isolated from pig sera. The specific binding properties of the antibodies were then analyzed using immunostaining and immunoassays. Lastly, SARS-CoV-2 was detected in culture fluid using the surface plasmon resonance (SPR) biosensor with the immobilized antibodies.
Results and Discussion
Purification of Anti-SARS-CoV-2 Antibodies from Pig Sera
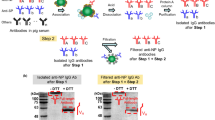
The anti-SARS-CoV-2 antibodies were purified using NPs, which were immobilized on magnetic beads as shown in Figure 1(a). After incubation of the magnetic beads with immobilized NP in pig sera, the bound proteins were dissociated using an acidic treatment. The dissociated proteins included antibodies against SARS-CoV-2 NP as well as other kinds of proteins. We used the protein-A column to separate the IgG fraction from the dissociated proteins. The antibody fraction was used for further assays. The final yield of the purified antibodies against SARS-CoV-2 NP was estimated to be 0.26 ± 0.05 % (absolute amount of 143.4 ± 25.2 ng, n=5) from 1 mL pig sera, using the bicinchoninic acid (BCA) assay method for total protein analysis12.
The purified antibodies against SARS-CoV-2 NP were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). As the immunoglobulin G (IgG) has four peptide chains connected with disulfide bonds, two heavy chains and two light chains could be released by reduction of disulfide bonds with a reducing agent of dithiothreitol (DTT). As shown in Figure 1(b), the protein bands from the heavy (molecular weight of 55 kDa) and light (molecular weight of 25 kDa) chains were observed. For isolated antibodies without the reduction with DTT, full-length antibodies were observed. However, no protein bands from the heavy and light chains were observed. These results showed that the purified protein, using magnetic beads with immobilized NPs and the protein-A column was a part of the antibody fraction (IgG’s)12, 13.
Binding Properties of Purified Anti-SARS-CoV-2 Antibodies to NP
The specific binding property of the isolated antibodies against SARS-CoV-2 NP to NP was estimated using an immunoassay and compared across various immobilized proteins on the microplate through immunostaining the magnetic beads with immobilized NP. The immobilized NPs were used for the estimation of the specific binding of isolated antibodies. First, five kinds of proteins (SARS-CoV-2 NP, Influenza A/New Caledonia/20/99, Influenza B/Florida/07/04, Influenza B/Tokio/53/99, Hepatitis B virus surface antigen (HBsAg)) were immobilized on a microplate (96-well) and incubated with isolated antibodies against SARS-CoV-2 NP. The number of bound antibodies to the NP was measured colorimetrically using secondary antibodies labeled with horseradish peroxidase (HRP) and 3,3’, 5,5’-tetramethylbenzidine (TMB) as the chromogenic agents. As shown in Figure 2(a), Influenza B (Florida/Tokio) and HBsAg showed 30 % greater binding to the isolated antibodies against SARS-CoV-2 NP than against NP-coated microplates. Influenza A (New Caledonia) showed no binding to the isolated antibodies against SARS-CoV-2 NP. These results demonstrated that the isolated antibodies against SARS-CoV-2 NP were specifically binding to NPs.
To confirm specific binding, we immunostained the antibodies using magnetic beads with immobilized NPs. The secondary anti-pig IgG labeled with fluorescein was treated. As shown in Figure 2(b), the magnetic beads with immobilized NP showed a significantly higher fluorescence intensity in comparison with the BSA coated magnetic beads. These results demonstrated that the isolated antibodies against SARS-CoV-2 NP were specific to NP.
Quantitative Analysis of SARS-CoV-2 NP Using Anti-SARS-CoV-2 Antibodies Derived from Pig Sera
The specific binding properties of the isolated antibodies against SARS-CoV-2 NP were used for the quantitative analysis of NPs using SPR biosensor. The isolated antibodies against SARS-CoV-2 NP were immobilized on the SPR biosensor through covalent binding. The gold surface of the SPR biosensor was modified to incorporate a carboxylic acid using mercaptoundecanoic acid. The isolated antibodies were covalently bound using the N′-ethylcarbodiimide/ N-Hydroxysuccinimide (EDC/NHS) reagent as shown in Figure 3(a). The SPR signal for each sample was calculated as the change in signal-baseline prior to and after sample incubation. The signal-baseline was established from the averaged SPR signals following three washes.
SPR biosensor for the immunoassay of NP using the isolated antibodies against SARS-CoV-2 NP from pig sera. (a) Covalent immobilization of NP to the modified surface of SPR chip. (b) SPR sensorgram for the treatments of the isolated anti-SARS-CoV-2 and anti-CRP antibodies. (c) SPR sensor responses for the treatments of the isolated anti-SARS-CoV-2 NP and anti-CRP antibodies. (d) Dose-response curve from SPR measurements at different concentrations of isolated antibodies against SARS-CoV-2 NP from pig sera.
The SPR biosensors (Figure 3(b)) were prepared to separately inject samples of the purified anti-SARS-CoV-2 and anti-CRP antibodies. Larger SPR signals were observed in samples with SARS-CoV-2 antibodies than in samples with anti-CRP antibodies. As shown in Figure 3(c), the SPR signals from the antibodies at 100 ng/mL and 1 μg/mL were estimated to be 48.3 and 111.3 arbitrary units (AU), respectively. In the case of anti-CRP antibodies at the concentration of 100 ng/mL and 1 μg/mL, the SPR signals were estimated to be 3.2 and 11.0 AU, respectively. These results showed that the specific binding of the isolated purified anti-SARS-CoV-2 antibodies to NP could be detected using the SPR biosensor.
We used the SPR biosensor to detect NP by immobilizing the purified anti-SARS-CoV-2 antibodies on a modified gold surface. As shown in Figure 3(d), the SPR signal was measured for samples of NP at the concentration of 2, 5, 10, 20, 50, 100, 200, 500, 1000 ng/mL. The SPR signal according to the concentration of NPs was calculated as mentioned above. The dose-response curve for the samples with NPs was carried out by the isotherm model:
where R (Rmax) represented the SPR signal (maximum SPR signal), [Ag] represented the NP concentration and Kd represented affinity constant of the isolated purified anti-SARS-CoV-2 antibodies to the NP. The SPR analysis of NP was possible at the concentration range of 2–1000 ng/mL, and the limit of detection was estimated to be 1.02 pM from the three standard values of averaged baseline signal. From the curve fitting based on the isotherm model, the affinity constant (Kd) was estimated to be 185 pM. These results represented that the isolated purified anti-SARS-CoV-2 antibodies from pig sera could specifically bind to NP. The quantitative analyses of NP were feasible using the SPR biosensors.
Immunoassay of SARS-CoV-2 Using Anti-SARS-CoV-2 Antibodies from Pig Sera
The feasibility of the medical diagnosis of SARS-CoV-2 was tested using the isolated anti-SARS-CoV-2 NP antibodies and culture fluid of SARS-CoV-2. As the culture fluid of SARS-CoV-2 included the intact virus and NP from the disrupted virus. The SPR biosensor was prepared by immobilizing the purified anti-SARS-CoV-2 NP antibodies on the modified gold surface of SPR biosensor as shown in Figure 4(a). And then, the immunoassay of SARS-CoV-2 in culture fluid was carried out using the SPR biosensor with the immobilized isolated antibodies as shown in Figure 4(b). Culture fluids of SARS-CoV-2 with dilution factors of 100 (1 %), 333 (0.3 %), 1000 (0.1 %), 3333 (0.03 %), 10000 (0.01 %) were used as standards (Figure 4(b)). For the comparison of the sensor response with other types of CoVs, the diluted culture fluid of MERS-CoV and CoV strain 229E were used as negative samples. For the cutoff establishment for the medical diagnosis, the lateral flow immunoassay for SARS-CoV-2 antigen detection from SD Biosensors Co was used as a reference method. The cutoff for the determination of a positive SARS-CoV-2 test was the culture fluid at a dilution factor of 927 (0.11 %). As shown in Figure 4(b), the response from the SPR biosensor was plotted for the SARS-CoV-2 culture fluid at a dilution factor of 10000 (0.01 %) - 100 (1 %). The cutoff level for the SPR biosensor was estimated to be 171 RU for a known dilution factor of 927 (0.11 %). These results showed that the SPR biosensor with purified antibodies against SARS-CoV-2 NP can be used for the detection of SARS-CoV-2 in culture fluid at the cutoff level for medical diagnoses. As shown in Figure 4(c), the dose-response curves for the culture fluid from SARS-CoV-2, MERS-CoV, and the CoV strain 229E were compared to estimate selectivity. The SARS-CoV-2 culture fluid had a higher sensitivity than the other two strains. The response also showed a quantitative increase according to the concentration of SARS-CoV-2 culture fluid. The detection range in the SARS-CoV-2 culture fluid was established at dilution factors within the range of 10000 (0.01 %)–100 (1 %). The detection limit was estimated to be at a dilution factor of 8333 (0.012 %). For MERS-CoV and CoV strain 229E, the sensor response was observed to be at a baseline level. This low sensitivity to MERS-CoV and CoV strain 229E can be explained by the amino acid homology. In MERS-CoV, NP had an identity (similarity) of 48 % to that of SARS-CoV-2 NP. CoV strain 229E, an alpha-type CoV, had a relatively low identity (similarity) of 29 % to SARS-CoV-2 NP. These results showed that the isolated antibodies against SARS-CoV-2 NP from pig sera could be used for the selective detection of this virus.
SPR biosensor for the immunoassay of SARS-CoV-2 in culture fluid. (a) Immobilization of the isolated antibodies against SARS-CoV-2 NP from pig sera to the modified SPR biosensor surface. (b) SPR sensorgram for the treatments of SARS-CoV-2 culture fluids at different dilution factors. The lateral flow immunoassay for SARS-CoV-2 antigen detection from SD Biosensors Co. was used as a reference method. (c) The dose-response curves of the SPR biosensor for the culture fluid of SARS-CoV-2, MERS-CoV, CoV strain 229E for the estimation of selectivity.
Conclusions
In this study, anti-SARS-CoV-2 NP (nucleoprotein) was isolated from pig sera without immune-stimulation through the injection of antigen. The basic idea of this work was begun from the fact that the amino acid homology between SARS-CoV-2 and the piginfecting beta-CoV such as PHEV, TGEV, PEDV had a high homology in amino acid sequence. The anti-SARS-CoV-2 antibodies were isolated using NP and immobilized on magnetic beads. After the isolation of antibody fraction using protein-A column, the final yield of the purified antibodies against SARS-CoV-2 NP was estimated to be 0.26 ± 0.05 % (absolute amount of 143.4 ± 25.2 ng, n=5) from 1 mL of pig sera. The specific binding properties of purified antibodies against SARS-CoV-2 NP were estimated using immunoassay and immunostaining. They were also quantified using the SPR biosensor. The SPR biosensor was feasible within the concentration range of 2–1000 ng/mL. The detection limit was estimated to be 1.02 pM from three standard values of averaged baseline signals. From the curve fitting based on the isotherm model, the affinity constant (Kd) was estimated to be 185 pM. These results represented that the isolated antibodies against SARS-CoV-2 NP from pig sera had specific binding activities to NP. Lastly, we used the SPR biosensor with the isolated antibodies to detect SARS-CoV-2, MERS-CoV, and CoV strain 229E in culture fluids. The SPR biosensor showed that the isolated antibodies against SARS-CoV-2 NP from pig sera could be used for selective detection of SARS-CoV-2 against MERS-CoV and CoV strain 229E. These results showed that the anti-SARS-CoV-2 NP could be isolated from pig sera without immune-stimulation and the isolated antibody from pig sera could be effectively used for the medical diagnosis of anti-SARS-CoV-2 with the selectivity against other corona virus such as MERS-CoV and CoV strain 229E.
Materials and Methods
Materials
Bovine serum albumin (BSA) and other chemicals were purchased from Sigma-Aldrich Korea (Seoul, Korea). The anti-pig IgG antibodies labeled with horseradish peroxidase (anti-pig(HRP)) or fluorescein (ab9611) were purchased from Abcam (Cambridge, UK). The bicinchonic acid (BCA) assay kit, pig sera (porcine serum), 3,3’,5,5’-tetramethylbenzidine (TMB) reagentwere purchased from Thermo Fisher Scientific (Waltham, MA, USA). The tosyl-activated magnetic beads (diameter of 2.8 µm) were purchased from Invitrogen Co (Carlsbad, CA, USA). The SARS-CoV-2 nucleoprotein (SARS-CoV-2 NP) with the His-tag was supplied from Genbody Inc (Cheonan, Korea) and Optolane Inc (Seongnam, Korea), respectively. The influenza viruses (R86380 (A/New caledonia/20/99), R01247 (B/Florida/07/04), R86250 (B/ Tokio/53/99)) were produced from Meridian Life Science Inc (Memphis, TN, USA). Culture fluids of SARS-CoV-2, MERS-CoV, CoV strain 229E were purchased from Zeptometrix (Buffalo, NY, USA).
Purification of Anti-SARS-CoV-2 Antibodies
The isolation of antibodies against SARS-CoV-2 NP from pig sera was carried out using magnetic beads coated with NP via the following steps as shown in Fig. 1(a): (1) The pig sera (1 mL) was incubated with magnetic beads coated with NP (10 mg of beads) for 1 h under mild mixing conditions (15 rpm) using a wheel-rotating mixer (RT-10, Daehan Scientific Co, Wonju, Korea). The NPs were immobilized to the tosylactivated magnetic beads by incubation of magnetic beads at the amount of 10 mg and NP (800 μg/mL, 250 μL) for 16 h at 37 °C. (2) The bound proteins were dissociated from the magnetic beads via treatment with 1 mL of 0.1 M glycine-HCl buffer (pH 2.7) for 10 s. Then, the reaction was neutralized by the addition of 50 μL of 1 M Tris-HCl buffer (pH 10.5). (3) For the isolation of antibody fraction, the dissociated proteins were loaded onto the protein-A column and washed with 10 mL of PBS to any unbound proteins. The isolated antibodies against SARS-CoV-2 NP were eluted by loading 2 mL of 0.1 M glycine-HCl buffer (pH 2.7) and directly collected in a microcentrifuge tube containing 100 μL of 1 M Tris-HCl buffer (pH 10.5). Lastly, the antibodies were concentrated via Amicon centrifugal filtration (molecular weight cutoff: 3 kDa). The yield of antibodies was defined as the amount of protein separated from a unit volume (1 mL) of pig sera. The amount of protein was calculated using a BCA assay kit from Thermo Fisher Scientific (Waltham, MA, USA).
Binding Assay of Anti-SARS-CoV-2 Antibodies
The specific binding of the isolated antibodies against SARS-CoV-2 NP was estimated using a microplate coated with various proteins (Influenza A/New Caledonia/20/99, Influenza B/Florida/07/04, Influenza B/Tokio/53/99, Hepatitis B antigen (HBsAg)). The immobilization of proteins was achieved by incubating the protein solution (10 μg/mL, 100 μL) for 2 h at 37 °C in a 96-well microplate. Following this, the antibody solution (100 μL) at a concentration of 1 μg/mL was added to each well for 1 h. The secondary antibodies against pig IgG labeled with HRP (100 μL) at the concentration of 0.5 μg/mL were incubated for 1 h. At each change of reagent, the microbeads were washed using excess amount (120 μL) of 0.1 % Tween 20 in PBS. The colorimetric reaction was performed using the TMB reagent (100 μL) for 30 min. After quenching with 2 M sulfuric acid (100 μL), the optical density was measured at the wavelength of 450 nm using an ELISA reader (Versamax Microplate reader) from Molecular Devices Co (San Jose, CA, USA).
The immunostaining of magnetic beads with immobilized NP was performed to confirm the specific binding of the isolated antibodies against SARS-CoV-2 NP. For the immobilization of NP to magnetic beads, tosylactivated magnetic beads from Invitrogen Co. were incubated with NP at the concentration of 800 μg/mL for 16 h. For the binding of the isolated antibodies from pig sera, NP immobilized magnetic beads (1 mg) were incubated with the antibody solution (500 μL) at the concentration of 1 μg/mL for 1 h. The secondary antibodies against pig IgG labeled with fluorescein (500 μL) at a concentration of 5 μg/mL were then incubated for 1 h. At each change of reagent, the microbeads were washed using excess amounts of 0.1 % Tween 20 in PBS. The fluorescence images were taken using a fluorescence microscope (BX51) from Olympus (Tokyo, Japan)14,15.
SPR Measurement
An SPR biosensor from i-ClueBio (Sungman, Korea) was used for measurements. This biosensor used the SPR-chip, which was prepared by sputtering an adhesive layer of titanium (2 nm) and then gold (48 nm) onto BK-7 glass (10 × 10 mm2). The isolated antibodies against SARS-CoV-2 NP were immobilized on the SPR biosensor through covalent binding. The gold surface of SPR biosensor was modified to have carboxylic acid by incubating the SPR chip in 1 mM mercaptoundecanoic acid for 24 h. For the covalent binding of antibodies, the anti- antibodies at the concentration 10 μg/mL for 2 h after treatment of 0.4 M EDC/0.1 M NHS (pH 5.0) for 10 min. The SPR biosensor was equipped with a flow cell made with a capacity of 50 μL. The samples and the washing solutions (0.1 % Tween 20 in phosphate buffered saline (PBS)) were injected to the flow cell using an injection valve and a peristaltic pump. The pumping rate was set as 50 μL/min, and the flow of the solution was programmed to stop during the incubation step. SPR signal for each sample was calculated to be the change in signal-baselines before and after the sample incubation. The signal-baseline was established from averaged SPR signals following three washes16–18.
Estimation of the Limit of Detection
The SPR signals were fitted with the four parameter logistic equation19–22 described below:
where a and d are the maximum and minimum SPR signal (RU), respectively; c is the concentration of SARS-CoV-2 NP (pM); b is Hill’s slope of the curve. The limit of detection (LOD) was calculated as the concentration of NP at the point of three times of standard deviations (SD) above the average baseline signal.
References
Monchatre-Leroy, E., Boué, F., Boucher, J.M., Renault, C., Moutou, F., Ar Gouilh, M. & Umhang, G. Identification of alpha and beta Coronavirus in wildlife species in France: bats, rodents, rabbits, and hedgehogs. Viruses 9, 364 (2017).
Chen, Y., Liu, Q. & Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423 (2020).
Zeng, W., Liu, G., Ma, H., Zhao, D., Yang, Y., Liu, M., Mohammed, A., Changcheng Zhao, C., Yang, Y., Xie, J., Ding, C., Ma, X., Weng, J., Gao, Y., He, H. & Jin, T. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 527, 618–623 (2020).
Shang, B., Wang, X.-Y., Yuan, J.-W., Vabret, A., Wu, X.-D., Yang, R.-F., Tian, L., Ji, Y.-Y., Deubel, V. & Sun, B. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 336, 110–117 (2005).
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., Wang, W., Song, H., Huang, B., Zhu, N., Bi, Y., Ma, X., Zhan, F., Wang, L., Hu, T., Zhou, H., Hu, Z., Zhou, W., Zhao, L., Chen, J., Meng, Y., Wang, J., Lin, Y., Yuan, J., Xie, Z., Ma, J., Liu, W.J., Wang, D., Xu, W., Holmes, E.C., Gao, G.F., Wu, G., Chen, W., Shi, W. & Tan, W. Genomic characterisation and epidemiology of 2019 novel Coronavirus: implications for virus origins and receptor binding. The Lancet. 395, 565–574 (2020).
Liu, S.-J., Leng, C.-H., Lien, S.-P., Chi, H.-Y., Huang, C.-Y., Lin, C.-L., Lian, W.-C., Chen, C.-J., Hsieh, S.-L. & Chong, P. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine 24, 3100–3108 (2006).
Ahmed, S.F., Quadeer, A.A. & McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12, 254–268 (2020).
Narayanan, K., Chen, C.-J., Maeda, J. & Makino, S. Nucleocapsid-independent specific viral RNA packaging via viral envelope protein and viral RNA signal. J. Virol. 77, 2922–2927 (2003).
Tilocca, B., Soggiu, A., Sanguinetti, M., Musella, V., Britti, D., Bonizzi, L., Urbani, A. & Roncada, P. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microbes Infect. 22, 188–194 (2020).
Kim, J.-M., Chung, Y.-S., Jo, H. J., Lee, N.-J., Kim, M. S., Woo, S. H., Park, S., Kim, J. W., Kim, H. M. & Han, M.-G. Identification of Coronavirus isolated from a patient in Korea with COVID-19. Osong Public Health Res. Perspect. 11, 3–7 (2020).
Elfiky, A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 117477 (2020).
Bong, J.-H., Kim, J., Lee, G.-Y., Park, J.-H., Kim, T.-H., Kang, M.-J. & Pyun, J.-C. Fluorescence immunoassay of E. coli using anti-lipopolysaccharide antibodies isolated from human serum. Biosens. Bioelectron. 126, 518–528 (2019).
Brody, T. Multistep denaturation and hierarchy of disulfide bond cleavage of a monoclonal antibody. Anal. Biochem. 247, 247–256 (1997).
Tian, T., Cho, S. & Rhee, S.W. Microfluidic devices for eye irritation tests of cosmetics and cosmetic ingredients. Biochip. J. 13, 142–150 (2019).
Park, M. Orientation control of the molecular recognition layer for improved sensitivity: a Review. Biochip. J. 13, 82–94 (2019).
Kim, D.-H., Paek, S.-H., Choi, D.-Y., Lee, M.-K, Park, J.-N, Cho, H.-M. & Paek, S.-H. Real-time monitoring of biomarkers in serum for early diagnosis of target disease. Biochip. J. 14, 2–17 (2020).
Baek, J.M. & Ryu, Y.S. Surface sensitive analysis device using model membrane and challenges for biosensor-chip. Biochip. J. 14, 110–123 (2020).
Hasani, A., Do, H.H., Tekalgne, M., Hong, S.H., Jang, H.W. & Kim, S.Y. Recent progress of two-dimensional materials and metal-organic framework-based taste sensors. J. Korean Ceram. Soc. 57, 353–367 (2020).
Tijssen, P. Practice and theory of enzyme immunoassays, Elsevier, 1985.
Masdor, N.A., Altintas, Z. & Tothill, I.E. Sensitive detection of Campylobacter jejuni using nanoparticles enhanced QCM sensor. Biosens. Bioelectron. 78, 328–336 (2016).
Kim, H.-R., An, B.-G., Chang, Y. W., Kang, M.-J., Park, J.-G. & Pyun, J.-C. Characterization of in-situ synthesized CdSxSe1-x ternary alloy nanowire photosensor. J. Korean Ceram. Soc. 56, 308–316 (2019).
Son, Y.-J., Lee, I.-C., Jo, H.-H., Chung, T.-J. & Oh, K.-S. Setting behavior and drug release from brushite bone cement prepared with granulated hydroxyapatite and β-tricalcium phosphate. J. Korean Ceram. Soc. 56, 56–64 (2019).
Acknowledgements
This work was supported by the National Research Foundation of Korea [grant number: NRF-2020R1A2B5B01002187].
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of Interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Bong, JH., Kim, TH., Jung, J. et al. Pig Sera-derived Anti-SARS-CoV-2 Antibodies in Surface Plasmon Resonance Biosensors. BioChip J 14, 358–368 (2020). https://doi.org/10.1007/s13206-020-4404-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-020-4404-z